Abstract
Background
The male germ line in flowering plants is initiated within developing pollen grains via asymmetric division. The smaller cell then becomes totally encased within a much larger vegetative cell, forming a unique "cell within a cell structure". The generative cell subsequently divides to give rise to two non-motile diminutive sperm cells, which take part in double fertilization and lead to the seed set. Sperm cells are difficult to investigate because of their presence within the confines of the larger vegetative cell. However, recently developed techniques for the isolation of rice sperm cells and the fully annotated rice genome sequence have allowed for the characterization of the transcriptional repertoire of sperm cells. Microarray gene expression data has identified a subset of rice genes that show unique or highly preferential expression in sperm cells. This information has led to the identification of cis-regulatory elements (CREs), which are conserved in sperm-expressed genes and are putatively associated with the control of cell-specific expression.
Findings
We aimed to identify the CREs associated with rice sperm cell-specific gene expression data using in silico prediction tools. We analyzed 1-kb upstream regions of the top 40 sperm cell co-expressed genes for over-represented conserved and novel motifs. Analysis of upstream regions with the SIGNALSCAN program with the PLACE database, MEME and the Mclip tool helped to find combinatorial sets of known transcriptional factor-binding sites along with two novel motifs putatively associated with the co-expression of sperm cell-specific genes.
Conclusions
Our data shows the occurrence of novel motifs, which are putative CREs and are likely targets of transcriptional factors regulating sperm cell gene expression. These motifs can be used to design the experimental verification of regulatory elements and the identification of transcriptional factors that regulate sperm cell-specific gene expression.
Keywords: cis-regulatory elements, plant reproduction, male gamete, gene expression, Oryza sativa
Introduction
As in animals, flowering plant sperm cells are small cells that fuse with the egg during fertilization. The sperm cells produced within developing pollen remain enveloped by much larger vegetative cell. Typically, sperm cells occupy < 0.1% of the pollen grain volume. The germination of pollen leads to the extension of the vegetative cell wall to produce a pollen tube, which grows via tip elongation to deliver sperm cells to the embryo sac. Until recently, the condensed appearance of chromatin associated with its small cytoplasmic volume was considered to reflect transcriptional quiescence of sperm cells. Recent developments in techniques to isolate sperm cells from pollen [1] along with the availability of high-throughput genomic and transcriptomic tools have allowed for the analysis of gene expression in these small cells. The latest reports of gene expression studies in Arabidopsis [2], maize [3], Plumbago and lily [4] sperm cells have shown that the initial views regarding sperm cells as transcriptionally quiescent were not correct [5]. Recent investigations have revealed that sperm cells have highly distinct expression profiles from vegetative stages. Our recent microarray investigations of sperm cells in Oryza sativa exposed distinct expression profiles for many genes in male germ line cells, including GEX1, GEX2, and GCS1/HAP2, a set of genes whose sperm specificity is conserved in Arabidopsis, lily, and rice. These expression studies exhibit the conserved and sophisticated control of molecular mechanisms in sperm cell development.
Cellular signaling pathways often consist of interacting loops of transcription factors and cis-regulatory DNA elements that direct the expression of target genes. We aimed to take a computational approach to investigate candidate cis-regulatory elements (CREs) in a cluster of co-expressing genes in rice sperm cells. In this paper, we discuss the functions of these CREs in plant biology, including the regulation of cell cycle and reproductive development. This study will accelerate the functional characterization of CREs and their interacting transcription factors and will serve as a step forward in exploring systems biology networks in transcriptional regulation.
Transcriptional regulation of organ-specific and cell-specific gene expression is mediated by the recruitment of transcription factors to CREs. Transcription factors interact with specific DNA elements, other factors and the basal transcriptional machinery to regulate the expression of target genes. In plants, transcriptional regulation is mediated by more than 1500 transcriptional factors; each of these factors controls the expression of tens or even thousands of target genes in complex signaling networks [6]. Along with transcription factors, CREs are functional DNA motifs or elements that establish conspicuous temporal and spatial transcriptional activity. Identifying and understanding the functions of such CREs are essential for elucidating the mechanism by which cells perceive and correctly respond to their environment and participate in organism growth and development.
Microarray gene expression data can help to identify groups of co-expressed genes. Clusters of such co-expressed genes are assumed to be co-regulated and upstream sequences of these genes are likely to share common DNA motifs. Presumed upstream regulatory regions of arbitrary length can be used to identify candidate DNA motifs. Because of their importance, we studied multiple motifs over-represented in the promoters of a co-expressed gene cluster. This type of investigation can illustrate the utility of the co-expression-driven prediction of CREs as a means to begin deciphering transcriptional networks. A number of algorithms and bioinformatics tools have been developed to identify potential CREs in the regulatory sequences of co-expressed genes. Most computational approaches assume that co-regulated genes should contain similar CREs in their upstream regulatory regions at statistically significant levels. The transcriptional control of gene expression depends on a balance between activating and repressing regulatory components in upstream regulatory regions. Hence, CREs play a central role in regulating gene expression by integrating signals at the DNA-level upstream of a target gene. Our study presents an in silico analysis of 1-kb upstream promoter sequences that regulate sperm cell-expressing genes in rice.
Materials and methods
One Kb upstream DNA sequences of top 40 genes that showed high expression in sperm cells of rice http://www.ricechip.org were mined from TIGR (The Institute of Genome Research) database release 6.1 ftp site ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_6.1/all.dir/all.1kUpstream.
Analysis of one Kb upstream promoter sequences for locating known CREs was conducted using SIGNALSCAN program available in Plant cis-Regulatory DNA Elements (PLACE), and database of cis-regulatory element motif http://www.dna.affrc.go.jp/PLACE/[7,8]. The database contains mainly plant motifs extracted from the published reports in the literature.
CREs for more extensive analysis were selected based on the frequency of their occurrence. These results were further refined to find CREs present in 80% of the gene dataset. A comprehensive map depicting precise locations of these abundantly occurring CREs was also prepared by placing them on scales of upstream promoter sequences manually.
Unknown novel CREs in the sequence dataset were also detected using MEME (Multiple EM for Motif Elicitation) tool version 4.4.0 http://meme.sdsc.edu/meme/intro.html MEME analyses sequences for similarities and produces a description for each pattern or motif it discovers [9]. A motif is a sequence pattern that occurs repeatedly in a group of related protein or DNA sequences. MEME represents motifs as position-dependent letter-probability matrices that describe the probability of each possible letter at each position in the pattern. Individual MEME motifs do not contain gaps.
To complement searches for novel CREs detection, we also used the Mclip program developed by Frickey and Weiller (2007), which we operated online at: http://bioinfoserver.rsbs.anu.edu.au/utils/mclip/ modifying parameters by using E-values of 1e-13 as cut-off values for motif matching and motif alignment (rather than the default 1e-3). End gaps were allowed to permit longer motifs to be considered. For validation, we selected sperm cell probe sets reported as three unanimous presence calls ("PPP") in sperm microarray results, accompanied by corresponding unanimous absence calls ("AAA") in pollen (vegetative) cells and seedlings. The selected sperm probe sets were matched to loci. Loci annotated as containing transposable element sequences were omitted as a precaution to reduce the likelihood of finding sequences that may be disproportionately be amplified owing to transposable element activity. For the remaining loci after screening, 1 K upstream sequences were obtained as FASTAs. We considered the occurrence of all possible 8-nt motifs by constructing a matrix of all finds. To the extent that a "found" motif can be considered as a discrete event, the Poisson distribution is appropriate to determine if expression is distorted from that occurring by chance and thus it is possible to assign a calculated p value. The motifs that were found through Mclip were then screened against known CREs, and only novel sequences were considered.
Results and Discussion
The investigation and identification of cis-regulatory elements (CREs) in the promoters of high expressing sperm cell genes will aid in deciphering the function of these genes in male gametophyte and the sperm cell development in rice. In addition, the results can be used for further analysis of gene networks in male gametophyte development in plants.
Highly expressed transcripts in rice sperm cells
Some initial reports have described differentially expressed genes in pollens, anthers and male gamete cells in plants [10]. However, deciphering gene expression in plant sperm cells was not possible due to the unavailability of protocols allowing for their meaningful characterization. With the development of techniques to collect sperm cells and compare gene expression profiles between pollens, anthers, sperm cells and sporophytic tissues, sperm cells are now known to have distinct expression profiles. The sperm cell-expressed transcripts are functionally categorized for their involvement in DNA repair, cell cycle progression and ubiquitin-mediated proteolysis.
Conserved genes with especially enriched sperm cell expression in flowering plants include Arabidopsis GEX-1 (gamete expressed protein 1), which encodes a transmembrane domain-containing protein, and GEX-2 (gamete expressed protein 2) [5]. GCS-1 (generative cell specific 1), also known as HAP2, is expressed only in haploid sperms, contributes to pollen tube guidance and is required for fertilization [11].
We considered a dataset of CREs consisting of 1-kb upstream promoter sequences of 40 sperm cell-expressed genes in rice, which are located at different positions on 12 chromosomes. Information about the genes and their positions, descriptions and expression values are presented in Table 1. These genes are highly expressed in rice sperm cells as detected by microarrays with intensity values http://www.ricechip.org.
Table 1.
List of highly expressing sperm cell genes in rice.
| S. No | Probe Set ID | Gene ID | Location 5'-3' (Base pair) | Nucleotide Length (Base Pair) | Gene Product Description | Intensity log2 |
|---|---|---|---|---|---|---|
| 1 | Os.10737.1.S1_at | LOC_Os05g18730.1 | 10854579 - 10847943 | 2169 | generative cell specific-1, putative, expressed | 15.4436 |
| 2 | Os.54874.1.S1_at | LOC_Os09g27040.1 | 16444436 - 16446851 | 1920 | GEX1, putative, expressed | 12.808 |
| 3 | OsAffx.17894.1.S1_at | LOC_Os09g25650.1 | 15414547 - 15407935 | 3291 | GEX2, putative, expressed | 14.6951 |
| 4 | Os.41333.1.A1_at | LOC_Os01g42060.1 | 23845997 - 23849282 | 1908 | expressed protein | 12.4728 |
| 5 | Os.53049.1.S1_at | LOC_Os04g46490.1 | 27391780 - 27390795 | 810 | aquaporin protein, putative, expressed | 15.7476 |
| 6 | Os.26448.1.A1_at | LOC_Os03g08070.1 | 4125935 - 4119369 | 2658 | copper-transporting ATPase PAA1, putative, expressed | 11.0327 |
| 7 | Os.21018.1.S1_at | LOC_Os09g35720.1 | 20547711 - 20541270 | 2397 | generative cell specific-1, putative | 14.8986 |
| 8 | OsAffx.26224.1.S1_s_at | LOC_Os04g29090.1 | 17091173 - 17086877 | 1989 | FAD-binding and arabino-lactone oxidase domains containing protein, putative, expressed | 12.3096 |
| 9 | Os.18560.1.S1_at | LOC_Os05g01500.1 | 291425 - 296877 | 1641 | tubulin-specific chaperone E, putative, expressed | 11.2132 |
| 10 | OsAffx.3617.1.S1_at | LOC_Os03g55890.1 | 31822145 - 31818577 | 1950 | ternary complex factor MIP1, putative, expressed | 10.1043 |
| 11 | OsAffx.2553.1.S1_at | LOC_Os02g09580.1 | 4931298 - 4935969 | 1308 | OsFBX39 - F-box domain containing protein | 9.7882 |
| 12 | Os.51974.1.S1_at | LOC_Os06g20860.1 | 12052626 - 12049870 | 2580 | paramyosin, putative, expressed | 11.6793 |
| 13 | Os.50267.1.S1_at | LOC_Os08g34640.1 | 21757895 - 21754127 | 3339 | receptor-like protein kinase precursor, putative, expressed | 12.8324 |
| 14 | Os.55267.1.S1_at | LOC_Os03g44630.1 | 25129797 - 25129252 | 429 | plastocyanin-like domain containing protein, putative, expressed | 13.8559 |
| 15 | Os.9559.1.S1_at | LOC_Os03g37570.1 | 20838322 - 20836787 | 633 | expressed protein | 13.1792 |
| 16 | OsAffx.2680.1.S1_at | LOC_Os02g19180.1 | 11184386 - 11182943 | 915 | ZOS2-06 - C2H2 zinc finger protein | 13.3352 |
| 17 | Os.52171.1.S1_at | LOC_Os06g38950.1 | 23111994 - 23104560 | 2850 | ABC transporter, ATP-binding protein, putative, expressed | 15.1547 |
| 18 | Os.32737.1.S1_at | LOC_Os11g08440.1 | 4439113 - 4441939 | 1734 | DnaK family protein, putative, expressed | 12.267 |
| 19 | Os.38984.1.S1_at | LOC_Os01g23580.1 | 13236319 - 13244856 | 2322 | inorganic H+ pyrophosphatase, putative, expressed | 12.9446 |
| 20 | Os.23286.1.A1_at | LOC_Os10g02920.1 | 1190765 - 1188954 | 678 | cytochrome b561, putative, expressed | 12.3191 |
| 21 | OsAffx.26489.1.S1_at | LOC_Os04g46760.1 | 27543355 - 27541189 | 1050 | trehalose phosphatase, putative | 12.2378 |
| 22 | Os.53437.1.S1_at | LOC_Os03g45980.1 | 25991611 - 25992474 | 864 | expressed protein | 12.459 |
| 23 | Os.46544.1.A1_at | LOC_Os10g25060.1 | 12830509 - 12828658 | 588 | expressed protein | 13.4751 |
| 24 | Os.9431.1.A1_a_at | LOC_Os08g16610.1 | 10159371 - 10164170 | 2187 | Rad21/Rec8 like protein, putative, expressed | 15.1214 |
| 25 | OsAffx.12136.1.S1_at | LOC_Os02g20530.1 | 12106516 - 12110223 | 1812 | hypothetical protein | 12.4087 |
| 26 | OsAffx.24724.1.S1_x_at | LOC_Os02g44599.1 | 27021672 - 27021157 | 516 | expressed protein | 12.4125 |
| 27 | Os.4125.1.S1_at | LOC_Os12g38460.1 | 23577850 - 23572909 | 3399 | RNA recognition motif family protein | 12.8627 |
| 28 | Os.54137.1.S1_at | LOC_Os02g08080.1 | 4268788 - 4264756 | 1428 | expressed protein | 12.6933 |
| 29 | OsAffx.31616.1.S1_at | LOC_Os12g06480.1 | 3123385 - 3117268 | 2376 | PHD-finger family protein, expressed | 13.5878 |
| 30 | Os.52287.1.S1_at | LOC_Os02g02800.1 | 1067067 - 1068845 | 762 | AGAP001222-PA, putative, expressed | 12.4595 |
| 31 | OsAffx.14496.1.S1_at | LOC_Os05g02030.1 | 590266 - 591787 | 582 | OB-fold nucleic acid binding domain containing protein, putative | 11.1188 |
| 32 | Os.50552.1.S1_at | LOC_Os08g35700.1 | 22526032 - 22519854 | 2706 | Leucine Rich Repeat family protein, expressed | 13.9674 |
| 33 | OsAffx.28262.1.S1_at | LOC_Os07g04520.1 | 1993662 - 1991627 | 1554 | protein kinase, putative, expressed | 12.6217 |
| 34 | Os.56612.1.A1_x_at | LOC_Os05g11980.1 | 6852880 - 6843407 | 3624 | timeless protein, expressed | 11.097 |
| 35 | Os.34965.2.S1_s_at | LOC_Os06g07130.1 | 3407672 - 3417233 | 2043 | SHR5-receptor-like kinase, putative, expressed | 10.7422 |
| 36 | Os.54810.1.A1_at | LOC_Os08g28080.1 | 17129894 - 17119408 | 2514 | coatomer subunit delta, putative, expressed | 12.4403 |
| 37 | Os.52821.1.S1_at | LOC_Os11g37200.1 | 21516300 - 21519963 | 777 | transmembrane BAX inhibitor motif-containing protein, putative, expressed | 14.25 |
| 38 | Os.54486.1.S1_at | LOC_Os05g03320.1 | 1352666 - 1348957 | 1686 | expressed protein | 12.624 |
| 39 | Os.10491.1.S1_at | LOC_Os03g04690.1 | 2227622 - 2226738 | 885 | expressed protein | 10.5236 |
| 40 | Os.23535.1.A1_at | LOC_Os08g05820.1 | 3124469 - 3119480 | 1803 | monocopper oxidase, putative, expressed | 15.4961 |
Identification of Known CREs
CREs were extracted from the input dataset based on previously published reports and databases of regulatory elements and motifs. The PLACE database for cis-acting regulatory DNA elements and the SIGNALSCAN search tool were used to estimate the mode of gene regulation and to find the regulatory and other pertinent regions in regulatory promoter sequences in genes highly expressed in rice sperm cells.
Abundant CREs
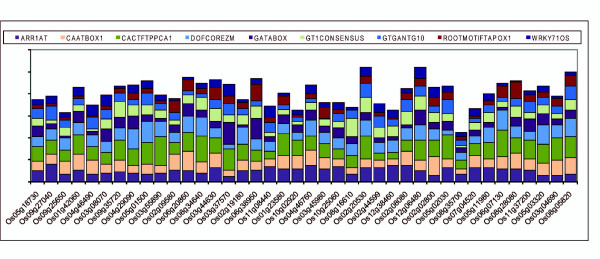
Evaluation of 1-kb upstream promoter regions (-1 to -1000) of rice sperm cell-expressing genes using SIGNALSCAN resulted in the identification of 223 types of CREs. Nine of the CREs were found in all of the 40 genes: ARR1AT (5'-NGATT-3'), CAATBOX1 (5'-CAAT-3'), CACTFTPPCA1 (5'-CACGTG-3'), DOFCOREZM (5'-AAAG-3'), GATABOX (5'-GATA-3'), GT1CONSENSUS (5'-GRWAAW-3'), GTGANTG10 (5'-GTGA-3'), ROOTMOTIFTAPOX1 (5'-ATATT-3') and WRKY71OS (5'-TGAC-3'). The duplication frequency of these CREs in all 40 genes is depicted in Figure 1 and the numbers are given in Additional file 1. Among these nine CREs, CACTFTPPCA1 is the most abundant CRE, with duplications in the range of 2 to 26 in the 1-kb upstream region of each highly up-regulated sperm cell promoter, followed by DOFCOREZM, ARR1AT and CAATBOX1 with 595, 460, 417 and 416 duplications, respectively, in all 40 sequences.
Figure 1.
Duplication frequency of 9 most abundant CREs in top 40 highly expressed genes in sperm cell.
Recently, it has been experimentally explained that AGO5 promoter drives gene specifically expressed in Arabidopsis sperm cells [12] during gametophyte development. All 9 most abundant CREs discussed above were also found in Arabidopsis AGO5 promoter 1 Kb upstream region when analyzed with SIGNALSCAN function of PLACE. For instance there are 6 ARR1AT, 12 CACTFTPPCA1, 12 GT1CONSENSUS and 22 DOFCOREZM.
CACTFTPPCA1, the most duplicated CRE, is a tetranucleotide motif responsible for mesophyll-specific gene expression of C4 phosphoenolpyruvate carboxylase gene in C4 plants [13]. It is a key component of Mem1 (mesophyll-expression module 1) in Flaveria trinervia but might have a different role in C3 plants, such as rice.
DOFCOREZM is the target binding site of Dof proteins, which are specific DNA-binding proteins associated with the expression of multiple genes in plants [14]. Dof proteins also differentially regulate diverse promoters in a variety of plant tissues [15]. Binding sites for Dof transcription factors have been recorded in the upstream sequences of GEX1 and GEX2, two genes showing sperm-specific expression in Arabidopsis [5]. Interestingly, rice homologues of both of these genes are represented among the 40 highly expressed genes of rice sperm cells.
ARR1AT (ARR1-binding element) is found in both Arabidopsis and rice. ARR1 and ARR2 are cytokinin response regulators that function as transcriptional activators [16]. AGATT has also been reported to be in the promoter of the rice non-symbiotic haemoglobin-2 (NSHB) gene [17]. CAATBOX1, the CAAT promoter consensus sequence is responsible for the tissue-specific promoter activity of the pea legumin gene LegA [18].
GATABOX, which are GATA motifs, are known to be required for high level, light regulated and tissue specific gene expression. GATA transcription factors are a group of DNA-binding proteins distinguished by a zinc finger motif, which have been implicated in light and nitrate-dependent transcription control [19]. The zinc finger transcription factor genes are among the genes showing the highest expression level in Arabidopsis sperm cells [2]. However, the G-Box and GATA elements are reported to occur several times on average in every potential upstream regulatory region [20]. GATA transcription factors are reported to bind the CaMV 35S promoter and are conserved in cab promoters as well [21].
GT1CONSENSUS recognizes GT-1 proteins, which have tri-helix DNA-binding domains, are conserved in plant nuclear genes and have diverse functions [22,23]. GT elements are ubiquitously expressed and show complex regulatory features of plant gene transcription [24]. GTGANTG10, is a GTGA motif found in the promoter of tobacco late pollen gene g10. The tobacco gene g10 is preferentially and maximally expressed in mature pollen, shows homology to pectate lyases, and is the putative homologue of the tomato gene lat56 [25]. ROOTMOTIFTAPOX1 is a motif found in the rolD promoter of Agrobacterium rhizogenes. The rolD-gus genes were found to have a distinctive expression pattern in roots [26].
WRKY71OS is a binding site of rice WRKY71, a transcriptional repressor of the gibberellin signaling pathway [27]. It is a core of TGAC-containing W-box of the Amy32b promoter within PR-10 genes [28]. Because WRKY ESTs are highly abundant in plant cDNA libraries generated from floral and embryonic material, WRKY transcription factors are presumed to have vital functions in these tissues [29]. Family members of WRKY transcription factors appear to be involved in the regulation of various physiological programs that are unique to plants, including pathogen defense, senescence, trichome development plant growth and development. The rice WRKY gene superfamily has also been implicated in the regulation of abscisic acid signaling in aleurone cells [30].
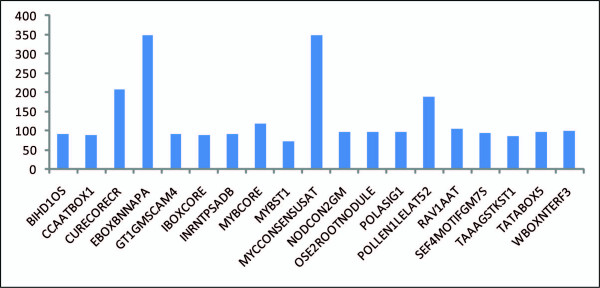
The other 19 CREs present in almost 80% of genes and abundant in distribution were BIHD1OS (5'-TGTCA-3'), CCAATBOX1 (5'-CCAAT-3'), CURECORECR (5'-GTAC-3'), EBOXBNNAPA (5'-CANNTG-3'), GT1GMSCAM4 (5'-GAAAAA-3'), IBOXCORE (5'-GATAA-3'), INRNTPSADB (5'-YTCANTYY-3'), MYBCORE (5'-CNGTTR-3'), MYBST1 (5'-GGATA-3'), MYCCONSENSUSAT (5'-CANNTG-3'), NODCON2GM (5'-CTCTT-3'), OSE2ROOTNODULE (5'-CTCTT-3'), POLASIG1 (5'-AATAAA-3'), POLLEN1LELAT52 (5'-AGAAA-3'), RAV1AAT (5'-CAACA-3'), SEF4MOTIFGM7S (5'-RTTTTTR-3'), TAAAGSTKST1 (5'-TAAAG-3'), TATABOX5 (5'-TTATTT-3') and WBOXNTERF3 (5'-TGACY-3'). Duplication numbers of these CREs are delineated in Figure 2, and the values are given in Additional file 2. Taken together, these CREs represent some of the major categories abundantly distributed and duplicated in the 1-kb upstream regulatory sequences of sperm cell-specific genes in O. sativa. The most over-represented CREs were EBOXBNNAPA, MYCCONSENSUSAT, CURECORECR, POLLEN1LELAT52 and MYBCORE. All these 19 CREs were also found to be present in AGO5 promoter sequence, which induces sperm cell specific expression in Arabidopsis [12].
Figure 2.
Duplication numbers of other abundant 19 CREs in 80% of top 40 highly expressed genes in sperm cell.
EBOXBNNAPA is an E-box sequence, which has been reported in Brassica napus [31]. Also known as an RRE element, this CRE is responsible for light responsiveness and is regulated by bHLH and the MYB-transcription factor in directing tissue-specific expression [32]. MYCCONSENSUSAT is a MYC recognition site found in the promoters of the dehydration-responsive gene rd22 and many other genes in Arabidopsis [33]. This CRE also regulates the transcription of Arabidopsis genes under cold conditions by a MYC-like bHLH transcriptional activator [34]. Few bHLH transcription factors have been detected in Arabidopsis sperm cells and none have been found in pollen, suggesting that they might play a specific role in male gamete formation [2]. CURECORECR (GTAC) is located in the core of a CuRE (copper-response element), which is found in the Cyc6 and Cpx1 genes in Chlamydomonas. This CRE is also involved in oxygen-response of these genes [35]. POLLEN1LELAT52 is a regulatory element responsible for pollen-specific activation of the tomato lat52 gene. This CRE has also been found in the promoter of tomato endo-beta-mannanase gene during late stages of anther development [36]. MYBCORE is a binding site for two plant MYB proteins, AtMYB1 and AtMYB2, which were isolated from Arabidopsis. AtMYB2 is involved in the regulation of genes responsive to water stress [37]. MYB-type transcription factors are reported to have relatively high expression in sperm cells [2]. Furthermore, MYB-proteins play crucial roles in cell proliferation and differentiation. Moreover, the MYB DNA-binding domains are relatively similar to those of transcription factors containing zinc finger, basic region/leucine zipper and basic region/helix-loop-helix domains [38].
BIHD1OS is a binding site of OsBIHD1, which is rice BELL homeodomain transcription factor [39] present in the nucleus, whose induction is associated with the resistance response in rice. Homeodomain transcription factors in Arabidopsis have also shown substantial expression in sperm cells [2]. CCAATBOX1 (CCAAT Box) and CONSTANS form a binding complex, which contains a functionally important domain and regulates flowering in Arabidopsis [40]. The CCAAT box is also found in the promoters of heat shock protein genes [41]. RAV1AAT is a consensus sequence of the Arabidopsis transcription factor target domain [42]. The RAV1 protein contains Ap2-like and B3-like domains [43]. AP2 is a member of the transcription factor family unique to plants and is a key regulator of several developmental processes in the plant life cycle including floral organ identity determination [44].
Positionally biased CREs
Comprehensive mapping for the above 28 CREs and their locations in the 1-kb upstream region of each sperm cell specific gene regulatory sequence is shown in Additional file 3. The CREs seem randomly dispersed in the promoters with no particular pattern detected in their occurrence with respect to positions on 1-kb upstream sequences. But an interesting speculation is for some of the entries; comparatively large numbers of CREs are located on antisense strand as compared to sense strand. This observation is held true for Os05g18730.1, Os09g27040.1, Os11g08440.1, Os01g23580.1, Os05g11980.1 and Os08g08080.1.
Low abundant CREs
In addition, 35 other unique CREs were only found in one of the 40 sperm cell-specific genes with just one or two duplications. These 35 CREs are presented in Additional file 4. Twenty-one genes contain these 35 unique CREs. In contrast to the 19 CREs found in 80% of sperm specific genes, 44 CREs are represented in about 5-10% of sperm-specific genes. These CREs are listed in Additional file 5.
Identification of unknown CREs using MEME
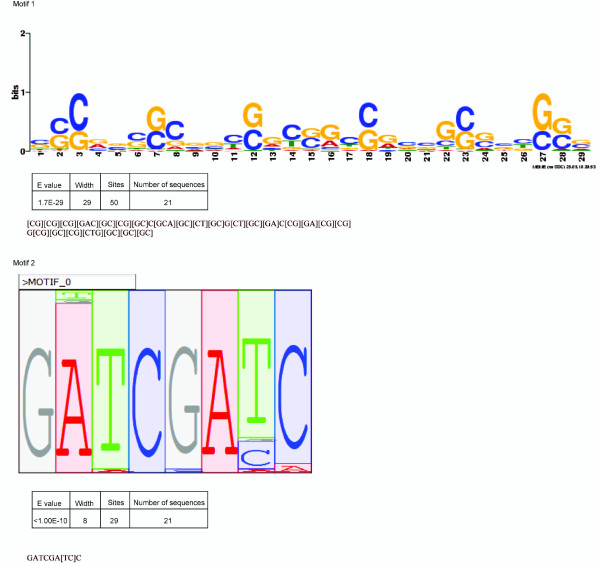
Tools can be used for detecting unknown motifs in related DNA sequences. For example, MEME uses statistical modeling techniques to choose the width, number of occurrences and descriptions for each motif. Using MEME, the same dataset of high sperm cell-expressing genes produced one novel motif. The motif is found to present in 21 genes. These results are illustrated in Figure 3. Motif 1, which is present in 21 of the 40 genes, illustrates CG enrichment similar to that found in regulatory elements related to DNA methylation, which may be copied during replication. CpG islands are a common motif by which sequences are normally repressed. The prevalence of this motif suggests that sperm-restricted functions may function in repression within somatic tissue.
Figure 3.
Highest scoring novel motifs found by MEME and Mclip.
Some reports about CRE and promoter duplications indicate that the duplication numbers of CREs might participate in leveling the mRNA concentration or gene expression [44]. Thus, these investigations may reveal some clues about synchronous expression of this subset of genes in sperm cells.
Cis-regulatory elements of genes are closely related to spatiotemporal gene expression. CREs have been reported to control tissue-specific gene expression and condition-dependent gene expression similar to how heat shock elements (HSEs) control heat shock induction of gene expression [45].
Duplication and distribution of CREs might play a role in up- or down-regulation of specific genes. This data may be useful for understanding various phenomena in sperm cell development and will shed light on sperm cell development and expression. This study may facilitate our understanding of the molecular aspects of male gamete differentiation and function in flowering plants, such as rice.
Search for novel CREs using Mclip
To complement searches for unknown transcription factor cis-regulatory sequences, we also conducted searches of the same 40 highly transcribed sperm loci using Mclip [46]. One-kb upstream sequences of these loci were used as seeds for the discovery of unknown conserved motifs, which were then screened against known cis-regulatory elements. Our results are illustrated in Figure 3. Interestingly, the program identified an expressed AT-rich sequence that matched a promoter sequence isolated from the upstream region of the Plumbago gene PzIPT [10]. The canonical TATA box was also represented in this sequence, but the deletion of the matched AT-rich cis-regulatory region resulted in a loss of expression. AT-rich motifs have also been reported in upstream regions of GEX1 and GEX2 [5] and were over-represented in 1-kb upstream sequences of sperm-expressed genes.
Novel motifs in the form of GATCGATC were present in 15 genes with 54 duplications, of which TCGA was most highly conserved. Similar sequences (ATCGATCG, TCGATCGA and CGATCGTA) were also represented frequently as 8-nt motifs and far exceeded random 8-nt motifs (ranging from 4.89E-21 to 3.3E-28).
In summary, we have revealed sequences (CREs) that may be responsible for driving high expression in male germ line cells. Though these sequence patterns require experimental validation (e.g. insertion deletion experiments), nevertheless, our current findings may open new avenues for studying the regulation of gene expression in male gametes of flowering plants.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Conceived and designed the study: MS PB NS. Performed computational analysis: NS SDR.
Wrote the paper: NS PB SDR MS. All authors have read and approved the manuscript.
Supplementary Material
Duplication numbers of 9 most abundant CREs in top 40 highly expressed genes in sperm cells of rice. Based on the results of PLACE database SIGNALSCAN searches for total of 223 CREs, 9 of them are found to present in 1 Kb upstream regions of all 40 genes. The duplication numbers of those CREs are represented in this table. Frequency graph is also plotted for this distribution as shown in Figure 1.
Duplication numbers of other 19 abundant CREs present in 80% of sperm cell expressing genes of rice. Results of SIGNALSCAN searches gave 19 other CREs abundantly present in the dataset. The duplication numbers of these CREs are represented in this table. Frequency graph is also plotted for this distribution as shown in Figure 2.
A map of 28 abundant CREs and their positions within 1 Kb upstream sequences. SIGNALSCAN program of PLACE database identified the positions of CREs in the upstream regions of top 40 highly expressed genes in sperm cell of rice. These selected CREs were subjected to further extensive analysis for their duplication numbers and distribution across the upstream regions. The figure shows exact location of CREs present in 80% of the gene dataset. The blue bars above the horizontal black line indicate CREs on sense strand and the blue bars below the black line designates CREs on anti-sense strand.
Unique CREs. The analysis exhibited some unique CREs present in only one of the 40 sperm cell expressing genes with one or two duplications. These CREs were found in these specific sperm cell expressing genes.
Peculiar CREs. Besides abundant CREs present in 80% of the gene dataset, there are few others present in just 5-10% of rice sperm cell expressing genes.
Contributor Information
Niharika Sharma, Email: shn@student.unimelb.edu.au.
Scott D Russell, Email: srussell@ou.edu.
Prem L Bhalla, Email: premlb@unimelb.edu.au.
Mohan B Singh, Email: mohan@unimelb.edu.au.
Acknowledgements
We thank the Australian Research Council for financial support. NS acknowledges the receipt of the University of Melbourne postgraduate scholarship during the course of this study. We also thank Dr. Annie Wong for providing valuable feedback on the manuscript and advice regarding the design of figures.
References
- Gou XP, Xu Y, Tang L, Yan F, Chen F. Representative cDNA library from isolated rice sperm cells. Act Bot Sin. 2001;43(10):1093–1096. [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 2008;148(2):1168–1181. doi: 10.1104/pp.108.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel ML, Chaboud A, Dumas C, McCormick S. Sperm cells of Zea mays have a complex complement of mRNAs. Plant J. 2003;34(5):697–707. doi: 10.1046/j.1365-313X.2003.01761.x. [DOI] [PubMed] [Google Scholar]
- Okada T, Bhalla PL, Singh MB. Expressed sequence tag analysis of Lilium longiflorum generative cells. Plant Cell Physiol. 2006;47(6):698–705. doi: 10.1093/pcp/pcj040. [DOI] [PubMed] [Google Scholar]
- Engel ML, Holmes-Davis R, McCormick S. Green sperm. Identification of male gamete promoters in Arabidopsis. Plant Physiol. 2005;138(4):2124–2133. doi: 10.1104/pp.104.054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290(5499):2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestridge DS. SIGNAL SCAN: A computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci. 1991;7(2):203–206. doi: 10.1093/bioinformatics/7.2.203. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006. pp. W369–W373. [DOI] [PMC free article] [PubMed]
- Gou X, Yuan T, Wei X, Russell SD. Gene expression in the dimorphic sperm cells of Plumbago zeylanica: transcript profiling, diversity, and relationship to cell type. Plant J. 2009;60(1):33–47. doi: 10.1111/j.1365-313X.2009.03934.x. [DOI] [PubMed] [Google Scholar]
- Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat Cell Biol. 2006;8(1):64–71. doi: 10.1038/ncb1345. [DOI] [PubMed] [Google Scholar]
- Borges F, Pereira PA, Slotkin RK, Martienssen RA, Becker JD. MicroRNA activity in Arabidopsis male germline. J Exp Bot. 2011;62(5):1611–1620. doi: 10.1093/jxb/erq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Burscheidt J, Akyildiz M, Schlue U, Koczor M, Streubel M, Westhoff P. cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell. 2004;16(5):1077–1090. doi: 10.1105/tpc.019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999;17(2):209–214. doi: 10.1046/j.1365-313X.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 2000;21(3):281–288. doi: 10.1046/j.1365-313x.2000.00685.x. [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 2000;24(6):703–711. doi: 10.1046/j.1365-313x.2000.00909.x. [DOI] [PubMed] [Google Scholar]
- Ross EJ, Stone JM, Elowsky CG, Arredondo-Peter R, Klucas RV, Sarath G. Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J Exp Bot. 2004;55(403):1721–1731. doi: 10.1093/jxb/erh211. [DOI] [PubMed] [Google Scholar]
- Shirsat A, Wilford N, Croy R, Boulter D. Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Mol Gen Genet. 1989;215(2):326–331. doi: 10.1007/BF00339737. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Muro-Pastor MI, Florencio FJ. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004;134(4):1718–1732. doi: 10.1104/pp.103.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest HD, Filichkin SA, Mockler TC. Cis-regulatory elements in plant cell signalling. Curr Opin Plant Biol. 2009;12(5):643–649. doi: 10.1016/j.pbi.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Lam E, Chua NH. ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell. 1989;1(12):1147–1156. doi: 10.1105/tpc.1.12.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villain P, Mache R, Zhou DX. The mechanism of GT element-mediated cell type-specific transcriptional control. J Biol Chem. 1996;271(51):32593–32598. doi: 10.1074/jbc.271.51.32593. [DOI] [PubMed] [Google Scholar]
- Le Gourrierec J, Li YF, Zhou DX. Transcriptional activation by Arabidopsis GT-1 may be through interaction with TFIIA-TBP-TATA complex. Plant J. 1999;18(6):663–668. doi: 10.1046/j.1365-313x.1999.00482.x. [DOI] [PubMed] [Google Scholar]
- Zhou DX. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci. 1999;4(6):210–214. doi: 10.1016/S1360-1385(99)01418-1. [DOI] [PubMed] [Google Scholar]
- Rogers HJ, Bate N, Combe J, Sullivan J, Sweetman J, Swan C, Lonsdale DM, Twell D. Functional analysis of cis-regulatory elements within the promoter of the tobacco late pollen gene g10. Plant Mol Biol. 2001;45(5):577–585. doi: 10.1023/A:1010695226241. [DOI] [PubMed] [Google Scholar]
- Elmayan T, Tepfer M. Evaluation in tobacco of the organ specificity and strength of the rolD promoter, domain A of the 35S promoter and the 35S2 promoter. Transgenic Res. 1995;4(6):388–3896. doi: 10.1007/BF01973757. [DOI] [PubMed] [Google Scholar]
- Zhang ZL, Xie Z, Zou X, Casaretto J, Ho TH, Shen QJ. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004;134(4):1500–1513. doi: 10.1104/pp.103.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. The EMBO J. 1999;18(17):4689–4699. doi: 10.1093/emboj/18.17.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5(5):199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005;137(1):176–189. doi: 10.1104/pp.104.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stålberg K, Ellerstöm M, Ezcurra I, Ablov S, Rask L. Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta. 1996;199(4):515–519. doi: 10.1007/BF00195181. [DOI] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol. 2005;57(2):155–171. doi: 10.1007/s11103-004-6910-0. [DOI] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signalling. Plant Cell. 2003;15(1):63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17(8):1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JM, Barraco P, Eriksson M, Merchant S. Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J Biol Chem. 2000;275(9):6080–6089. doi: 10.1074/jbc.275.9.6080. [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Leonard JM, Monteros A, Liu PP, Nonogaki H. A novel endo-beta-mannanase gene in tomato LeMAN5 is associated with anther and pollen development. Plant Physiol. 2004;134(3):1080–1087. doi: 10.1104/pp.103.035998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5(11):1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B, Eisenman RN. New light on Myc and Myb. Part II. Myb. Genes Dev. 1990;4(12):2235–2241. doi: 10.1101/gad.4.12b.2235. [DOI] [PubMed] [Google Scholar]
- Luo H, Song F, Goodman RM, Zheng Z. Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biol. 2005;7(5):459–468. doi: 10.1055/s-2005-865851. [DOI] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18(11):2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralampidis K, Milioni D, Rigas S, Hatzopoulos P. Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiol. 2002;129(3):1138–1149. doi: 10.1104/pp.004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999;27(2):470–478. doi: 10.1093/nar/27.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379(6):633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Hatorangan MR, Sentausa E, Wijaya GY. In silico identification of cis-regulatory elements of phosphate transporter genes in rice (Oryza sativa L.) J Crop Sci Biotechnol. 2009;12(1):25–30. doi: 10.1007/s12892-008-0054-8. [DOI] [Google Scholar]
- Cohn B, Manque P, Lara AM, Serrano M, Sheth N, Buck G. Putative cis-regulatory elements associated with heat shock genes activated during excystation of Cryptosporidium parvum. PLoS One. 2010;5(3):e9512. doi: 10.1371/journal.pone.0009512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey T, Weiller G. Mclip: motif detection based on cliques of gapped local profile-to-profile alignments. Bioinformatics. 2007;23(4):502–503. doi: 10.1093/bioinformatics/btl601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Duplication numbers of 9 most abundant CREs in top 40 highly expressed genes in sperm cells of rice. Based on the results of PLACE database SIGNALSCAN searches for total of 223 CREs, 9 of them are found to present in 1 Kb upstream regions of all 40 genes. The duplication numbers of those CREs are represented in this table. Frequency graph is also plotted for this distribution as shown in Figure 1.
Duplication numbers of other 19 abundant CREs present in 80% of sperm cell expressing genes of rice. Results of SIGNALSCAN searches gave 19 other CREs abundantly present in the dataset. The duplication numbers of these CREs are represented in this table. Frequency graph is also plotted for this distribution as shown in Figure 2.
A map of 28 abundant CREs and their positions within 1 Kb upstream sequences. SIGNALSCAN program of PLACE database identified the positions of CREs in the upstream regions of top 40 highly expressed genes in sperm cell of rice. These selected CREs were subjected to further extensive analysis for their duplication numbers and distribution across the upstream regions. The figure shows exact location of CREs present in 80% of the gene dataset. The blue bars above the horizontal black line indicate CREs on sense strand and the blue bars below the black line designates CREs on anti-sense strand.
Unique CREs. The analysis exhibited some unique CREs present in only one of the 40 sperm cell expressing genes with one or two duplications. These CREs were found in these specific sperm cell expressing genes.
Peculiar CREs. Besides abundant CREs present in 80% of the gene dataset, there are few others present in just 5-10% of rice sperm cell expressing genes.