Abstract
Cytogenetics play a major role in determining the prognosis of patients with AML. However, the existing cytogenetics classifications were developed on chemotherapy-treated patients and may not be optimal for patients undergoing allogeneic hematopoietic cell transplantation (HCT). We studied 821 adult patients reported to the CIBMTR who underwent HCT for AML in first or second CR between 1999 and 2004. We compared the ability of the 6 existing classifications to stratify patients by overall survival (OS). We then defined a new schema specifically applicable to HCT patients using this patient cohort. Under this CIBMTR schema, inv(16) is favorable, complex karyotype (4+ abnormalities) is adverse, and all other classified abnormalities are intermediate in predicting survival after HCT (5y OS 64%, 18%, and 50%, respectively, p=0.0001). This schema stratified patients into 3 groups with similar non-relapse mortality, but significantly different incidences of relapse, overall and leukemia-free survival. It applied to patients regardless of their disease status (CR1 or CR2), donor type (MRD or URD), or conditioning intensity (myeloablative or reduced intensity). This transplantation-specific classification could be adopted for prognostication purposes and to stratify patients with AML and karyotypic abnormalities entering HCT clinical trials.
Keywords: AML, cytogenetics, stem cell transplantation
INTRODUCTION
The importance of cytogenetics in determining the outcome of patients with acute myeloid leukemia (AML) is well established(1, 2). Several collaborative groups have proposed classification schemata to stratify patients by leukemia karyotype, based on retrospective studies of large independent patient cohorts. The most commonly used groupings are those of the MRC(3, 4) (now the National Cancer Research Institute (NCRI)), the CALGB(5), the SWOG/ECOG groups(6), and the EORTC/GIMEMA groups(7). More recently, Breems and colleagues have shown that a monosomal karyotype (MK) is a predictor of very poor outcome in AML(8). However, most patients in the above studies were treated with conventional chemotherapy rather than allogeneic hematopoietic cell transplantation (HCT). HCT is an important treatment modality for selected patients with AML, which underscores the necessity of understanding the role of cytogenetics in determining HCT outcome. While it appears that cytogenetics retain their prognostic relevance in the setting of HCT(6, 7, 9–11), there is no validated HCT-specific cytogenetics classification schema. Since a large part of the benefit of HCT depends on an immunologic graft-versus-tumor effect, it is conceivable that a given karyotypic subtype of AML might behave differently after HCT than after conventional chemotherapy. Therefore, classifications based on series of patients treated with chemotherapy may not apply optimally to transplanted patients. An HCT-specific cytogenetics grouping schema would be useful for prognostication, to help guide treatment decisions, to stratify patients entering clinical trials, and to compare reported HCT outcomes across studies or centers. We have previously proposed such a classification schema based on a cohort of patients transplanted at Dana-Farber Cancer Institute/Brigham and Women’s Hospital (henceforth referred to as the DFCI schema)(11), but this schema has not been validated on an independent cohort. We therefore performed a retrospective study through the Center for International Blood and Marrow Transplant Research (CIBMTR) with two aims: to validate the previous report by comparing the performance of the 6 existing classifications on an independent cohort of HCT patients; and to derive a new grouping schema based on this multicenter cohort.
METHODS
Data Source
The CIBMTR is a research organization comprising more than 500 transplant centers worldwide that contribute detailed data on consecutive allogeneic HCT. Patients are followed longitudinally, with yearly follow-up. Computerized checks for errors, physician reviews of submitted data, and on-site audits of participating centers ensure data quality. All patients registered with the National Marrow Donor Program (NMDP) were retrospectively contacted, and informed consent was obtained from surviving patients in accordance with the Declaration of Helsinki for participation in the NMDP research program. Informed consent was waived by the NMDP Institutional Review Board for all deceased patients. Surviving patients who did not provide signed informed consent to allow analysis of their clinical data were excluded. To adjust for the potential bias introduced by exclusion of non-consenting surviving patients, a corrective action plan (CAP) modeling process randomly excluded the same percentage of deceased patients using a biased coin randomization with exclusion probabilities based on characteristics associated with not providing consent for use of the data in survivors.
Patient Selection
We included all patients 18 years and older transplanted for AML in 1st or 2nd complete remission (CR1 or CR2) between 1999 and 2004 at CIBMTR centers that treated more than 20 eligible patients during this period. CR was defined as absence of blasts in peripheral blood, <5% blasts in marrow, with cellular marrow and normal CBC. No information was available on CRp or CRi status. Patients with acute promyelocytic leukemia or pre-existing myelodysplastic syndromes (MDS) were excluded (since cytogenetics for these patients may be better grouped according to an MDS-specific classification(11, 12)), as were patients with t(9;22) AML (given the difficulty of distinguishing this disease from blast crisis chronic myelogenous leukemia in a registry setting). Patients who had received a previous autologous progenitor cell transplantation for AML were excluded, as were patients transplanted from syngeneic or cord blood donors. Patients transplanted at Dana-Farber/Brigham and Women’s Hospital were ineligible (since they were used to derive the DFCI schema). Finally, we excluded all patients on whom cytogenetic information was missing or not obtainable.
Cytogenetics
We reviewed all available primary cytogenetics reports for patients with abnormal karyotypes (available for 92% of patients), with the help of an expert cytogeneticist (P.D.C.). Patients who were reported as having a normal karyotype were not reviewed. We used for this analysis the latest available tumor karyotype (i.e., at diagnosis for patients in CR1 and at relapse for patients in CR2 when available). Cytogenetics were classified according to each of the existing schemata (Supplementary Table 1), with the following modification: patients with t(8;21) or inv(16) were classified as favorable under the MK schema, even though this schema does not specifically define a favorable subgroup.
Study End points, Definitions and Statistical Analysis
The primary endpoint for this analysis was overall survival (OS) after HCT, defined as the time from transplantation to death, with surviving patients censored at the last time reported alive. Leukemia-free survival (LFS) was defined as the time from transplantation to death or relapse, with surviving patients censored at last time reported alive and leukemia-free. Non-relapse mortality (NRM) was defined as death without evidence of leukemia recurrence. OS and LFS were calculated using the Kaplan-Meier method. The log-rank test was used for comparisons of Kaplan-Meier curves. Cumulative incidence curves for non-relapse death and relapse were constructed reflecting time to relapse and time to NRM as competing risks. The difference between cumulative incidence curves in the presence of a competing risk was tested using the Gray method(13). Potential prognostic factors for OS, LFS, relapse, and NRM were examined in the proportional hazards model as well as in the competing risks regression model(14). Proportional hazards assumption for each variable of interest was tested. Interaction terms including interaction with time were examined in the proportional hazards regression model. In order to compare the performance of classification systems, we calculated the difference in Akaike Information Criterion (AIC) between the full model and a model that did not include cytogenetics. We also calculated the c-index(15) for censored OS data and Integrated Brier Score (IBS)(16) for models using each of the various grouping schemata. The analyses were conducted using Matlab v6.5 (Mathworks, Natick, MA), SAS version 9.2 (SAS Institute, Cary, NC) and R version 2.18.1.
RESULTS
Patient Characteristics and Cytogenetics
The baseline characteristics of the 821 patients are shown in Table 1. The median age was 41 years (range, 18–74). Four percent of patients had therapy-related leukemia. Sixty-one percent were transplanted in CR1 and 39% in CR2; 48% had a matched related donor (MRD), and 24% a well-matched unrelated donor (URD)(17); nearly two thirds received peripheral blood stem cells, and three quarters received a myeloablative conditioning regimen. Graft-versus-host disease (GVHD) prophylaxis regimens for the most part (94%) consisted of a calcineurin inhibitor-based regimen. Median follow-up was 61 months for survivors. Cytogenetics are shown in Table 2. Fifty-six percent of patients transplanted in CR1 and 67% of patients transplanted in CR2 had a normal karyotype. Six percent of CR1 and 16% of CR2 patients had core-binding factor (CBF) AML; 13% of CR1 and 9% of CR2 patients had a complex karyotype with at least 3 abnormalities.
Table 1.
Baseline patient characteristics
| Variable | N. (%)a |
|---|---|
| Number of patients | 821 |
| Number of centers | 49 |
|
| |
| Age (years) (median, range) | 41 (18–74) |
| <40 | 366 (45) |
| 40–49 | 220 (27) |
| 50–59 | 167 (20) |
| 60–64 | 48 (6) |
| 65+ | 20 (2) |
|
| |
| Gender | |
| Male | 457 (56) |
| Female | 364 (44) |
|
| |
| Therapy-related diseaseb | 36 (4) |
|
| |
| WBC at diagnosisc | |
| <20 | 411 (50) |
| 20–50 | 138 (17) |
| >50 | 174 (21) |
| Data missing | 98 (12) |
|
| |
| Disease Status at HCT | |
| CR1 | 498 (61) |
| CR2 | 322 (39) |
|
| |
| Duration of 1st CRd | |
| <6 months | 67 (21) |
| 6–12 months | 93 (29) |
| >12 months | 110 (34) |
| Data missing | 52 (16) |
|
| |
| Karnofsky performance score at SCT | |
| ≥90% | 597 (73) |
| <90% | 171 (21) |
| Data missing | 53 (6) |
|
| |
| Donor categorye | |
| MRD | 390 (48) |
| Non-MRD | 431 (52) |
| Well-matched URD | 194 (24) |
| Partially matched URD | 110 (13) |
| Mismatched URD | 110 (13) |
| URD matching unknown | 50 (6) |
| Mismatched relative | 40 (5) |
|
| |
| Graft source | |
| PB | 537 (65) |
| BM | 284 (35) |
|
| |
| Conditioning | |
| Myeloablative | 612 (75) |
| Non-myeloablative/Reduced intensity | 209 (25) |
|
| |
| GVHD prophylaxis | |
| Calcineurin inhibitor-based | 768 (94) |
| T-cell depletion | 32 (4) |
| Other | 21 (3) |
|
| |
| CMV serostatusf | |
| Recipient or donor + | 619 (75) |
|
| |
| Gender matchingg | |
| Female to male | 180 (22) |
| Male to female | 202 (25) |
| Female to female | 161 (20) |
| Male to male | 277 (34) |
|
| |
| Year of HCT (median, range) | 2002 (1999–2004) |
|
| |
| Months of follow-up (median, range) | 61 (3–122) |
Percentages may not add to 100 because of rounding
Data missing on 4 patients.
Units are x109/l.
For patients in CR2.
Classified according to reference (17).
Data missing on 21 patients.
Data missing on 3 patients.
WBC, white blood cell count; HCT, allogeneic hematopoietic stem cell transplantation; CR, complete remission; MRD, matched related donor; URD, unrelated donor; PB, peripheral blood; BM, bone marrow; GVHD, graft-versus-host disease; CMV, cytomegalovirus.
Table 2.
Cytogenetics
| Abnormality | CR1 patients N. (%)a |
CR2 patients N. (%)a |
|---|---|---|
| Number of patients | 499 | 322 |
|
| ||
| Normal | 281 (56) | 215 (67) |
|
| ||
| t(8;21) | 17 (3) | 24 (7) |
| Isolated | 11 | 12 |
| With del(9q) or complex karyotype | 2 | 8 |
| With other abnormalities | 4 | 4 |
|
| ||
| inv(16)-related (inv(16) or t(16;16)) | 15 (3) | 29 (9) |
| Isolated | 7 | 20 |
| With complex karyotype | 6 | 4 |
| With other abnormalities | 2 | 5 |
|
| ||
| Abnormal 7b | 33 (7) | 12 (4) |
| −7 | 17 | 5 |
| del(7q) | 10 | 5 |
| der(1;7)(q10;p10) | 2 | 0 |
| Other 7q abnormality | 4 | 2 |
|
| ||
| Abnormal 5c | 28 (6) | 3 (1) |
| −5 | 7 | 0 |
| del(5q) | 17 | 3 |
| Other 5q abnormality | 4 | 0 |
|
| ||
| 11q23 abnormality | 37 (7) | 10 (3) |
| t(6;11) | 6 | 3 |
| t(9;11) | 14 | 2 |
| t(10;11) | 2 | 0 |
| t(11;19) | 5 | 1 |
| Other 11q23 abnormality | 10 | 4 |
|
| ||
| 3q abnormality | 11 (2) | 3 (1) |
| t(3;3) | 1 | 0 |
| inv(3q) | 2 | 0 |
| t(3;5) | 0 | 1 |
| Other 3q abnormality | 8 | 2 |
|
| ||
| Trisomy/Tetrasomy | ||
| trisomy 8 | 31 (6) | 13 (4) |
| trisomy 21 | 6 (1) | 4 (1) |
| Other trisomy | 24 (5) | 9 (3) |
| tetrasomy 8 | 2 (0) | 2 (1) |
| tetrasomy 21 | 1 (0) | 0 (0) |
|
| ||
| Monosomy | ||
| Autosomal (except 5, 7 or 17) | 29 (6) | 7 (2) |
| -X | 3 (1) | 1 (0) |
| -Y | 5 (1) | 7 (2) |
|
| ||
| 17p abnormality | 18 (4) | 3 (1) |
| del(17p) | 1 | 0 (0) |
| Other 17p abnormality (including monosomy 17) | 17 | 3 (1) |
|
| ||
| 12p abnormality | 10 (2) | 4 (1) |
| del(12p) | 2 | 2 |
| add(12p) | 2 | 1 |
| t(4;12) | 0 | 1 |
| t(5;12) | 1 | 0 |
|
| ||
| Other 12p abnormality | 5 | 0 |
| Other | ||
| t(1;19) | 1 (0) | 0 (0) |
| t(6;9) | 7 (1) | 2 (1) |
| inv(6) | 1 (0) | 1 (0) |
| t(8;16) | 3 (1) | 0 (0) |
| del(9q) | 4 (1) | 8 (2) |
| dup(11q) | 0 (0) | 0 (0) |
| del(11q23) | 8 (2) | 0 (0) |
| del(16q22) | 1 (0) | 0 (0) |
| del(20q) | 2 (0) | 2 (1) |
| del(21q) | 1 (0) | 0 (0) |
| Tetraploid | 0 (0) | 1 (0) |
|
| ||
| Monosomal karyotyped | 31 (6) | 6 (2) |
|
| ||
| Complex karyotype (≥3 abnormalities) | 67 (13) | 29 (9) |
| 3 abnormalities | 16 (3) | 9 (3) |
| 4 abnormalities | 17 (3) | 7 (2) |
| 5 abnormalities | 34 (7) | 13 (4) |
| # of abnormalities (median, range) | 5 (3–15) | 4 (3–9) |
Percentages may not add to 100 because of rounding and because some patients have more than one abnormality (see Methods).
Of the patients with monosomy 7, 4 had no other abnormalities, 4 had 1 other abnormality, and the remainder had a complex karyotype (3–15 abnormalities).
Other than for 2 patients who had isolated del(5q), abnormalities of chromosome 5 were in all case associated with other abnormalities; patients with monosomy 5 all had complex karyotypes (5–15 abnormalities).
defined as ≥2 autosomal monosomies or 1 single autosomal monosomy with ≥1 structural abnormality (clonal deletion, addition, inversion or translocation).
Model Comparison for Overall Survival
Five-year OS for the entire cohort was 48% (95% confidence interval 44–51%). We built Cox models for OS using cytogenetics (grouped according to the MRC schema), considering the following covariates: year of transplant, age, patient and donor gender, disease status at HCT, CR1 duration (for patients in CR2), therapy-related disease, white blood cell count at diagnosis, Karnofsky performance score (KPS) at HCT, donor category(17), cytomegalovirus (CMV) serostatus, conditioning intensity, GVHD prophylaxis regimen, graft source, and CD34 cell dose. Among these factors, cytogenetics, therapy-related disease, KPS (above or below 90%), duration of CR1, donor category, donor gender and graft source were significant for OS (not shown). The results did not change significantly if cytogenetics were categorized using another of the existing schemata. We compared the ability of the 6 existing cytogenetics grouping schemata to stratify patients for OS, using 3 different metrics for this comparison: the Akaike Information Criterion (AIC) difference, which measures the improvement in model fit by the addition of the covariate of interest (in this case, cytogenetics risk group), with a better fit indicated by a higher difference; the c-index, which is a generalization of the area under the curve (AUC) for censored survival data, using the cytogenetics risk assignment as a predictor, with a better predictor indicated by a higher proportion (and an expected value of 0.5 for random prediction); and the Integrated Brier Score (IBS), which is a measure of prediction error with a better predictor indicated by a lower score. The results are shown in Table 3. The DFCI schema provided the highest AIC difference, c-index and lowest IBS (although the differences were small and likely insignificant). Of note, in the Cox model, cytogenetics were the most influential factor (highest HR for mortality) for OS.
Table 3.
Comparison of existing cytogenetic schemata on entire cohort
| Grouping schema | Number of patients not classifiable (%) | Model fit scorea | c-indexb | IBSc | HR for favorable (p value)d | HR for adverse (p value)d |
|---|---|---|---|---|---|---|
| MRCe | 0 (0) | 56.3 | 0.53 | 0.154 | 0.8 (0.13) | 1.8 (0.0001) |
| CALGB | 68 (8) | 58.5 | 0.52 | 0.155 | 0.6 (0.02) | 1.5 (0.001) |
| SWOG/ECOG | 51 (6) | 49.0 | 0.53 | 0.154 | 0.7 (0.19) | 1.4 (0.010) |
| EORTC/GIMEMA | 0 (0) | 50.0 | 0.53 | 0.153 | 0.8 (0.4) | 1.5 (0.003)f |
| DFCI | 5 (1) | 60.0 | 0.55 | 0.153 | 0.7 (0.080) | 1.8 (<0.0001) |
| MK | 0 (0) | 51.3 | 0.52 | 0.155 | 0.8 (0.2) | 1.9 (0.001) |
Comparisons were performed on the subset of 749 patients classifiable under all schemata, in a multivariable model that included patient age, gender, therapy-related disease, WBC at diagnosis, disease status, duration of 1st CR, conditioning intensity, GVHD prophylaxis regimen, Karnofsky score, graft source, donor category, and CMV serostatus.
Defined as the difference in Akaike Information Criterion between model without covariates and model with covariates (the higher the difference, the better the model fit.
Defined as the generalization of the area under the curve for censored survival data using as a predictr the cytogenetics risk group assignment, with a better fit indicated by a higher proportion.
Integrated Brier Score, which measures prediction error; the lower the score, the better the prediction ability.
Intermediate risk is the reference group for both comparisons. HR denotes hazard ratio for mortality.
The model fit using the revised MRC classification(4) was inferior to the one using the original classification(3) (not shown).
For the very adverse group. Adverse group in this schema was comparable to the intermediate group (HR=1.1, p=0.6).
HR denotes hazard ratio for mortality; other abbreviations are as in Tables 1–3.
Derivation of a CIBMTR Schema
We then used the present cohort to derive a new grouping schema. For this, we excluded 13 patients with 2 abnormalities (to avoid making assumptions about the hierarchy of different abnormalities) and only included karyotypic abnormalities present in at least 20 patients, thus restricting the analysis to 787 patients. We determined the hazard ratio for mortality for each cytogenetic abnormality relative to normal karyotype, in the same Cox model used above for model comparison. In order to better define the outcome of patients with complex karyotype, we split them into those with 3, 4, and 5 or more abnormalities. The results, shown in Table 4, allowed us to define 3 distinct risk groups: a favorable group with inv(16) (5% of patients), an adverse group with complex karyotype (with 4 or more abnormalities) (9% of patients), and an intermediate group comprising normal karyotype and all other classified abnormalities (86% of patients). Even though the p value associated with 4 abnormalities did not reach statistical significance for OS, the hazard ratio for mortality was 1.5, and the LFS of those patients was significantly worse than those with normal karyotype (HR=1.6, p=0.046), justifying its place in the adverse group. The OS of patients with t(6;9) was significantly worse than that of patients with normal karyotype (HR=2.7, p=0.049), but we did not include this in the final classification given the small number of patients with this abnormality. In this analysis, neither isolated t(8;21) nor t(8;21) in conjunction with other abnormalities was significantly different from normal karyotype. In addition, within the complex karyotype group, no specific abnormality appeared to affect outcome with the exception of inv(16), which in the presence of a complex karyotype (4+ abnormalities) behaved like an intermediate-risk abnormality (HR relative to normal karyotype 1.0, p=1.0). Finally, within the adverse group, an MK genotype was not associated with significantly worse outcome (HR=1.2, p=0.5). Using the same method to derive a grouping schema for LFS yielded the same results.
Table 4.
CIBMTR Cytogenetics Grouping Schema
| Group | Abnormality | HRb | p value |
|---|---|---|---|
| Favorable | Inv(16) (without complex karyotype) | 0.5 | 0.030 |
| Normal | 1.0 | Reference | |
| t(8;21) | 1.1 | 0.8 | |
| 11q23 abnormality | 1.1 | 0.7 | |
| Intermediate | Trisomy/tetrasomy 8 | 1.2 | 0.5 |
| Abnormal 5 or 7 | 1.3 | 0.3 | |
| Complex (3 abnormalities) | 1.1 | 0.7 | |
| Inv(16) with complex karyotype | 1.0 | 1.0 | |
| Other abnormalitiesa | 1.1 | 0.7 | |
| Adverse | Complex (4 abnormalities) | 1.5 | 0.10 |
| Complex (5+ abnormalities) | 2.3 | <0.0001 | |
Except for abn12p, abn3q, del(9q), t(1;19), t(9;22), t(6;9), which are not classified in this schema
Compared to normal karyotype
Performance of the CIBMTR Schema
We then tested the performance of the new CIBMTR cytogenetics grouping schema in comparison with the existing schemata, again restricting the comparison to patients classifiable under all schemata (727 patients). The new schema overall outperformed all others (AIC difference 67.6, compared to next best score of 61.5 for DFCI schema, IBS 0.151 compared to 0.153 for DFCI, c-index 0.54, compared to 0.54 for DFCI). Because there were too few patients in this dataset to perform internal validation studies, we used our previous (and entirely independent) DFCI cohort(11) as a validation group, restricting it to the 189 patients who met eligibility criteria for the present study. The CIBMTR schema could stratify patients into three groups with significantly different OS (5y OS 76%, 41% and 23% for favorable, intermediate and adverse cytogenetics, respectively; p=0.019).
The OS, LFS, cumulative incidence of relapse and NRM for all classifiable patients based on this new CIBMTR schema are shown in Figure 1. The 5-year (5y) OS for patients in the favorable group was 64% (95% confidence interval [95CI], 47–77%), compared to 50% (95CI, 46–54%) in the intermediate group and 18% (95CI, 9–28%) in the adverse group (p=0.0001). In the multivariable model, the HR for mortality of the favorable group, compared to the intermediate one, was 0.7 (p=0.03), while the HR of the adverse group compared to intermediate was 1.9 (p<0.0001). Abnormal karyotypes included in the intermediate group had a similar outcome to normal karyotype (5y OS 46% versus 51%, p=0.5). The 5-year LFS for patients in the 3 groups were 58% (95CI, 40–72%), 45% (95CI, 41–49%) and 16% (95CI, 8–26%) (p<0.0001). The cumulative incidence of relapse (CIR) was also significantly different (19%, 29% and 51%, p=0.0003), while NRM was not significantly different (23%, 25%, and 33%, p=0.30). The significant association of cytogenetic risk group with LFS and CIR was confirmed in multivariable models: the HR for death or relapse of favorable compared to intermediate was 0.6 (p=0.055), and of adverse compared to intermediate 1.9 (p<0.0001). The HR for relapse of favorable compared to intermediate was 0.5 (p=0.067), and of adverse compared to intermediate 2.0 (p<0.0001). Conversely, cytogenetics risk group did not significantly affect NRM (HR of favorable compared to intermediate 0.9, p=0.8; of adverse compared to intermediate 1.2, p=0.4).
Figure 1. Transplantation outcomes for patients classified by the CIBMTR grouping schema.
(A) Overall survival; (B) Leukemia-free survival; (C) Cumulative incidence of relapse; (D) Cumulative incidence of non-relapse mortality.
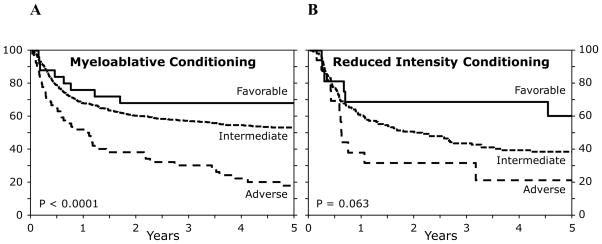
As shown in Figures 2–4, the CIBMTR schema applied to patients regardless of disease status (CR1 versus CR2, see Figure 2), donor category (MRD versus URD, see Figure 3), and conditioning intensity (ablative versus reduced intensity (RIC), see Figure 4). The log-rank p value in the RIC subgroup did not reach statistical significance (p=0.063), but this is likely because of small patient numbers, and the differences in LFS remained significant (log-rank p=0.025).
Figure 2. Overall survival of patients classified according to the CIBMTR schema, stratified by disease status.
(A) Patients transplanted in CR1; (B) Patients transplanted in CR2.
Figure 4. Overall survival of patients classified according to the CIBMTR schema, stratified by conditioning intensity.
(A) Patients transplanted with myeloablative conditioning; (B) Patients transplanted with reduced intensity conditioning.
Figure 3. Overall survival of patients classified according to the CIBMTR schema, stratified by donor type.
(A) Patients transplanted from a matched sibling donor; (B) Patients transplanted from an unrelated donor.
DISCUSSION
We propose a new cytogenetics grouping schema specifically applicable to patients with AML undergoing HCT in 1st or 2nd CR. This schema separates patients with abnormal karyotypes into 3 groups: a favorable group (inv(16)), an adverse group (complex karyotype, which may be optimally defined in the HCT context as 4 or more abnormalities), and an intermediate group comprising all other classified abnormalities, which has an outcome similar to normal karyotype.
Several limitations of this work must be considered. This is a retrospective study subject to selection bias of patients chosen to receive a HCT. This is particularly relevant for patients with CBF AML, for whom HCT is not commonly performed in 1st CR. There were not significantly more patients with WBC > 20 × 109/l at diagnosis in the t(8;21) or inv(16) subgroups (compared to normal karyotype); but it is quite possible that the patients in our study had other undefined adverse features associated with the decision for transplantation (e.g., leukemia cutis, number of regimens needed to reach CR, etc). It is nonetheless reassuring that a favorable subgroup could still be identified, and that the classification is applicable to both patients in CR1 and in CR2 (see Figure 2), since HCT may be considered standard therapy for CBF AML in CR2. Moreover, the retrospective nature of this study has the advantage that the observational database of the CIBMTR provides realistic data reflecting current practice and allows for a variety of heterogeneous factors to be potentially neutralized(18).
We stress that this schema should not be interpreted as a direct comparison of HCT and conventional chemotherapy outcomes. While it is tempting to conclude from this study that patients with abnormal karyotypes that are considered adverse under most conventional schemata, but intermediate under the CIBMTR schema (such as abnormal 5 or 7) should be considered for HCT in CR1, our data cannot directly address this inference. In this respect it should be remembered that the survival curves shown here depict the outcomes of transplanted patients, and are not comparable to intent-to-treat curves for cohorts of patients with newly diagnosed AML.
This work could not incorporate any molecular information, such as c-Kit, FLT3, NPM-1 status, etc, minimal residual disease (MRD) status, or cytogenetic remission information, which was not available in our dataset. The relevance of karyotypically silent mutations is now beyond dispute(19), although their relevance in the HCT setting is not well understood. Certainly the present schema could be refined in the future by the incorporation of mutation status, which would be a very important step forward in defining prognostic subgroups for patients with AML undergoing HCT. Gene-gene interactions involving both mutations and karyotype may also be important in the biology of leukemia(20); their relevance in the HCT setting may be addressed when a large transplant population with complete karyotypic and molecular information is available.
The present cohort was not large enough to allow an independent validation cohort. However, the CIBMTR schema performed well in the (independent) cohort that we used previously to establish the DFCI schema. It is also worth noting the very close similarity of the CIBMTR and the DFCI schemata (weighted Kappa statistic 0.73). The only differences are the assignment of isolated t(8;21) (intermediate here and favorable in DFCI), and the definition of complex karyotype (4+ abnormalities here versus 3+ in DFCI schema; but it should be noted that the optimal definition of complex karyotype was not studied in the DFCI schema). Those schemata (CIBMTR and DFCI) were derived on 2 entirely independent patient cohorts and yielded essentially the same results. This provides an external validation of the CIBMTR schema, with the exception of isolated t(8;21), whose exact place will have to be confirmed in further studies. There is at least one study suggesting that t(8;21) is not as favorable as inv(16) after HCT, similar to the present results(21).
Finally, it is possible that a 3-group cytogenetics classification underestimates the complexity of the problem, and that a much larger study could subdivide karyotypes further, as well as elucidate the prognostic importance of rare karyotypic abnormalities that were under-represented in our dataset. For example, it appears that a complex karyotype with 4 abnormalities is less adverse than 5 or more. However, given the challenges associated with reviewing primary cytogenetics data in a registry study, a much larger study may not be practical. It may also be of interest in the future to examine the prognostic impact of cytogenetic evolution. There were not sufficient data in our cohort to assess this rigorously. We therefore chose the abnormal karyotype closest to the time of transplant for analysis, since this is likely to be the best determinant of outcome.
An HCT-specific cytogenetics grouping schema, such as the one proposed here, may have several uses. First, it may help clinicians and patients better estimate the prognosis after HCT, which is often a critical part of the decision to pursue HCT. Second, it provides a system for stratifying adult patients entering clinical trials of HCT for AML. Indeed, in our cohort (as in a previous one(11)), cytogenetics was the single most important determinant of patient outcome, highlighting the importance of cytogenetics stratification in clinical trials. This highlights the need to stratify patients in clinical trials, and provides a means for doing so. Similarly, this schema could help to compare outcomes across different HCT studies or across institutions, which has become essential in the era of mandatory outcome reporting.
Supplementary Material
Acknowledgments
P.A. is supported by an ASH Scholar Award and an ASCO Career Development Award. This work was also supported by NIAID U19 AI 29530 and NHLBI PO1 HL 070149.
The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement U24-CA76518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 5U01HL069294 from NHLBI and NCI; a contract HHSH234200637015C with Health Resources and Services Administration (HRSA/DHHS); two Grants N00014-06-1-0704 and N00014-08-1-0058 from the Office of Naval Research; and grants from Allos, Inc.; Amgen, Inc.; Angioblast; Anonymous donation to the Medical College of Wisconsin; Ariad; Be the Match Foundation; Blue Cross and Blue Shield Association; Buchanan Family Foundation; CaridianBCT; Celgene Corporation; CellGenix, GmbH; Children’s Leukemia Research Association; Fresenius-Biotech North America, Inc.; Gamida Cell Teva Joint Venture Ltd.; Genentech, Inc.; Genzyme Corporation; GlaxoSmithKline; Kiadis Pharma; The Leukemia & Lymphoma Society; The Medical College of Wisconsin; Millennium Pharmaceuticals, Inc.; Milliman USA, Inc.; Miltenyi Biotec, Inc.; National Marrow Donor Program; Optum Healthcare Solutions, Inc.; Otsuka America Pharmaceutical, Inc.; Seattle Genetics; Sigma-Tau Pharmaceuticals; Soligenix, Inc.; Swedish Orphan Biovitrum; THERAKOS, Inc.; and Wellpoint, Inc. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, or any other agency of the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mrozek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 2.Grimwade D, Moorman A, Hills R, et al. Impact of karyotype on treatment outcome in acute myeloid leukemia. Ann Hematol. 2004;83 (Suppl 1):S45–48. doi: 10.1007/s00277-004-0849-8. [DOI] [PubMed] [Google Scholar]
- 3.Grimwade D, Walker H, Oliver F, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 4.Grimwade D, Hills RK, Moorman AV, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 5.Byrd JC, Mrozek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 6.Slovak ML, Kopecky KJ, Cassileth PA, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–4083. [PubMed] [Google Scholar]
- 7.Suciu S, Mandelli F, de Witte T, et al. Allogeneic compared with autologous stem cell transplantation in the treatment of patients younger than 46 years with acute myeloid leukemia (AML) in first complete remission (CR1): an intention-to-treat analysis of the EORTC/GIMEMAAML-10 trial. Blood. 2003;102:1232–1240. doi: 10.1182/blood-2002-12-3714. [DOI] [PubMed] [Google Scholar]
- 8.Breems DA, Van Putten WL, De Greef GE, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J Clin Oncol. 2008;26:4791–4797. doi: 10.1200/JCO.2008.16.0259. [DOI] [PubMed] [Google Scholar]
- 9.Ogawa H, Ikegame K, Kawakami M, et al. Impact of cytogenetics on outcome of stem cell transplantation for acute myeloid leukemia in first remission: a large-scale retrospective analysis of data from the Japan Society for Hematopoietic Cell Transplantation. Int J Hematol. 2004;79:495–500. doi: 10.1532/ijh97.03166. [DOI] [PubMed] [Google Scholar]
- 10.Burnett AK, Wheatley K, Goldstone AH, et al. The value of allogeneic bone marrow transplant in patients with acute myeloid leukaemia at differing risk of relapse: results of the UK MRC AML 10 trial. Br J Haematol. 2002;118:385–400. doi: 10.1046/j.1365-2141.2002.03724.x. [DOI] [PubMed] [Google Scholar]
- 11.Armand P, Kim HT, DeAngelo DJ, et al. Impact of cytogenetics on outcome of de novo and therapy-related AML and MDS after allogeneic transplantation. Biol Blood Marrow Transplant. 2007;13:655–664. doi: 10.1016/j.bbmt.2007.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armand P, Deeg HJ, Kim HT, et al. Multicenter validation study of a transplantation-specific cytogenetics grouping scheme for patients with myelodysplastic syndromes. Bone Marrow Transplant. 2009 doi: 10.1038/bmt.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray R. A class of K-sample tests for comparing the cumulative incidence of a competing risk. The Annals of Statistics. 1988;16:1140–1154. [Google Scholar]
- 14.Fine J, Gray R. A proportional hazards model for the subdistribution of a competing risk. Journal of the American Statistical Association. 1999;94:496–509. [Google Scholar]
- 15.Harrell FE, Jr, Lee KL, Califf RM, Pryor DB, Rosati RA. Regression modelling strategies for improved prognostic prediction. Stat Med. 1984;3:143–152. doi: 10.1002/sim.4780030207. [DOI] [PubMed] [Google Scholar]
- 16.Graf E, Schmoor C, Sauerbrei W, Schumacher M. Assessment and comparison of prognostic classification schemes for survival data. Stat Med. 1999;18:2529–2545. doi: 10.1002/(sici)1097-0258(19990915/30)18:17/18<2529::aid-sim274>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 17.Weisdorf D, Spellman S, Haagenson M, et al. Classification of HLA-matching for retrospective analysis of unrelated donor transplantation: revised definitions to predict survival. Biol Blood Marrow Transplant. 2008;14:748–758. doi: 10.1016/j.bbmt.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale RP, Eapen M, Logan B, Zhang MJ, Lazarus HM. Are there roles for observational database studies and structured quantification of expert opinion to answer therapy controversies in transplants? Bone Marrow Transplant. 2009;43:435–446. doi: 10.1038/bmt.2008.447. [DOI] [PubMed] [Google Scholar]
- 19.Motyckova G, Stone RM. The role of molecular tests in acute myelogenous leukemia treatment decisions. Curr Hematol Malig Rep. 2010;5:109–117. doi: 10.1007/s11899-010-0049-7. [DOI] [PubMed] [Google Scholar]
- 20.Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol. 2011;29:475–486. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 21.Kuwatsuka Y, Miyamura K, Suzuki R, et al. Hematopoietic stem cell transplantation for core binding factor acute myeloid leukemia: t(8;21) and inv(16) represent different clinical outcomes. Blood. 2009;113:2096–2103. doi: 10.1182/blood-2008-03-145862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.