Abstract
Molecular imaging allows clinicians to visualize disease specific molecules, thereby providing relevant information in the diagnosis and treatment of patients. With advances in genomics and proteomics and underlying mechanisms of disease pathology, the number of targets identified has significantly outpaced the number of developed molecular imaging probes. There has been a concerted effort to bridge this gap with multidisciplinary efforts in chemistry, proteomics, physics, material science, and biology; all essential to progress in molecular imaging probe development. In this review, we will discuss target selection, screening techniques and probe optimization with the aim of developing clinically relevant molecularly targeted imaging agents.
Keywords: molecular imaging, probe discovery, screening, target
Development of molecular imaging agents
Molecular imaging relies on appropriate targets or biomarkers and the ability to generate probes that are selective and specific for those biomarkers. With the genomics and proteomics eras as well as increased understanding of the etiology of disease, targets are being rapidly identified, but the number of imaging agents to those targets has not kept pace. Bridging this gap has many far-reaching implications for the future directions of molecular imaging and radiological sciences including the early detection of disease (e.g. atherosclerosis, cancer, diabetes, etc.) by the identification of early molecular signatures, accurate disease staging and treatment stratification, and the evaluation of various treatments and therapeutics (i.e. efficacy and dosing).
In general, there are three aspects of probe development, although sometimes these can be combined. The first is to identify a biological target for the condition of interest. The second is to generate a compound that binds to this target. The third is to convert the binding compound to a probe and to optimize in-vitro and in-vivo characteristics such as affinity, pharmacokinetics, toxicity, and elimination, with validation. In this review, we will discuss target selection, screening techniques for identifying the targeting moiety, and probe optimization with the aim of developing clinically relevant molecularly targeted imaging agents
Targets
The outpouring of information about aberrant molecular pathways and the proliferation of tools (such as genechips and other microarrays) have led to a great deal of information about the etiology of human disease. It is becoming easier and easier to select a list of proteins that are relevant to any biological problem from a literature search. Unfortunately, not every protein is useful for imaging. A good target has several characteristics: 1) It is expressed differently in diseased tissue than surrounding normal tissue – preferentially not at all in the normal with high levels in the diseased tissue. 2) The target is highly expressed, although the exact density of receptors required varies with the sensitivity of the imaging modality. For example, in prostate cancer, the bombesin receptor is expressed on the order of 10,000 copies/cell (1). This target has proven adequate for SPECT imaging using Tc99; (2, 3). However, similar probes proved inadequate to image the same condition by MRI using iron oxide nanoparticles for contrast(4). 3) It has to be accessible. The endothelium lining of blood vessels forms a tangible barrier. Therefore, unless the target is an endothelial cell, a probe must be able to extravasate out of the blood vessels and into the tissue of interest. In many diseases, such as cancer or atherosclerosis, the endothelial barrier is compromised and the lymphatics impaired, leading to the enhanced permeability and retention (EPR) effect. Many non-targeted and targeted imaging agents take advantage of this circumstance, allowing delivery and accumulation to the diseased area. This is especially important for nanoparticles large enough to stay in the circulation: the fenestrations in normal vessels exclude extravasation into those tissues but the enlarged pores from the disease conditions allow selective perfusion in those areas. Similarly, the plasma membrane is another barrier that must be overcome if the desired target is a cytoplasmic or nuclear protein (fig 1). Currently, there are few effective mechanisms to breach the plasma membrane and ensure homogenous distribution throughout the cellular compartments, therefore, almost all probes bind to extracellular or membrane associated targets. 4) To increase the sensitivity and contrast, it is preferable for a cell surface target to be internalized upon binding and quickly recycled to the cell surface. This pumps the targeted agent into the cell to gain a measure of amplification relative to the bloodstream concentration (5).
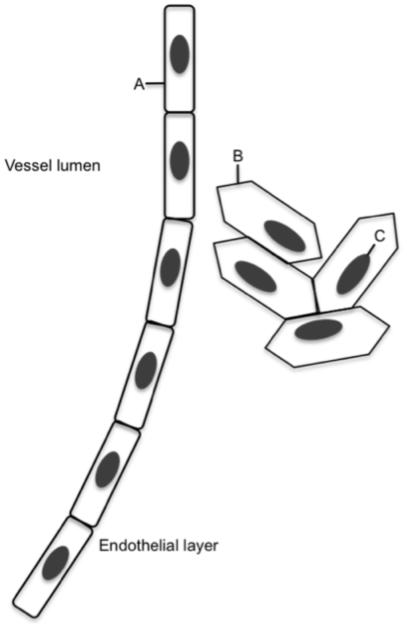
Fig 1. Physical barriers to target.
A- Target is on the surface of the endothelium and accessible from the bloodstream. B- Target is at cell surface, and is accessible from the bloodstream only if the endothelial layer is compromised. C- Target is inside the cell. Any probe binding to this target must cross both the endothelium and the cell membrane.
Although genomics and proteomics have given investigators a wealth of potential targets, there is information to be gained by performing a no a priori knowledge screen using one of the combinatorial techniques described below (6-9). For example, a novel marker of pancreatic cancer has been identified using phage display and diseased tissue (6). In this example, the protein, plectin1 is on the cell surface for pancreatic cancer whereas in normal cells it is strictly cytoplasmic; taking advantage of the plasma membrane barrier to allow exquisite selectivity between normal and diseased tissue. With this type of screen, it is not always trivial to determine the exact receptor the probe binds to, but it is essential in this case to determine what other tissues, if any, express the mystery protein on their cell surface.
Targeting Moiety Identification
Peptides
Peptides have many characteristics that make them useful as imaging agents. They have a short blood lifetime, are non-immunogenic, are relatively inexpensive to synthesize using standard conditions on commonly available automated machines, and easy to chemically modify. While using a synthetic protein from a known protein-protein interaction to generate an imaging agent is feasible, these tend to have long blood lifetimes if they don't trigger an immune response, are expensive, and may lose their binding ability when modified. Truncating a known protein binding partner into a peptide, however, can resolve these problems while retaining much of the affinity of the original protein-protein interaction (for example (10-12)). These peptide fragments can be transformed into an imaging agent (2, 3, 13), but a project such as this is an immense amount of work requiring biological details including structure that are presently unknown for many targets.
Generating libraries of possible targeting compounds offers a way to circumvent the limitations of systematic design. With the 20 natural amino acids forming the entire palette from which proteins are made, the diversity of a library of peptides containing every combination of 7 amino acids (1.3 × 109 sequences) will usually have a few members that bind with a target with desired affinity. Libraries can be synthesized chemically, which allows the simple addition of non-natural amino acids and access to derivatives, or biologically, on phage, yeast, or bacteria; allowing the researcher to harness the power of biological enzyme systems to speed up and simplify the screening process. Both methods can generate either linear or disulfide constrained libraries. Due to entropic effects, disulfide constrained libraries tend to generate fewer leads with better affinities compared to linear peptide libraries (14) but chemically modifying the peptides afterwards can be challenging.
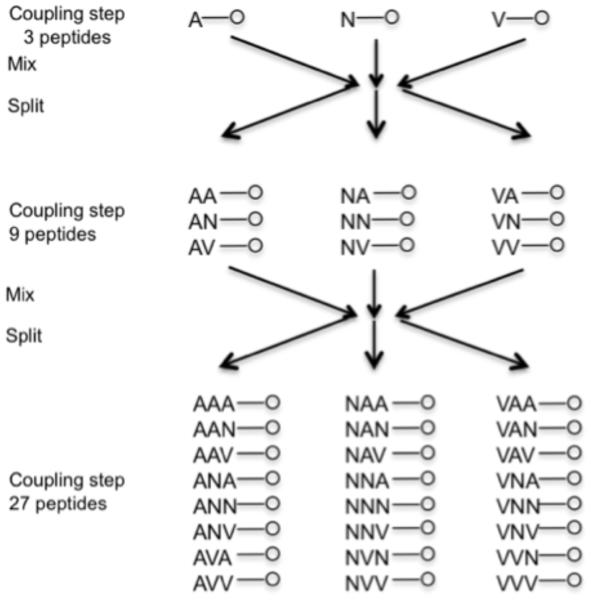
As synthesizing and screening 109 peptides one by one is unfeasible with present technology, there have been many methods developed to synthesize, screen, and deconvolute peptide libraries, including pin synthesis (15, 16), library encoding (17), various positionally encoded methods (18, 19), and affinity chromatography methods (20). The most comprehensively developed chemically synthesized system appears to be the one bead one compound methodology (21-25). This system uses a pool and split technique to generate random peptides on 100μm polystyrene beads. The beads are split into 19 separate pools; each pool has a different amino acid added to it using standard chemistry (cystine is not used outside defined locations to avoid random crosslinking). All the separate reactions are mixed together, and again split into 19 reactions (see Fig 2). This is continued until the peptides are long enough to provide the desired diversity. The peptides on any one bead are identical, but for all but the shortest peptides, no two beads will have the same sequence (23).
Fig. 2.
Pool and split library synthesis scheme
Screening of proteins for binding peptide sequences consists of mixing the biotin labeled protein with the beads, then adding streptavidin-alkaline phosphatase (23, 25). The chromogenic substrate bromo-chloro-indoyl-phosphate is added, and the beads that turn visibly blue are collected using microdisection tools. The bound receptor and streptavidin are removed, and the beads reprobed with streptavidin-alkaline phosphatase to remove the peptides that bind to streptavidin rather than the intended protein. Each bead is individually sequenced by automated Sanger degredation to recover the peptide sequences (23, 25).
Recovering sequences that bind to cells uses a slightly more complex protocol (21, 25). Trypsanized cells are mixed with the on-bead library and allowed to sit for 48 hours. If the cells have surface markers that bind to a peptide, they grow on the bead containing that sequence, which can be identified under a microscope. It is possible to use a negative cell line, by labeling those cells with Calcein AM, a green fluorescent dye. To restrict the screen to a specific protein on the cell membrane a third screen is run on the beads that bound using an antibody to the desired protein to block binding (21, 25).
One bead one compound allows rapid screening using common equipment. There is great flexibility in the library structure; standard peptide synthesis permits D-amino acids and non-natural amino acids, and strategic positioning of cystines allow for cyclic peptides (21, 23, 24). The potential pitfalls relate to the size of the beads used. As the number of possible sequences grows exponentially with peptide length, it becomes very difficult to cover the complete diversity space with peptides longer than about 5 amino acids (24). A second challenge is in-vivo assays; these beads are too large to inject into a living animal without causing an embolism.
Phage Display
Researchers have harnessed the power of biological methods to make and screen libraries. Peptide sequences have been encoded on a plasmid attached to the peptide (26) with the library expressed in E-coli, peptides attached to bacterial polysomes (27, 28), displayed on yeast (29-31) or bacteria and on bacteriophage.
Phage display has been the workhorse technique with thousands of publications for screening peptides with desired binding characteristics. It has been used to screen for peptides that bind to protein, such as streptavidin (32-39), small molecules (40), cells (for example prostate cancer (41-46)), inorganic compounds (for example, platinum metal (47-51)), and even substrates such as paint chips (titanium dioxide (52)). It has been used in-vivo in animals (7, 53-58) and humans (59), and can be engineered to display non-natural amino acids (60, 61) although with more difficulty than chemical synthesis. Phage display has not been able to find peptide sequences that bind to specific nucleotide sequences however, as the screens yield sequences enriched in postive amino acids that bind indiscriminately to the ribose or deoxyribose backbone (for example (62) –hits are essentially polyarginine).
While there are a very large number of phage screening libraries for peptide binding, including different bacterophage (63, 64) and expression of various valency levels on different coat proteins (14), most of the literature uses one of the M13 bacterophage libraries commercialized by New England Biolabs. These are filimentous phage with the peptide library expressed on the five PIII protein on one end of the viron, one peptide per protein. The various libraries differ in the length of the peptides of the library and disulfide constraint. Longer peptide sequences are best for selection against targets where the binding will be relatively weak, but repeating, such as inorganic crystals. The tradeoff is in peptide diversity; the typical phage display protocol uses around 1010 viron particles, enough to completely cover a 7 amino acid library, but the commercial 12 amino acid library has 1015 possible combinations. Most proteins bind to other proteins through relatively small areas (65) (hotspots) which, tend to be areas that phage peptide sequences also bind (65, 66). Constrained libraries have peptides that are forced into a limited number of configurations; an example of which is the disulfide constrained libraries. Another way to confer confirmation restraints involves histidine complexation to metal ions (67). None of these libraries are long enough to form a stable tertiary structures.
The protocol for panning for clones that bind to a target, while subject to the creativity of the researcher and can be modified to emphasize different factors of importance, is rather basic (14). Phage are incubated on the target, often immobilized on a plate or some other geometry. The non-binding phage are washed off, and the bound phage eluted, titered, and amplified. This is repeated for a total of 2-5 rounds, after which clones are picked and sequenced. Many protocols include a subtraction step at the first panning to remove phage that bind to similar but undesired targets in addition to the selection (40, 45).
Elution of the phage from the target has been an area of great creativity. The M13 bacteriophage is very stable, allowing researchers to use harsh non-specific elution conditions to weaken the peptide-target binding, such as high salt concentrations, denaturing conditions, pH extremes, or even proteases (14). Phage that bind to specific sites on a target can be eluted by displacement with the natural ligand (for example (32)) or an antibody to the target (68, 69). If the phage-target binding is extremely strong, it may be necessary to amplify the phage without elution (70, 71).
An important nuance allows the differentiation between phage that are internalized into a cell, presumably by binding to a recepter that is internalized, and phage that are confined to the surface. Once all the phage bound to the cell are eluted, cell lysis will free all the clones that were able to enter the cell membrane (72). As the cell is constantly sampling the media by pinocytosis and turning over receptors, the time the cells are exposed to phage is an important parameter for selectivity; ideally this would be similar to the turnover time of the receptor targeted (72). Unfortunately, this information is difficult to discover, especially for a screen against an unknown target.
In-vivo phage display screening is slightly different than in-vitro. The phage is usually injected i.v. and allowed to circulate long enough for blood clearance (about 15 min to an hour (56)). The exposure of the phage to all the surfaces of the endothelium remove undesired clones and can function as a subtraction step (5). Phage are recovered by excising the tissue of interest, homogenizing it, and adding it to bacteria for amplification. As with the in-vivo screens, this is repeated as needed, usually for 2-4 rounds.
The biodistribution of unlabeled phage give one of the limits of in-vivo screening. In mice, the virons tend to accumulate in the spleen, liver, lung, and, to a lesser extent, the kidney, with some variation with mouse breed (56). This can cause problems in isolating phage from these organs, due to the high number of non-specific clones. For example, an in-vivo screen for lung binding peptides yielded 3 clones that bound out of a total of 143 that were isolated in 5 rounds of panning (73). The second limitation is the ability of the phage to reach desired tissue. Intravenously injected phage are limited in their ability to exit the endothelium unless it is compromised (as in cancer or inflamation) (72). An in-vivo screen against cardiomyocytes using an i.v. injection for example will be challenging as the phage may have difficulty gaining access to the cardiomyocytes.
While phage display is the main method in the literature for discovering peptide sequences that bind to a target, there are a number of papers where bacterial display is utilized. This technique consists of a random peptide sequenced engineered to be expressed on an extracellular protein of the bacterium. The first and most common modification is the peptide sequence placed into a disulfide constrained loop of E. coli thioredoxin (trxA) which is inserted into an E. coli flagellum protein (fliC) (74, 75), commercialized by Invitrogen under the name FliTrx, although it does not presently appear to be available. Other proteins used are FhuA (76), OmpA (77-79) and FimH (80). Bacteria other than E coli have also been used (81). The differences between the different platforms are not obvious from the literature with the exception of OmpA (78), which allows for linear peptide sequences free at either the C terminus or the N-terminus. However, unlike phage display, this technology does not appear to be as well developed. The main application is panning against immobilized (39, 74, 75, 82, 83) and dissolved proteins (77, 79) with some work involving cell binding (42, 76, 78, 84) and metal ions (80, 85, 86).
The advantages of bacteria versus phage display are a greater potential diversity (approx 10x that of phage (77)), simpler lab workup and culture (39), and the ability to use antibiotics with resistance genes to limit contamination with wild-type bacteria (39). The downsides are that bacteria have many extraneous proteins that can bind to the target (74) and the inability to do in-vivo selection due to immune response. The commercial bacteria display system appears to have quality control issues, with some researchers finding the peptide insert is a different length than expected (39, 80). One paper comparing phage to bacteria display (39) states that bacteria display is inferior to phage display; however their data suggests part of the issue may be unfamiliarity with the technology.
Screening is very similar to phage display. Bound bacteria are eluted by vortexing to tear them from their flagella, titered using absorbance, and expanded. If the screen was on cells, they can be lysed with pure water to collect the bacteria (76) or just vortexed as with immobilized proteins (42, 84). Internalization into cells can be selected for by eliminating all non-internalizing clones with gentamicin (76). The peptide expressed by the binding bacteria is determined by sequencing the appropriate part of the genome. An alternative panning protocol is to attach a biotin onto a target protein and conduct the binding in solution, using PE-streptavidin and FACS to isolate the binding bacteria (77, 78).
Aptamers
RNA and single stranded DNA (ssDNA), due to their specific interactions, will form complex secondary and tertiary structures that are not seen with peptide libraries (87). The advantages of using aptamers is that it is easy to generate extremely large starting libraries of 1015 different compounds, achieving greater diversity than with peptides (88-92) and the techniques (PCR, translation of RNA to DNA and back) are commonly used in many labs. RNA also has very good pharmokinetics for imaging with a blood half life of around 2-3 min (93, 94). The primary elimination mechanism is blood degredation by endonucleases followed by renal excretion, both of which can be modified for longer lifetimes.
Multiple labs described the aptamer library generation and screening process at about the same time, one screeing RNA oligomers to find compounds that bind to selected dyes (88), another using the same technique to find RNA ligands that bind to an RNA binding protein (95), (naming the process systematic evolution of ligands by exponential enrichment (SELEX)). A third paper used similar technology to generate catalytic sequences that would cleave a DNA substrate (96). Since then there have been thousands of papers examining these aptamers for binding proteins (89, 91, 94, 97-99), small molecules (88, 91, 100, 101), even chemical weapons (102, 103). Several of these compounds have progressed to human clinical trials (99, 104-106) with one compound FDA approved to treat macular degeneration (97, 107). The affinities can be in the picomolar range, leading to the nickname chemical antibodies.
The initial library is a chemically synthesized random RNA chain between 15 and 100 nucleotides long (108) flanked by fixed primer sequence handles for reverse transcription and PCR amplification. The primer sequences do not influence binding (108), and are excised in the final probe. There are a number of chemically modified non-natural nucleotide bases accepted by RNA enzymes that can be used for additional diversity (90, 91, 109), but they do not appear to enjoy wide usage. For in-vivo applications, such as imaging agents, the polymer chain needs to be stabilized against nucleases. An elegent method of doing this is to select against the mirror image of the target, such as a peptide or protein made of d- amino acids. Once the selection is completed, the aptamer is chemically synthesized with levorotary bases (110). These mirror image aptamers, called spiegelmers, are stable to nucleases, but the inavailability of proteins constructed of d-amino acids limits the technique to chemically synthesized targets and small molecules. For systems where this approach is not available, modifications at the 2′ position on the ribose are used to stabilize the final compound, as enzymes tolerant of these substitutions are available. Amines, fluorines, and methoxy groups have been used (92, 106, 109). The amino group on pyrimidines was first tried and proved effective in prolonging the life of the RNA in the presence of nucleases (111), but the sequences were expensive and difficult to synthesize. A solution to the problems of expense and synthesis was found in 2′ fluoro pyrimidines. This chemistry was used in pegaptinib sodium, the first aptamer approved by the FDA (97, 107). Further modifications have included 2′ methoxy groups with the rationale that this is a common post-translational modification biologically and should be well tolerated (92, 106, 109). The methoxy substitutions also have the advantage of being less expensive than the fluoro or amino substitution. It is possible to make these modifications after screening, but as substitutions can drastically change the affinity, it is much simpler and less time consuming to screen using the modified ribose backbone.
Screening is very similar to phage panning, with two exceptions. Aptamers do not show amplification bias and the washing step is less effective; leading to many more panning iterations, usually 10-20 or more. The protein or small molecule of interest is typically attached to a resin and packed into a column, similar to affinity chromatography, but can also be presented on nitrocelulose membrane. Temperature is mentioned in a few papers, with one (94) demonstrating an aptamer selected for high affinity at one temperature may lose that affinity when the temperature is changed. The literature of screening against cells is limited, but the basic approach is identical to phage display (106, 112). Internalization can be selected for using trypsin to cleave off the targeted proteins and their attached aptamers (113) . In-vivo selection has been reported once at this time (114), using a 2′ fluoro stabilized RNA library to isolate aptamers that bind to a mouse model of colon cancer metastases.
Small molecules
There is a long history of screening small molecules to discover drug leads, but a much shorter publication record for imaging. Assays have been developed to look at binding (115, 116), including fluorescence polarization, surface plasmon resonance, and calorimetry, but these techniques are not high throughput. The two solutions in the literature to avoiding expensive compound by compound screening are to immobilize the compounds on a slide or to use a tag on the small molecule to simplify the screen.
The success of gene chips and other microarray technology has lead to a number of papers attempting to extend the idea into other areas, including small molecules. Both attachment (115, 117-128); and detection (115, 123, 125, 129) are facets that have been extensively published. An array of small molecules is bonded to a solid surface, with the identity of each small molecule positionally encoded. Protein, cell lysates, homogenized tissue, or other material to be tested is washed over the microarray, and adherence of proteins measured. The detection is typically fluorescence or biotin on the protein, but antibodies (123) and surface plasmon resonance (129) are also used.
A common approach to identifying small molecules that can be used for imaging is to make libraries of compounds that already have the imaging moiety attached (8, 130, 131). In one example, magnetic nanoparticles used for MRI contrast were labeled with a fluorescent tag and a small molecule. Binding of the small molecule to a cellular or protein target is monitored either directly by the fluorescent tag or an antibody to it. The advantage of this technique is that a hit is the agent, attachment of a binding moiety to the nanoparticle is already completed. The disadvantage is that each small molecule is tested individually, limiting the diversity of the library. The literature studies referenced here used an MRI probe. Some other imaging techniques would be very expensive to do this way, such as F18 labeling for PET.
Other groups have used a polynucleotide tag to label each small molecule, giving them a unique identifier that can be amplified by PCR. The basic idea has been in use for almost two decades (132), where a peptide sequence (or any other polyamide) was encoded in the attached DNA sequence. The basic protocol is similar to phage panning. The mix of small molecules is exposed to either a cell or an immobilized protein, and the non-binding samples washed away. The samples that bind may be re-panned, often with PCR amplification and library resynthesis. Another protocol is to mix the DNA labeled small molecules with the protein(s) in solution, remove unbound material by size exclusion, and to detect by complement binding on a gene chip (133). Other variations have the DNA sequence encoding the reactions to make the small molecule in a split and pool type combinatorial synthesis, either adding a section of DNA when the reactions are run (134), or the DNA synthesiszed first using combinatorial methods and used to direct the synthesis (135, 136). Very large libaries can be constructed by these methods, and as the DNA indicates the reactions, biological amplification techniques can be used. A final variant has the DNA enclosed in a phage (137, 138); a variant that will protect the tag during the various synthetic and screening steps.
One final note on small molecule screening. Most small molecules can only be modified at certain locations on their scaffold, or the modification will interfere with binding (for example, see (139)). If a small molecule is attached to a plate at position required for binding, it will not bind its target, even if there is high affinity in solution. Adding complexity, some sites will be permissive, but with size or charge limitations (140).
Antibodies
Antibodies were the first compounds developed that could bind specificly to a known protein target. It is possible to find commercially available antibodies that bind to almost any desired disease state, even discriminating between single molecules such as phosphates on specific amino acid residues. Further, hybridoma and other technologies have facilitated the generation of antibodies on a large scale. There is a large repertoire of reliable chemical modifications based on N hydroxy succinimide (NHS) ester chemistry to link them to chelators, peptides, or nanoparticles for imaging. As can be imagined, there is an extensive literature using antibodies to target imaging agents. However, antibodies are expensive, have very long blood lifetimes (141), can elicit an immune response if made in a different species (142), and labeling can affect the affinity (143) although this must be determined empirically as it varies from antibody clone to antibody clone. Attempts to use antibody fragments give better pharmacokinetics, but this often yields reduced binding and tumor uptake (141). There are several reviews on using antibodies as imaging agents (144, 145) with a cohort clinically approved (146-148).
Optimization/development
Optimization and development consists of determining the binding partner of the targeting compound (if not already known), optimizing the affinity, converting it into an imaging agent, and improving its in-vivo characteristics. Selectivity is optimized in the screen that gave the lead if the screens were designed properly. Optimization is too broad a subject to cover as one section of a review paper, so only the most common problems and solutions will be given.
A good imaging agent will have a blood lifetime long enough for the compound to bind but short enough to allow imaging soon after administration, can reach the target (either penetrate the endothelial wall or bind to a protein on the endothelium), will bind strongly to the target and not accumulate anywhere that will interfere with imaging, and will be completely non-toxic in the dosages used. Ideally, there will be an amplification mechanism, such as receptor mediated internalization or signal amplification (149, 150). These are different design endpoints than those used in developing drugs. Drugs, by design, will perturb a process in the body, while imaging agents should have minimal physiological effect. Off target binding in drugs is not desired, but may be an acceptable tradeoff, whereas off target binding of imaging agents causes false positives and high background, rendering the compound difficult to use. Side effects in a therapeutic agent may be tolerated, depending on the condition being treated. The same side effects in an imaging agent are intolerable, as some fraction of the people dosed with the compound will be healthy. Ideally, drugs will remain in the bloodstream for long periods of time to minimize dosing requirements, but good imaging agents are cleared quickly to reduce background. A drug may be administered for years for some conditions, while an imaging agent will be dosed at most a few times, so chronic effects are examined differently. While both drugs and imaging agents optimize pharmacokinetics and pharmacodynamics, the goalposts are very different.
If the exact protein target of the identified compound is unknown, it should be identified before further work is done to help determine off target binding and possible side effects. Unfortunately, this is a difficult task. The most common method with peptides and aptamers is affinity chromatography (114, 151). While this method often works well, if the target protein is membrane stabilized, isolating the protein will denature it, abrogating the binding. Some researchers have used bioinformatics tools, such as BLAST (57, 59, 152-154) to identify what protein the peptide is mimicking, but there are serious limitiations to that approach. First, the peptides identified are not likely to match exactly the protein that binds to the same site they are using. Often only a few amino acids in the sequence are essential for binding; the rest are essentially random, leading to poor matches. The second limitation is that a bioinformatics approach assumes that the protein that binds will do so along one linear section of its amino acid sequence. Often, the binding site will consist of a point where a protein is folded, bringing areas that are distant in the primary structure of the protein in close proximity (66). A peptide mimicking a protein with multiple folds will not show sequence alignment. Therefore, it is often necessary to use protein-protein binding techniques, such as yeast two hybrid, to solve this problem.
Once the binding partner is known, the affinity is determined and improved if needed. All things being equal, the higher the affinity of the compound the better, as this allows lower doses, which can helpt to reduce off target binding. In addition, it may be necessary to trade off affinity to optimize another parameter such as specificity or selectivity during optimization. The easiest way to improve the affinity of an aptamer, peptide or small molecule consists of determining the moieties that are essential for binding, building a new library with the essential components unchanged and repeating the screening in a process called affinity maturation (66, 155-157). For peptides, a series are synthesized, each one differing from the original targeting peptide by the mutation of one position for an alanine. The affinity of each peptide is determined, and the peptides with significantly poorer affinity indicate the essential residues. The target is then rescreened with a new library holding these amino acids constant. A similar process is done with aptamers, but due to their length and the lack of an innocuous nucleotide, it is much more expensive and time consuming. A simpler method is to make several libraries with different sections randomized, and rescreen (90). It's possible to do this work as part of the initial screen in both phage display and aptamer screening by randomly mutating the compounds between selections (96, 155). Small molecule affinity is a more difficult process involving chemical synthesis. The idea is to systematically modify the molecule and look at structure-activity relationships (SARs) to develop heuristics about how such features as physical size, hydrophobicity and charge at different portions of the molecule affect the affinity. Changing any of the structures can change such features as specificity and toxicity, which is why this process is done early in the drug development timeline. This is an entire field of chemistry in itself; as much art as science. Different experienced groups starting with the same lead molecule often arrive at very different results.
If it is impossible to improve the affinity sufficiently by these techniques, avidity affects can compensate on the final agent. If an agent with multiple copies of a binding sequence has one that binds to a receptor, the remaining copies will be positioned to bind, leading to an artificially improved affinity. In effect the three dimentional tracking problem has been reduced to two dimentions (158). This will not make a millimolar binding affinity into a working agent, but can make a borderline affinity workable.
The next step is to modify the binding moiety to make it more resistant to enzymes that will cleave it. Both peptides and aptamers are usually degraded in the blood by endogenous enzymes and removed by the kidneys too quickly to be effective imaging agents. The ways to prevent this for aptamers, i.e. 2′ substitutions and 3′ capping, were mentioned in the screening section, as these modifications will affect binding. These can be added at this stage, but a mutation experiment is needed to see which residues are sensitive to the substitution (159). It is much less work to include these substitutions in the original screen. For peptides, capping the n-terminius (160) and substituting d-amino acids at non-essential residues will greatly reduce blood degredation, but care must be taken as the n-terminus is sometimes important for binding. Modifying the sequence to avoid cleavage sites of common proteases, if possible without sacrificing excessive affinity and selectivity, will also lengthen the blood lifetime (161).
Developing the agent with the optimized binding moiety is the next step, and is dependent on the imaging modality used. The targeting ligand is attached to a contrast segment, and the agent optimized. The contrast segment varies with imaging modality. For SPECT, this is usually a chelator binding Tc99m or In111(162). For PET, it's usually F18(163). MRI uses chelated Gd for T1 imaging or magnetic nanoparticles for T2 contrast (164). X-ray and CT techniques use heavy atoms (usually iodine, occasionally gold) to provide contrast, either attached directly or as a nanoparticle. Optical methods use fluorescent dyes. While this is not a complete list of contrast species for these imaging techniques, they are the ones most commonly used clinically. Each imaging moiety has its own associated chemistry and requirements for functional groups on the targeting ligand. Most if not all of these imaging moities can be utilized with any of the targeting compounds mentioned in this review. With few exceptions, the preference of the researcher and the achievable affinity and selectivity, rather than the exact chemical species (aptamer, peptide, small molecule, or antibody) used will be definitive. An example of a rare poor choice would be using a short half life isotope such as F18 with a long circulating nanoparticle. When the imaging agent has cleared from the blood to a level that gives low background, the radioactivity would have decayed as well.
Once the first iteration of the probe is made, it should be tested in cell culture to see if the affinity and specificity are still adequate, and in an animal model to look at target/background, off target binding, toxicity, and optimal dosing/timecourse. If any of these parameters are insufficient, the probe must be modified.
Blood lifetime is governed by probe degredation, elimination, and uptake by the immune system. Degredation in the blood should have been addressed before the synthesis of the probe. Elimination is caused by phagocytosis by immune cells, renal clearance, and various pathways of liver clearance. For volatile compounds, lung clearance will also be significant (165), but this is unusual for molecular imaging probes. All these elimination mechanisms will act on anything injected into the bloodstream. Due to a number of poorly understood active transport phenomenon, it is difficult to tell a priori exactly how a compound will be removed and where the agent will collect non specificly. However, there are heruistics about how compounds are eliminated which can be used to skew the dominant mechanism. Nanoparticles and compounds of similar size are usually predominantly opsonized and removed by phagocytes. Mobile phagocytes, such as macrophages and neutrophils will bring the probe to the lymph nodes, while stationary phagocytes, such as kupffer cells of the liver, will simply degrade them where they are injested. Reducing opsonization with polymer coatings will greatly slow this process down, but by removing a major elimination mechanism, the blood lifetime will be increased, often considerably (166). Hydrophillic compounds smaller than ~6 nm in diameter can be filtered by the kidneys; for compounds below 4 nm in diameter, this removal is very rapid. Hydrophobic compounds bind to serum albumin in the blood and will be cleared by the liver through the bile duct to the intestines or by partitioning into the blood plasma for renal excretion (165). It is common for hydrophobic compounds to be oxidized or otherwise rendered more hydrophillic by the cytochrome P450 liver enzymes followed by renal or hepatobillary excreation (165). These heuristics can be used as a basis for modifying a probe to influence the blood lifetime and redirect it away from areas to be imaged. For instance, a small hydrophillic probe may be eliminated renally too quickly to be a good agent. To increase blood lifetime, the probe can be made more hydrophobic to increase binding to albumin (decreasing renal clearance and increasing hepatic clearance, and incidentally often improving the affinity and reducing specificity), or by increasing the size of the compound by adding either a PEG chain or a protein (slowing but not necessarily eliminating renal clearance) (93, 106).
Conclusions
Much of the technology to generate an imaging agent is new and still developing. Advances in molecularly targeted probe development will likely increase the ability to diagnose patients and perform preclinical and basic research.
Acknowledgements
The authors would like to thank Frederick Epstein for critical reading of the manuscript. KAK is funded by: Johnson & Johnson Corporate Office of Science and Technology, Wallace H. Coulter Foundation, AACR-PanCAN career development award, the Department of Defense CDMRP, NIBIB EB010023, and NCI RO1 CA137071.
References
- 1.Reile H, Armatis PE, Schally AV. Characterization of high-affinity receptors for bombesin/gastrin releasing peptide on the human prostate cancer cell lines PC-3 and DU-145: internalization of receptor bound 125I-(Tyr4) bombesin by tumor cells. Prostate. 1994;25:29–38. doi: 10.1002/pros.2990250105. [DOI] [PubMed] [Google Scholar]
- 2.De Vincentis G, Remediani S, Varvarigou AD, et al. Role of 99mTc-bombesin scan in diagnosis and staging of prostate cancer. Cancer Biother Radiopharm. 2004;19:81–4. doi: 10.1089/108497804773391711. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Schuhmacher J, Waser B, et al. DOTA-PESIN, a DOTA-conjugated bombesin derivative designed for the imaging and targeted radionuclide treatment of bombesin receptor-positive tumours. Eur J Nucl Med Mol Imaging. 2007;34:1198–208. doi: 10.1007/s00259-006-0347-4. [DOI] [PubMed] [Google Scholar]
- 4.Josephson L, Reynolds F. Unpublished results. [Google Scholar]
- 5.Kelly KA, Nahrendorf M, Yu AM, Reynolds F, Weissleder R. In vivo phage display selection yields atherosclerotic plaque targeted peptides for imaging. Mol Imaging Biol. 2006;8:201–7. doi: 10.1007/s11307-006-0043-6. [DOI] [PubMed] [Google Scholar]
- 6.Kelly KA, Bardeesy N, Anbazhagan R, et al. Targeted nanoparticles for imaging incipient pancreatic ductal adenocarcinoma. PLoS Med. 2008;5:e85. doi: 10.1371/journal.pmed.0050085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton JR, Kelly KA, Mahmood U, Weissleder R, Deutscher SL. In vivo selection of phage for the optical imaging of PC-3 human prostate carcinoma in mice. Neoplasia. 2006;8:772–80. doi: 10.1593/neo.06331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly KA, Shaw SY, Nahrendorf M, et al. Unbiased discovery of in vivo imaging probes through in vitro profiling of nanoparticle libraries. Integr Biol (Camb) 2009;1:311–7. doi: 10.1039/b821775k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly K, Alencar H, Funovics M, Mahmood U, Weissleder R. Detection of invasive colon cancer using a novel, targeted, library-derived fluorescent peptide. Cancer Res. 2004;64:6247–51. doi: 10.1158/0008-5472.CAN-04-0817. [DOI] [PubMed] [Google Scholar]
- 10.Coy DH, Heinz-Erian P, Jiang NY, et al. Progress in the development of competitive bombesin antagonists. Ann N Y Acad Sci. 1988;547:150–7. doi: 10.1111/j.1749-6632.1988.tb23883.x. [DOI] [PubMed] [Google Scholar]
- 11.Gargosky SE, Wallace JC, Upton FM, Ballard FJ. C-terminal bombesin sequence requirements for binding and effects on protein synthesis in Swiss 3T3 cells. Biochem J. 1987;247:427–32. doi: 10.1042/bj2470427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng JG, Heavner GA, McEver RP. Lectin domain peptides from selectins interact with both cell surface ligands and Ca2+ ions. J Biol Chem. 1992;267:19846–53. [PubMed] [Google Scholar]
- 13.Reynolds PR, Larkman DJ, Haskard DO, et al. Detection of vascular expression of E-selectin in vivo with MR imaging. Radiology. 2006;241:469–76. doi: 10.1148/radiol.2412050490. [DOI] [PubMed] [Google Scholar]
- 14.Smith GP, Petrenko VA. Phage Display. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 15.Geysen HM, Meloen RH, Barteling SJ. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc Natl Acad Sci U S A. 1984;81:3998–4002. doi: 10.1073/pnas.81.13.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeji NJ, Bray AM, Geysen HM. Multi-pin peptide synthesis strategy for T cell determinant analysis. J Immunol Methods. 1990;134:23–33. doi: 10.1016/0022-1759(90)90108-8. [DOI] [PubMed] [Google Scholar]
- 17.Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature. 1991;354:84–6. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 18.Falsey JR, Renil M, Park S, Li S, Lam KS. Peptide and small molecule microarray for high throughput cell adhesion and functional assays. Bioconjug Chem. 2001;12:346–53. doi: 10.1021/bc000141q. [DOI] [PubMed] [Google Scholar]
- 19.Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–73. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 20.Songyang Z, Margolis B, Chaudhuri M, Shoelson SE, Cantley LC. The phosphotyrosine interaction domain of SHC recognizes tyrosine-phosphorylated NPXY motif. J Biol Chem. 1995;270:14863–6. doi: 10.1074/jbc.270.25.14863. [DOI] [PubMed] [Google Scholar]
- 21.Lam KS, Zhao ZG. Targeted therapy for lymphoma with peptides. Hematol Oncol Clin North Am. 1997;11:1007–19. doi: 10.1016/s0889-8588(05)70476-7. [DOI] [PubMed] [Google Scholar]
- 22.Kumaresan PR, Lam KS. Screening chemical microarrays: methods and applications. Mol Biosyst. 2006;2:259–70. doi: 10.1039/b602004f. [DOI] [PubMed] [Google Scholar]
- 23.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–4. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 24.Lam KS, Lebl M, Krchnak V, et al. Discovery of D-amino-acid-containing ligands with selectide technology. Gene. 1993;137:13–6. doi: 10.1016/0378-1119(93)90245-x. [DOI] [PubMed] [Google Scholar]
- 25.Aina OH, Sroka TC, Chen ML, Lam KS. Therapeutic cancer targeting peptides. Biopolymers. 2002;66:184–99. doi: 10.1002/bip.10257. [DOI] [PubMed] [Google Scholar]
- 26.Cull MG, Miller JF, Schatz PJ. Screening for receptor ligands using large libraries of peptides linked to the C terminus of the lac repressor. Proc Natl Acad Sci U S A. 1992;89:1865–9. doi: 10.1073/pnas.89.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattheakis LC, Bhatt RR, Dower WJ. An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc Natl Acad Sci U S A. 1994;91:9022–6. doi: 10.1073/pnas.91.19.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattheakis LC, Dias JM, Dower WJ. Cell-free synthesis of peptide libraries displayed on polysomes. Methods Enzymol. 1996;267:195–207. doi: 10.1016/s0076-6879(96)67013-x. [DOI] [PubMed] [Google Scholar]
- 29.Peelle BR, Krauland EM, Wittrup KD, Belcher AM. Design criteria for engineering inorganic material-specific peptides. Langmuir. 2005;21:6929–33. doi: 10.1021/la050261s. [DOI] [PubMed] [Google Scholar]
- 30.Krauland EM, Peelle BR, Wittrup KD, Belcher AM. Peptide tags for enhanced cellular and protein adhesion to single-crystalline sapphire. Biotechnol Bioeng. 2007;97:1009–20. doi: 10.1002/bit.21341. [DOI] [PubMed] [Google Scholar]
- 31.Silverman AP, Levin AM, Lahti JL, Cochran JR. Engineered cystine-knot peptides that bind alpha(v)beta(3) integrin with antibody-like affinities. J Mol Biol. 2009;385:1064–75. doi: 10.1016/j.jmb.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giebel LB, Cass RT, Milligan DL, Young DC, Arze R, Johnson CR. Screening of cyclic peptide phage libraries identifies ligands that bind streptavidin with high affinities. Biochemistry. 1995;34:15430–5. doi: 10.1021/bi00047a006. [DOI] [PubMed] [Google Scholar]
- 33.Devlin JJ, Panganiban LC, Devlin PE. Random peptide libraries: a source of specific protein binding molecules. Science. 1990;249:404–6. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- 34.Katz BA. Binding to protein targets of peptidic leads discovered by phage display: crystal structures of streptavidin-bound linear and cyclic peptide ligands containing the HPQ sequence. Biochemistry. 1995;34:15421–9. doi: 10.1021/bi00047a005. [DOI] [PubMed] [Google Scholar]
- 35.Katz BA. Streptavidin-binding and -dimerizing ligands discovered by phage display, topochemistry, and structure-based design. Biomol Eng. 1999;16:57–65. doi: 10.1016/s1050-3862(99)00036-4. [DOI] [PubMed] [Google Scholar]
- 36.Katz BA, Johnson C, Cass RT. Structure-based design of high affinity streptavidin binding cyclic peptide ligands containing thioether crosslinks. J Am Chem Soc. 1995;117:8541–7. [Google Scholar]
- 37.Avrantinis SK, Stafford RL, Tian X, Weiss GA. Dissecting the streptavidin-biotin interaction by phage-displayed shotgun scanning. Chembiochem. 2002;3:1229–34. doi: 10.1002/1439-7633(20021202)3:12<1229::AID-CBIC1229>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 38.Meyer SC, Gaj T, Ghosh I. Highly selective cyclic peptide ligands for NeutrAvidin and avidin identified by phage display. Chem Biol Drug Des. 2006;68:3–10. doi: 10.1111/j.1747-0285.2006.00401.x. [DOI] [PubMed] [Google Scholar]
- 39.Lunder M, Bratkovic T, Doljak B, et al. Comparison of bacterial and phage display peptide libraries in search of target-binding motif. Appl Biochem Biotechnol. 2005;127:125–31. doi: 10.1385/abab:127:2:125. [DOI] [PubMed] [Google Scholar]
- 40.Kelly KA, Carson J, McCarthy JR, Weissleder R. Novel peptide sequence (“IQ-tag”) with high affinity for NIR fluorochromes allows protein and cell specific labeling for in vivo imaging. PLoS One. 2007;2:e665. doi: 10.1371/journal.pone.0000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romanov VI, Durand DB, Petrenko VA. Phage display selection of peptides that affect prostate carcinoma cells attachment and invasion. Prostate. 2001;47:239–51. doi: 10.1002/pros.1068. [DOI] [PubMed] [Google Scholar]
- 42.Zitzmann S, Mier W, Schad A, et al. A new prostate carcinoma binding peptide (DUP-1) for tumor imaging and therapy. Clin Cancer Res. 2005;11:139–46. [PubMed] [Google Scholar]
- 43.Zitzmann S, Kramer S, Mier W, et al. Identification and evaluation of a new tumor cell-binding peptide, FROP-1. J Nucl Med. 2007;48:965–72. doi: 10.2967/jnumed.106.036699. [DOI] [PubMed] [Google Scholar]
- 44.Pola R, Pechar M, Ulbrich K, Fres AF. Polymer doxorubisin conjugate with a synthetic peptide ligand targeted on prostate tumor. Journal of bioactive and compatible polymers. 2007;22:602–20. [Google Scholar]
- 45.Kelly KA, Setlur SR, Ross R, et al. Detection of early prostate cancer using a hepsin-targeted imaging agent. Cancer Res. 2008;68:2286–91. doi: 10.1158/0008-5472.CAN-07-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newton-Northup JR, Figueroa SD, Quinn TP, Deutscher SL. Bifunctional phage-based pretargeted imaging of human prostate carcinoma. Nucl Med Biol. 2009;36:789–800. doi: 10.1016/j.nucmedbio.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flynn CE, Lee SW, Peelle BR, Belcher AM. Viruses as vehicles for growth, organization, and assembly of materials. Acta Materialia. 2003;51:5867–80. [Google Scholar]
- 48.Seker UO, Wilson B, Dincer S, et al. Adsorption behavior of linear and cyclic genetically engineered platinum binding peptides. Langmuir. 2007;23:7895–900. doi: 10.1021/la700446g. [DOI] [PubMed] [Google Scholar]
- 49.Bassindale AR, Codina-Barrios A, Frascione N, Taylor PG. An improved phage display methodology for inorganic nanoparticle fabrication. Chem Commun (Camb) 2007:2956–8. doi: 10.1039/b702650a. [DOI] [PubMed] [Google Scholar]
- 50.Seker UO, Wilson B, Sahin D, Tamerler C, Sarikaya M. Quantitative affinity of genetically engineered repeating polypeptides to inorganic surfaces. Biomacromolecules. 2009;10:250–7. doi: 10.1021/bm8009895. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Whyburn GP, Huang Y. Specific peptide regulated synthesis of ultrasmall platinum nanocrystals. J Am Chem Soc. 2009;131:15998–9. doi: 10.1021/ja907235v. [DOI] [PubMed] [Google Scholar]
- 52.Chen H, Su X, Neoh KG, S CW. Probing the interaction between peptides and metal ozides using point mutants of a TiO2 binding peptide. Langmuir. 2008;24:6852–7. doi: 10.1021/la800314p. [DOI] [PubMed] [Google Scholar]
- 53.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–6. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 54.Kennel SJ, Mirzadeh S, Hurst GB, et al. Labeling and distribution of linear peptides identified using in vivo phage display selection for tumors. Nucl Med Biol. 2000;27:815–25. doi: 10.1016/s0969-8051(00)00149-9. [DOI] [PubMed] [Google Scholar]
- 55.Rafii S, Avecilla ST, Jin DK. Tumor vasculature address book: identification of stage-specific tumor vessel zip codes by phage display. Cancer Cell. 2003;4:331–3. doi: 10.1016/s1535-6108(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 56.Zou J, Dickerson MT, Owen NK, Landon LA, Deutscher SL. Biodistribution of filamentous phage peptide libraries in mice. Mol Biol Rep. 2004;31:121–9. doi: 10.1023/b:mole.0000031459.14448.af. [DOI] [PubMed] [Google Scholar]
- 57.Kolonin MG, Sun J, Do KA, et al. Synchronous selection of homing peptides for multiple tissues by in vivo phage display. FASEB J. 2006;20:979–81. doi: 10.1096/fj.05-5186fje. [DOI] [PubMed] [Google Scholar]
- 58.Yu Y, Wang Z, Du T. Mouse thymus targeted peptide isolated by in vivo phage display can inhibit bioactivity of thymus output in vivo. Journal of biomolecular screening. 2008;13:968–74. doi: 10.1177/1087057108326537. [DOI] [PubMed] [Google Scholar]
- 59.Arap W, Kolonin MG, Trepel M, et al. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8:121–7. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 60.Tian F, Tsao ML, Schultz PG. A phage display system with unnatural amino acids. J Am Chem Soc. 2004;126:15962–3. doi: 10.1021/ja045673m. [DOI] [PubMed] [Google Scholar]
- 61.Sandman KE, Benner JS, Noren CJ. Phage display of selenopeptides. J Am Chem Soc. 2000;122:960–1. [Google Scholar]
- 62.Tan R, Frankel AD. A novel glutamine-RNA interaction identified by screening libraries in mammalian cells. Proc Natl Acad Sci U S A. 1998;95:4247–52. doi: 10.1073/pnas.95.8.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castagnoli L, Zucconi A, Quondam M, et al. Alternative bacteriophage display systems. Comb Chem High Throughput Screen. 2001;4:121–33. doi: 10.2174/1386207013331174. [DOI] [PubMed] [Google Scholar]
- 64.Malys N, Chang DY, Baumann RG, Xie D, Black LW. A bipartite bacteriophage T4 SOC and HOC randomized peptide display library: detection and analysis of phage T4 terminase (gp17) and late sigma factor (gp55) interaction. J Mol Biol. 2002;319:289–304. doi: 10.1016/S0022-2836(02)00298-X. [DOI] [PubMed] [Google Scholar]
- 65.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–17. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 66.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–83. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 67.De Ciechi PA, Devine CS, Lee SC, Howard SC, Olins PO, Caparon MH. Utilization of multiple phage display libraries for the identification of dissimilar peptide motifs that bind to a B7-1 monoclonal antibody. Mol Divers. 1996;1:79–86. doi: 10.1007/BF01721322. [DOI] [PubMed] [Google Scholar]
- 68.Hall PR, Hjelle B, Njus H, et al. Phage display selection of cyclic peptides that inhibit Andes virus infection. J Virol. 2009;83:8965–9. doi: 10.1128/JVI.00606-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rao WH, Camp RD. Novel cyclic and linear oligopeptides that bind to integrin beta1 chain and either inhibit or costimulate T lymphocytes. Int Immunopharmacol. 2003;3:435–43. doi: 10.1016/S1567-5769(03)00041-9. [DOI] [PubMed] [Google Scholar]
- 70.Noppe W, Plieva F, Galaev IY, Pottel H, Deckmyn H, Mattiasson B. Chromato-panning: an efficient new mode of identifying suitable ligands from phage display libraries. BMC Biotechnol. 2009;9:21. doi: 10.1186/1472-6750-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dharmasena MN, Jewell DA, Taylor RK. Development of peptide mimics of a protective epitope of Vibrio cholerae Ogawa O-antigen and investigation of the structural basis of peptide mimicry. J Biol Chem. 2007;282:33805–16. doi: 10.1074/jbc.M707314200. [DOI] [PubMed] [Google Scholar]
- 72.Kelly KA, Allport JR, Tsourkas A, Shinde-Patil VR, Josephson L, Weissleder R. Detection of vascular adhesion molecule-1 expression using a novel multimodal nanoparticle. Circ Res. 2005;96:327–36. doi: 10.1161/01.RES.0000155722.17881.dd. [DOI] [PubMed] [Google Scholar]
- 73.Wu M, Pasula R, Smith PA, Martin WJn. Mapping alveolar binding sites in vivo using phage peptide libraries. Gene Ther. 2003;10:1429–36. doi: 10.1038/sj.gt.3302009. [DOI] [PubMed] [Google Scholar]
- 74.Lu Z, Murray KS, Van Cleave V, LaVallie ER, Stahl ML, McCoy JM. Expression of thioredoxin random peptide libraries on the e. coli cell surface as functional fusions to flagellin: A system designed for exploring protein protein interactions. Nature biotechnology. 1995;13:366. doi: 10.1038/nbt0495-366. [DOI] [PubMed] [Google Scholar]
- 75.Lu Z, Lavallie ER, McCoy JM. Using biopanning of FLITRX peptide libraries displayed on e. coli cell surface to study protein protein interactions. Methods in Molecular Biology. 2008;205:267. doi: 10.1385/1-59259-301-1:267. [DOI] [PubMed] [Google Scholar]
- 76.Taschner S, Meinke A, von Gabain A, Boyd AP. Selection of peptide entry motifs by bacterial surface display. Biochem J. 2002;367:393–402. doi: 10.1042/BJ20020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bessette PH, Rice JJ, Daugherty PS. Rapid isolation of high-affinity protein binding peptides using bacterial display. Protein Eng Des Sel. 2004;17:731–9. doi: 10.1093/protein/gzh084. [DOI] [PubMed] [Google Scholar]
- 78.Dane KY, Chan LA, Rice JJ, Daugherty PS. Isolation of cell specific peptide ligands using fluorescent bacterial display libraries. J Immunol Methods. 2006;309:120–9. doi: 10.1016/j.jim.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 79.Rice JJ, Schohn A, Bessette PH, Boulware KT, Daugherty PS. Bacterial display using circularly permuted outer membrane protein OmpX yields high affinity peptide ligands. Protein Sci. 2006;15:825–36. doi: 10.1110/ps.051897806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kjaergaard K, Schembri MA, Klemm P. Novel Zn(2+)-chelating peptides selected from a fimbria-displayed random peptide library. Appl Environ Microbiol. 2001;67:5467–73. doi: 10.1128/AEM.67.12.5467-5473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Z, Liu Q, Wang Q, Zhang Y. Novel bacterial surface display system based on outer membrane anchoring elements from the marine bacterium Vibro Anguillarum. Appl Environ Microbiol. 2008;74:4359–65. doi: 10.1128/AEM.02499-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao S, Lee EY. A protein phosphatase-1-binding motif identified by the panning of a random peptide display library. J Biol Chem. 1997;272:28368–72. doi: 10.1074/jbc.272.45.28368. [DOI] [PubMed] [Google Scholar]
- 83.Xin ZT, Liu C, Gao YP, et al. Identification of mimotopes by screening of a bacterially displayed random peptide library and its use in eliciting an immune response to native HBV-preS. Vaccine. 2003;21:4373–9. doi: 10.1016/s0264-410x(03)00436-5. [DOI] [PubMed] [Google Scholar]
- 84.Brown CK, Modzelewski RA, Johnson CS, Wong MK. A novel approach for the identification of unique tumor vasculature binding peptides using an E. coli peptide display library. Ann Surg Oncol. 2000;7:743–9. doi: 10.1007/s10434-000-0743-0. [DOI] [PubMed] [Google Scholar]
- 85.Dong J, Liu C, Zhang J, et al. Selection of novel nickel-binding peptides from flagella displayed secondary peptide library. Chem Biol Drug Des. 2006;68:107–12. doi: 10.1111/j.1747-0285.2006.00421.x. [DOI] [PubMed] [Google Scholar]
- 86.Yang W, Luo D, Wang S, et al. TMTP1, a novel tumor-homing peptide specifically targeting metastasis. Clin Cancer Res. 2008;14:5494–502. doi: 10.1158/1078-0432.CCR-08-0233. [DOI] [PubMed] [Google Scholar]
- 87.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–5. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 88.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 89.Famulok M, Mayer G, Blind M. Nucleic acid aptamers-from selection in vitro to applications in vivo. Acc Chem Res. 2000;33:591–9. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 90.Eaton BE, Gold L, Hicke BJ, et al. Post-SELEX combinatorial optimization of aptamers. Bioorg Med Chem. 1997;5:1087–96. doi: 10.1016/s0968-0896(97)00044-8. [DOI] [PubMed] [Google Scholar]
- 91.Famulok M. Exploring chemical space with aptamers. J Med Chem. 2009;52:6951–7. doi: 10.1021/jm9014789. [DOI] [PubMed] [Google Scholar]
- 92.Bunka DH, Stockley PG. Aptamers come of age - at last. Nat Rev Microbiol. 2006;4:588–96. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 93.Dougan H, Lyster DM, Vo CV, Stafford A, Weitz JI, Hobbs JB. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl Med Biol. 2000;27:289–97. doi: 10.1016/s0969-8051(99)00103-1. [DOI] [PubMed] [Google Scholar]
- 94.Hicke BJ, Watson SR, Koenig A, et al. DNA aptamers block L-selectin function in vivo. Inhibition of human lymphocyte trafficking in SCID mice. J Clin Invest. 1996;98:2688–92. doi: 10.1172/JCI119092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 96.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–8. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 97.Ruckman J, Green LS, Beeson J, et al. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem. 1998;273:20556–67. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 98.Charlton J, Sennello J, Smith D. In vivo imaging of inflammation using an aptamer inhibitor of human neutrophil elastase. Chem Biol. 1997;4:809–16. doi: 10.1016/s1074-5521(97)90114-9. [DOI] [PubMed] [Google Scholar]
- 99.Dausse E, Da Rocha Gomes S, Toulme JJ. Aptamers: a new class of oligonucleotides in the drug discovery pipeline? Curr Opin Pharmacol. 2009;9:602–7. doi: 10.1016/j.coph.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 100.Patel DJ. Structural analysis of nucleic acid aptamers. Curr Opin Chem Biol. 1997;1:32–46. doi: 10.1016/s1367-5931(97)80106-8. [DOI] [PubMed] [Google Scholar]
- 101.Carothers JM, Oestreich SC, Szostak JW. Aptamers selected for higher-affinity binding are not more specific for the target ligand. J Am Chem Soc. 2006;128:7929–37. doi: 10.1021/ja060952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tang J, Xie J, Shao N, Yan Y. The DNA aptamers that specifically recognize ricin toxin are selected by two in vitro selection methods. Electrophoresis. 2006;27:1303–11. doi: 10.1002/elps.200500489. [DOI] [PubMed] [Google Scholar]
- 103.Bruno JG, Carrillo MP, Phillips T, Vail NK, Hanson D. Competitive FRET-aptamer-based detection of methylphosphonic acid, a common nerve agent metabolite. J Fluoresc. 2008;18:867–76. doi: 10.1007/s10895-008-0316-3. [DOI] [PubMed] [Google Scholar]
- 104.Gilbert JC, DeFeo-Fraulini T, Hutabarat RM, et al. First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation. 2007;116:2678–86. doi: 10.1161/CIRCULATIONAHA.107.724864. [DOI] [PubMed] [Google Scholar]
- 105.Majumder P, Gomes KN, Ulrich H. Aptamers: from bench side research towards patented molecules with therapeutic applications. Expert Opin Ther Pat. 2009;19:1603–13. doi: 10.1517/13543770903313746. [DOI] [PubMed] [Google Scholar]
- 106.Thiel KW, Giangrande PH. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides. 2009;19:209–22. doi: 10.1089/oli.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Apte RS. Pegaptanib sodium for the treatment of age-related macular degeneration. Expert Opin Pharmacother. 2008;9:499–508. doi: 10.1517/14656566.9.3.499. [DOI] [PubMed] [Google Scholar]
- 108.Cowperthwaite MC, Ellington AD. Bioinformatic analysis of the contribution of primer sequences to aptamer structures. J Mol Evol. 2008;67:95–102. doi: 10.1007/s00239-008-9130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Keefe AD, Cload ST. SELEX with modified nucleotides. Curr Opin Chem Biol. 2008;12:448–56. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 110.Eulberg D, Klussmann S. Spiegelmers: biostable aptamers. Chembiochem. 2003;4:979–83. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]
- 111.Lin Y, Qiu Q, Gill SC, Jayasena SD. Modified RNA sequence pools for in vitro selection. Nucleic Acids Res. 1994;22:5229–34. doi: 10.1093/nar/22.24.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shangguan D, Li Y, Tang Z, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103:11838–43. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xiao Z, Shangguan D, Cao Z, Fang X, Tan W. Cell-specific internalization study of an aptamer from whole cell selection. Chemistry. 2008;14:1769–75. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 114.Mi J, Liu Y, Rabbani ZN, et al. In vivo selection of tumor-targeting RNA motifs. Nat Chem Biol. 2010;6:22–4. doi: 10.1038/nchembio.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nicholson RL, Welch M, Ladlow M, Spring DR. Small-molecule screening: advances in microarraying and cell-imaging technologies. ACS Chem Biol. 2007;2:24–30. doi: 10.1021/cb600321j. [DOI] [PubMed] [Google Scholar]
- 116.Vegas AJ, Fuller JH, Koehler AN. Small-molecule microarrays as tools in ligand discovery. Chem Soc Rev. 2008;37:1385–94. doi: 10.1039/b703568n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sun YS, Landry JP, Fei YY, et al. Macromolecular scaffolds for immobilizing small molecule microarrays in label-free detection of protein-ligand interactions on solid support. Anal Chem. 2009;81:5373–80. doi: 10.1021/ac900889p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.MacBeath G, Koehler AN, Schreiber SL. Printing small molecules as microarrays and detecting protein-ligand interactions en masse. J Am Chem Soc. 1999;121:7967–8. [Google Scholar]
- 119.Hergenrother PJ, Depew KM, Schreiber SL. Small molecule microarrays: covalent attachment and screening of alcohol-containing small molecules on glass slides. J Am Chem Soc. 2000;122:7849–50. [Google Scholar]
- 120.Nicholson RL, Ladlow ML, Spring DR. Fluorous tagged small molecule microarrays. Chem Commun (Camb) 2007:3906–8. doi: 10.1039/b712906h. [DOI] [PubMed] [Google Scholar]
- 121.Kohn M, Wacker R, Peters C, et al. Staudinger ligation: a new immobilization strategy for the preparation of small-molecule arrays. Angew Chem Int Ed Engl. 2003;42:5830–4. doi: 10.1002/anie.200352877. [DOI] [PubMed] [Google Scholar]
- 122.Lee MR, Shin I. Fabrication of chemical microarrays by efficient immobilization of hydrazide-linked substances on epoxide-coated glass surfaces. Angew Chem Int Ed Engl. 2005;44:2881–4. doi: 10.1002/anie.200462720. [DOI] [PubMed] [Google Scholar]
- 123.Bradner JE, McPherson OM, Mazitschek R, et al. A robust small-molecule microarray platform for screening cell lysates. Chem Biol. 2006;13:493–504. doi: 10.1016/j.chembiol.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 124.Tort N, Salvador JP, Eritja R, et al. Fluorescence site-encoded DNA addressable hapten microarrays for anabolic androgenic steroids. Trends Anal Chem. 2009;26:718–28. [Google Scholar]
- 125.Kurosu M, Mowers WA. Small-molecule microarrays: development of novel linkers and an efficient detection method for bound proteins. Bioorg Med Chem Lett. 2006;16:3392–5. doi: 10.1016/j.bmcl.2006.04.057. [DOI] [PubMed] [Google Scholar]
- 126.Pohl NL. Fluorous tags catching on microarrays. Angew Chem Int Ed Engl. 2008;47:3868–70. doi: 10.1002/anie.200704801. [DOI] [PubMed] [Google Scholar]
- 127.Roska RL, Lama TG, Hennes JP, Carlson RE. Small molecule-based binding environments: combinatorial construction of microarrays for multiplexed affinity screening. J Am Chem Soc. 2009;131:16660–2. doi: 10.1021/ja9046944. [DOI] [PubMed] [Google Scholar]
- 128.Marsden DM, Nicholson RL, Ladlow M, Spring DR. 3D small-molecule microarrays. Chem Commun (Camb) 2009:7107–9. doi: 10.1039/b913665g. [DOI] [PubMed] [Google Scholar]
- 129.Kanoh N, Kyo M, Inamori K, et al. SPR imaging of photo-cross-linked small-molecule arrays on gold. Anal Chem. 2006;78:2226–30. doi: 10.1021/ac051777j. [DOI] [PubMed] [Google Scholar]
- 130.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat Biotechnol. 2005;23:1418–23. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 131.Sun EY, Josephson L, Kelly KA, Weissleder R. Development of nanoparticle libraries for biosensing. Bioconjug Chem. 2006;17:109–13. doi: 10.1021/bc050290e. [DOI] [PubMed] [Google Scholar]
- 132.Brenner S, Lerner RA. Encoded combinatorial chemistry. Proc Natl Acad Sci U S A. 1992;89:5381–3. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Winssinger N, Ficarro S, Schultz PG, Harris JL. Profiling protein function with small molecule microarrays. Proc Natl Acad Sci U S A. 2002;99:11139–44. doi: 10.1073/pnas.172286899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Clark MA, Acharya RA, Arico-Muendel CC, et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat Chem Biol. 2009;5:647–54. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 135.Halpin DR, Harbury PB. DNA display I. Sequence-encoded routing of DNA populations. PLoS Biol. 2004;2:E173. doi: 10.1371/journal.pbio.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wrenn SJ, Weisinger RM, Halpin DR, Harbury PB. Synthetic ligands discovered by in vitro selection. J Am Chem Soc. 2007;129:13137–43. doi: 10.1021/ja073993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Woiwode TF, Haggerty JE, Katz R, et al. Synthetic compound libraries displayed on the surface of encoded bacteriophage. Chem Biol. 2003;10:847–58. doi: 10.1016/j.chembiol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 138.Yin J, Liu F, Schinke M, Daly C, Walsh CT. Phagemid encoded small molecules for high throughput screening of chemical libraries. J Am Chem Soc. 2004;126:13570–1. doi: 10.1021/ja045127t. [DOI] [PubMed] [Google Scholar]
- 139.Norman BH, Srinivasan U, Dodge JA, Sato M. Wortmannin analogs for inhibiting bone loss or resorption. 1997;UK:26. [Google Scholar]
- 140.Yuan H, Luo J, Field S, Weissleder R, Cantley L, Josephson L. Synthesis and activity of C11-modified wortmannin probes for PI3 kinase. Bioconjug Chem. 2005;16:669–75. doi: 10.1021/bc049714f. [DOI] [PubMed] [Google Scholar]
- 141.Kobayashi H, Ogawa M, Alford R, Choyke PL, Urano Y. New strategies for fluorescent probe design in medical diagnostic imaging. Chem Rev. 2010;110:2620–40. doi: 10.1021/cr900263j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Scott DW, De Groot AS. Can we prevent immunogenicity of human protein drugs? Ann Rheum Dis. 2010;69:i72–i76. doi: 10.1136/ard.2009.117564. [DOI] [PubMed] [Google Scholar]
- 143.Paik CH, Ebbert MA, Murphy PR, et al. Factors influencing DTPA conjugation with antibodies by cyclic DTPA anhydride. J Nucl Med. 1983;24:1158–63. [PubMed] [Google Scholar]
- 144.Wu AM, Olafsen T. Antibodies for molecular imaging of cancer. Cancer J. 2008;14:191–7. doi: 10.1097/PPO.0b013e31817b07ae. [DOI] [PubMed] [Google Scholar]
- 145.Carter P, Merchant AM. Engineering antibodies for imaging and therapy. Curr Opin Biotechnol. 1997;8:449–54. doi: 10.1016/s0958-1669(97)80067-5. [DOI] [PubMed] [Google Scholar]
- 146.Han M, Partin AW. Current Clinical Applications of the In-capromab Pendetide Scan (ProstaScint(R) Scan, Cyt-356) Rev Urol. 2001;3:165–71. [PMC free article] [PubMed] [Google Scholar]
- 147.Breitz HB, Tyler A, Bjorn MJ, Lesley T, Weiden PL. Clinical experience with Tc-99m nofetumomab merpentan (Verluma) radioimmunoscintigraphy. Clin Nucl Med. 1997;22:615–20. doi: 10.1097/00003072-199709000-00007. [DOI] [PubMed] [Google Scholar]
- 148.Su WT, Brachman M, O'Connell TX. Use of OncoScint scan to assess resectability of hepatic metastases from colorectal cancer. Am Surg. 2001;67:1200–3. [PubMed] [Google Scholar]
- 149.Ronald JA, Chen JW, Chen Y, et al. Enzyme-sensitive magnetic resonance imaging targeting myeloperoxidase identifies active inflammation in experimental rabbit atherosclerotic plaques. Circulation. 2009;120:592–9. doi: 10.1161/CIRCULATIONAHA.108.813998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Moats RA, Fraser SE, Meade TJ. A “smart” magnetic resonance imaging agent that reports on specific enzymic activity. Angewandte Chemie, international edition in english. 1997;36:726–8. [Google Scholar]
- 151.Rajotte D, Ruoslahti E. Membrane dipeptidase is the receptor for a lung-targeting peptide identified by in vivo phage display. J Biol Chem. 1999;274:11593–8. doi: 10.1074/jbc.274.17.11593. [DOI] [PubMed] [Google Scholar]
- 152.Giordano RJ, Edwards JK, Tuder RM, Arap W, Pasqualini R. Combinatorial ligand directed lung targeting. Proc Am Thorac Soc. 2009;6:411–8. doi: 10.1513/pats.200903-014AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sadanandam A, Varney ML, Kinarsky L, Ali H, Mosley RL, Singh RK. Identification of functional cell adhesion molecules with a potential role in metastasis by a combination of in vivo phage display and in silico analysis. OMICS. 2007;11:41–57. doi: 10.1089/omi.2006.0004. [DOI] [PubMed] [Google Scholar]
- 154.Kelly KA, Allport JR, Yu AM, et al. SPARC is a VCAM-1 counter-ligand that mediates leukocyte transmigration. J Leukoc Biol. 2007;81:748–56. doi: 10.1189/jlb.1105664. [DOI] [PubMed] [Google Scholar]
- 155.Yu J, Smith GP. Affinity maturation of phage-displayed peptide ligands. Methods Enzymol. 1996;267:3–27. doi: 10.1016/s0076-6879(96)67003-7. [DOI] [PubMed] [Google Scholar]
- 156.Dwyer JJ, Dwyer MA, Kossiakoff AA. High affinity RNase S-peptide variants obtained by phage display have a novel “hot-spot” of binding energy. Biochemistry. 2001;40:13491–500. doi: 10.1021/bi011703b. [DOI] [PubMed] [Google Scholar]
- 157.Landon LA, Zou J, Deutscher SL. Effective combinatorial strategy to increase affinity of carbohydrate binding by peptides. Mol Divers. 2004;8:35–50. doi: 10.1023/b:modi.0000006897.40575.41. [DOI] [PubMed] [Google Scholar]
- 158.Kongsted J, Ryde U. An improved method to predict the entropy term with the MM/PBSA approach. J Comput Aided Mol Des. 2009;23:63–71. doi: 10.1007/s10822-008-9238-z. [DOI] [PubMed] [Google Scholar]
- 159.Green L, Waugh S, Binkley JP, Hostomska Z, Hostomsky Z, Tuerk C. Comprehensive chemical modification interference and nucleotide substitution analysis of an RNA pseudoknot inhibitor to HIV-1 reverse transcriptase. J Mol Biol. 1995;247:60–8. doi: 10.1006/jmbi.1994.0122. [DOI] [PubMed] [Google Scholar]
- 160.Orwig KS, Lassetter MR, Hadden MK, Dix TA. Comparison of N-terminal modifications on neurotensin(8-13) analogues correlates peptide stability but not binding affinity with in vivo efficacy. J Med Chem. 2009;52:1803–13. doi: 10.1021/jm801072v. [DOI] [PubMed] [Google Scholar]
- 161.Lee S, Xie J, Chen X. Peptides and peptide hormones for molecular imaging and disease diagnosis. Chem Rev. 2010;110:3087–111. doi: 10.1021/cr900361p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mariani G, Bruselli L, Kuwert T, et al. A review on the clinical uses of SPECT/CT. Eur J Nucl Med Mol Imaging. 2010;37:1959–85. doi: 10.1007/s00259-010-1390-8. [DOI] [PubMed] [Google Scholar]
- 163.Saif MW, Tzannou I, Makrilia N, Syrigos K. Role and cost effectiveness of PET/CT in management of patients with cancer. Yale J Biol Med. 2010;83:53–65. [PMC free article] [PubMed] [Google Scholar]
- 164.Tombach B, Reimer P. Soluble paramagnetic chelates and stabilized colloidal particle solutions of iron oxides as contrast agents for magnetic resonance imaging. Curr Med Chem. 2005;12:2795–804. doi: 10.2174/092986705774462932. [DOI] [PubMed] [Google Scholar]
- 165.Klaassen CD. Casarette & Doull's toxicology. McGraw Hill; New York: 1996. [Google Scholar]
- 166.Villaraza AJ, Bumb A, Brechbiel MW. Macromolecules, dendrimers, and nanomaterials in magnetic resonance imaging: the interplay between size, function, and pharmacokinetics. Chem Rev. 2010;110:2921–59. doi: 10.1021/cr900232t. [DOI] [PMC free article] [PubMed] [Google Scholar]