Abstract
Listeria monocytogenes lineages III and IV represent two uncommon lineages of the human and animal pathogen L. monocytogenes, characterized by occurrence of unusual phenotypic and genetic characteristics that differentiate them from the common lineages I and II. To gain further insights into the evolution of lineages III and IV, we amplified and sequenced housekeeping genes (i.e., gap, prs, purM, ribC, and sigB), internalin genes (i.e., inlA, inlB, inlC, inlG, inlC2, inlD, inlE, inlF, and inlH) and the virulence gene cluster containing prfA, plcA, hly, mpl, actA, and plcB for lineage III (n=7) and IV (n=4) isolates. Phylogenetic analyses of the sequences obtained along with previously reported sequence data for 40 isolates representing lineages I (n=18), II (n=21), and III (n=1), showed that lineages III and IV represent divergent and monophyletic lineages. The virulence gene cluster as well as the inlAB operon were present in all isolates, with inlF absent from all lineage III and IV isolates. While all lineage IV isolates contained only inlC (in addition to inlAB), lineage III isolates showed considerable diversity with regard to internalin gene presence, including presence of (i) only inlC (n=2), (ii) inlC and inlGC2DE (n=3), (iii) only inlGC2DE (n=2), and (iv) inlC and inlC2DE (n=1). In addition to evidence for horizontal gene transfer events, among lineage III and IV isolates, in prs, actA, plcB, mpl, inlA, inlB, inlG, inlD, and inlE, we also found significant evidence for positive selection in the hly promoter region and, along the lineage III and IV branches, for actA (including in sites recognized for interactions with proteins involved in actin tail polymerization). In conclusion, lineages III and IV represent two distinct monophyletic groups with contributions of intragenic recombination to the evolution of their internalin genes as well as contributions of positive selection to evolution of the virulence genes island.
Keywords: Listeria monocytogenes, molecular evolution, positive selection, recombination, virulence genes, internalins
1. Introduction
Listeria monocytogenes is a Gram-positive, facultative intracellular bacterium that can cause severe systemic infections in both humans and a variety of animal species. L. monocytogenes can often be found in natural and food processing environments, which can be sources for transmission of this pathogen to animals and humans, through the food and feed chain. This organism is recognized as a major problem in the food industry due to its ability to endure environmental stress better than many other foodborne-illness causing organisms (De Jesus and Whiting, 2003). An estimated 1,591 human listeriosis cases, including 255 deaths, occur annually in the United States (Scallan et al., 2011). Manifestations of invasive listeriosis in animals and humans include septicemia, meningitis, encephalitis, and/or spontaneous late-term abortions in pregnant hosts (Farber and Peterkin, 1991; Gellin and Broome, 1989; Jones, 1990; Lorber, 1997; Schuchat et al., 1991).
Three genetic lineages were initially defined for L. monocytogenes based on PCR-RFLP, sequence analyses and ribotyping (Nightingale et al., 2005; Rasmussen et al., 1995; Wiedmann et al., 1997). Lineage I strains have been reported to be overrepresented among strains associated with human listeriosis outbreaks as well as, at least in some countries, among isolates from human clinical cases (Gray et al., 2004; Hong et al., 2007; Kiss et al., 2006). Lineage II isolates, while also frequently isolated from human sporadic cases, have been reported to be statistically overrepresented among food isolates in some countries, including the United States (De Cesare et al., 2007; Gray et al., 2004; Handa et al., 2005; Klaeboe et al., 2006; Lukinmaa et al., 2004; Norton et al., 2001). Lineage III strains, on the other hand, have been predominantly isolated from ruminants and other non-primate mammals, but have also been occasionally isolated from human clinical cases (e.g. Jeffers et al., 2001; Liu et al., 2006; Roberts et al., 2006; Wiedmann et al., 1997). Recently, lineage III isolates were shown to belong to two divergent groups that were first called lineages IIIA/C (Roberts et al., 2006) and IIIB, but were recently designated lineages III and IV, respectively (den Bakker et al., 2010; Orsi et al., 2010; Ward et al., 2008). Isolates in these two lineages can show a number of unusual genetic and phenotypic characteristics and have been reported to show considerable genetic and phenotypic diversity (see review by Orsi et al., 2010), including, for example, slow rhamnose utilization (Roberts et al., 2006). While L. monocytogenes serotypes 4a and 4c are only found among lineage III and IV isolates, these lineages also contain isolates with serotype 4b, which is typically associated with lineage I (Nightingale et al., 2007). While both lineage III and IV strains have been isolated from human and animal clinical cases (Roberts et al., 2006; Wiedmann et al., 1999), isolates in these lineages show considerable virulence diversity with a number of virulence attenuated strains reported (Liu et al., 2006). The overall rare isolation, of lineage III and IV isolates, from human listeriosis cases may thus be due to a combination of factors, including, but not limited to, uncommon human exposure to these lineages (Ward et al., 2004); this hypothesis is consistent with the rare isolation of lineage III and IV strains from food and environmental sources as well as reports of their increased susceptibility to different stress conditions, including thermal inactivation (De Jesus and Whiting, 2003).
Despite a number of studies on phenotypic and genetic characterization of lineage III (and in a lesser extend lineage IV) isolates (De Jesus and Whiting, 2003; Liu et al., 2006; Roberts et al., 2006; Wiedmann et al., 1997), our understanding of the evolution of these lineages is still limited, particularly since lineage IV has only been recently identified. For example, while previous studies using sequence data on the prfA virulence gene cluster (Ward et al., 2004) and multilocus genotyping (Ward et al., 2008) data indicated that lineage I and lineage III are sister taxa, a recent MLST study suggested that lineages III and IV share a common ancestor and are not sister taxa to lineage I (den Bakker et al., 2010). In addition, while the relative contributions of different evolutionary processes (e.g., positive selection, recombination) to the evolution of L. monocytogenes lineages I and II have been well documented (Dunn et al., 2009; Nightingale et al., 2005; Orsi et al., 2008a; Orsi et al., 2007; Orsi et al., 2008b; Ragon et al., 2008; Tsai et al., 2006), our understanding of the role of these forces in the evolution of lineages III and IV is more limited. While housekeeping genes diversify slowly through the accumulation of neutral or nearly neutral changes, thus providing for more reliable reconstruction of phylogenies as compared to fast-evolving genes with potential for positive selection (e.g., bacterial virulence genes) (Maiden et al., 1998; Spratt and Maiden, 1999), analysis of virulence gene sequences is critical for our understanding of the evolution of virulence associated characteristics in bacterial pathogens. We thus sequenced and analyzed a set of housekeeping genes (i.e., gap, prs, purM, ribC, and sigB) and the main virulence gene cluster (i.e., the prfA cluster), as well as a number of genes encoding internalins (i.e., inlA, inlB, inlC, inlC2, inlD, inlE, inlF, inlG, and inlH), to probe the evolution of lineage III and IV isolates. Internalin genes were included as they represent surface or secreted proteins with known (e.g., inlA, inlB, inlC) or hypothesized roles in virulence (see Bierne et al., 2007 for a review of L. monocytogenes internalins).
2. METHODS
2.1. Bacterial isolates
A set of 11 L. monocytogenes isolates representing lineages III (n = 7) and IV (n = 4) (Table 1) were conveniently selected from isolates previously reported by Roberts et al. (2006) to represent different actA and sigB allelic types. Previously reported sequence data for another 40 isolates, including one lineage III isolate (Orsi et al., 2007; Tsai et al., 2006), were included in our analyses, yielding a total of 8 lineage III isolates. The lineage of each isolate was initially assigned based on ribotyping as well as actA and sigB allelic typing (Nightingale et al., 2005; Orsi et al., 2007; Roberts et al., 2006; Tsai et al., 2006). The 12 lineage III and IV isolates analyzed had been obtained from human (n=5) and animal (n=5) clinical cases as well as from foods (n=2) (Table 1). Bacterial lysates for PCR were prepared from overnight cultures in BHI broth using lysozyme and proteinase K as previously described (Furrer et al., 1991) with two modifications (i.e., use of 2 mg/ml of lysozyme and incubation for 1 h at 58°C).
Table 1.
Isolates used in this study.
| Isolate Name | Sero-type | Lineage (Previous lineage assignment) | Internalin genes present | Source |
|---|---|---|---|---|
| FSL F2-695a | 4a | III (IIIA) | inlAB, inlGC2DE | Human |
| FSL J2-074 | 4c | III (IIIA) | inlAB, inlC, inlGC2DE | Animal |
| FSL R2-128 | 4a | III (IIIA) | inlAB, inlC, inlGC2DE | Food |
| FSL F2-318 | NDb | III (IIIA) | inlAB, inlC | Animal |
| FSL J1-168 | 4a | III (IIIA) | inlAB, inlC, inlGC2DE | Human |
| FSL F2-501 | 4b | III (IIIA) | inlAB, inlC, inlC2DE | Human |
| FSL J2-071 | 4c | III (IIIA) | inlAB, inlGC2DE | Animal |
| FSL F2-270 | 4a | III (IIIC) | inlAB, inlC | Human |
| FSL F2-086 | 4a | IV (IIIB) | inlAB, inlC | Human |
| FSL J1-208 | 4a | IV (IIIB) | inlAB, inlC | Animal |
| FSL M1-001 | 4b | IV (IIIB) | inlAB, inlC | Animal |
| FSL R1-142 | 4a | IV (IIIB) | inlAB, inlC | Food |
FSL-F2-695 was previously analyzed with a set of other 39 isolates representing lineages I and II (Tsai et al., 2006; Orsi et al., 2007; Orsi et al., 2008a).
ND; not determined
2.2. Primer design and PCR amplification
Previously reported primers were used for PCR amplification and sequencing of housekeeping genes (Nightingale et al., 2005), internalin genes (Nightingale et al., 2005; Orsi et al., 2007; Tsai et al., 2006), and the prfA cluster (Orsi et al., 2008a), using previously described PCR conditions (Nightingale et al., 2005; Orsi et al., 2008a; Orsi et al., 2007; Tsai et al., 2006). PCR products were purified using Qiaquick PCR Purification Kit (Qiagin Inc, CA). DNA sequencing was performed by Cornell University’s Core Laboratories Center (Ithaca, NY) or Macrogen, Inc. (Seoul, Korea), using PCR primers and internal sequencing primers (Nightingale et al., 2005; Orsi et al., 2008a; Orsi et al., 2007; Tsai et al., 2006), Big Dye Terminator chemistry, and AmpliTaq-FS DNA Polymerase; sequencing reactions were run on an ABI 3730xl or on an ABI 3700 DNA analyzer.
2.3. Descriptive analysis
Sequences were proofread and assembled in Seqman (DNAStar, Lasergene, Madison, WI). Sequence alignments for each gene were generated using the Clustal W method in MegAlign (DNAStar, Lasergene, Madison, WI). Nucleotide diversity (π, average pairwise nucleotide difference per site), number of polymorphic sites, number of alleles, G+C content, number of synonymous mutations (S), and the number of nonsynonymous mutations (N) were assessed using DnaSP version 3.99 (Rozas and Rozas, 1999). Sequence types (STs), defined as unique combinations of alleles, were assigned manually.
2.4. Phylogenetic analysis
MODELTEST was used to identify the most appropriate DNA substitution model (Posada and Crandall, 1998) for each alignment used. PAUP (Swofford, 2002) was used to create Neighbor-Joining trees for (i) the prfA cluster alignment, (ii) a concatenated alignment of the housekeeping genes, (iii) a concatenated alignment of inlA and inlB genes, and (iv) a concatenated alignment of the prfA cluster, housekeeping genes, inlA and inlB. All phylogenetic trees were unrooted. Bootstrap replicates (n=5000) were performed to generate confidence measures for tree branch points. ClonalFrame (Didelot and Falush, 2007) was used to estimate the clonal relationship between the isolates taking into account the recombination events. ClonalFrame version 1.1 was run with default settings and a 33% consensus network was generated using the program SplitsTrees (Huson and Bryant, 2006).
2.5. Analysis for horizontal gene transfer
Sawyer’s test (Sawyer, 1989; Sawyer, 1999) was performed using GENECONV to identify recombination events, involving lineage III and IV isolates as donors or recipients, in the prfA cluster and internalin genes. GENECONV was initially performed allowing no mismatches within the recombinant fragment. Fragments that could be linked to identical 3′ or 5′ breakpoints were considered a single recombination event; if the breakpoints could not be linked, the fragments were considered independent recombination events (as described previously; see Nightingale et al., 2005; Orsi et al., 2007). Additional recombination analyses allowing mismatches were conducted in order to further determine if any of the actA recombination events represented the same event; allowing mismatches takes into account the possibility of point mutations after the recombination occurred. If point mutations did occur, these mutations would become the breakpoints of the recognized fragments, which would prevent linking similar recombination events. Fragments were verified visually to assign donors and recipients to events.
2.6. Analysis for intragenic positive selection
Intragenic positive selection was assessed using the Phylogenetic Analysis using Maximum Likelihood (PAML) package version 3.14 (Yang, 1997; Yang and Nielsen, 2000; Yang et al., 2005). Positive selection in coding regions can be estimated by calculating the ratio of the nonsynonymous substitution rate to the synonymous substitution rate (dN/dS, represented by the omega (ω) parameter in PAML). As the rate of synonymous substitution is assumed to be neutral, ω > 1 indicates that a coding region may have evolved by positive selection. Three Likelihood Ratio Tests (LRT) between nested null (M0, M1a, M7) and alternative (M3, M2a, M8) models were performed to assess the contribution of natural selection during evolution of the sequences. M0 versus M3 aims to identify variation in selection along a sequence, while comparing M1a to M2a and M7 to M8 tests for positive selection. These tests differ in that M1a and M2a contain two and three site classes for ω, respectively, whereas M7 and M8 approximate a continuous distribution of ω with ten classes (Yang and Nielsen, 2000). Moreover, M1a and M2a assume a class of sites with ω = 1, which makes the test between these models more conservative than the test between M7 and M8 that does not assume a site class with ω = 1. PAML also uses a Bayesian Empirical Method to identify amino acids sites potentially evolving by positive selection (Yang et al., 2005).
A branch-site model (Yang et al., 2005; Zhang et al., 2005) was used to assess whether specific branches in the phylogenetic tree had undergone positive selection. The branches of interest were tested using the alternative Model A in which the branches of interest are allowed to evolve by positive selection (ω > 1), and a nested null model in which the same branches are not allowed to evolve by positive selection (ω = 1). The branch-site model was used to identify positive selection events in lineage III or IV branches.
2.7. Analysis for intergenic positive selection
Analysis for intergenic positive selection in the prfA cluster was performed using EvoNC (Wong and Nielsen, 2004) as previously described (Orsi et al., 2008a). Briefly, in this analysis, the ratio of the rate of nucleotide substitution in the noncoding regions to the rate of synonymous substitution in the adjacent coding region is represented by the parameter ξ. When ξ for a noncoding fragment exceeds 1, there is evidence that the region evolved by positive selection. Positive selection in noncoding regions can be identified using three models implemented in EvoNC; the null model is compared to two alternative models, which either have two or three categories of ξ. The two category model allows for ξ < 1 or ξ ≥ 1, whereas the three category model allows for ξ < 1, ξ = 1, or ξ > 1. Each noncoding region in the pathogenicity island was tested separately for positively selected sites. Sites with probability ≥ 0.95 of being under positive selection were considered.
2.8. Data access
Access to detailed isolate information including DNA sequences is available through the Pathogen Tracker database [http://www.pathogentracker.net], using the isolates IDs (e.g. FSL F2-695). All sequences were also deposited on GenBank under the accession numbers: EU521577, DQ302593, DQ302614, DQ302669, DQ302710, DQ347706-DQ347818, JF12518-JF12529.
3. RESULTS
3.1. Descriptive analysis
All five housekeeping genes and the complete prfA cluster were successfully amplified and sequenced in all lineage III and IV isolates (Table 2). Among the internalin genes, only the two genes in the inlAB operon were detected in all isolates; inlF was not amplified in any of the lineage III and IV isolates. Among the other internalin genes tested here, only inlC was amplified in all lineage IV isolates. Lineage III isolates showed considerable diversity with regard to internalin gene amplification. Among the 8 lineage III isolates (including previously reported data for FSL F2-695, see Table 1), (i) only inlC was amplified in two isolates, (ii) inlC and genes in the inlGC2DE operon were amplified in three isolates, (iii) only genes in the inlGC2DE were amplified in two isolates, and (iv) inlC and genes in inlC2DE operon were amplified in one isolate (Table 1). inlC was thus found in all but two lineage III isolates and five of the eight lineage III isolates carried inlGC2DE cluster. Isolate FSL F2-270 (lineage IIIC) as well as all lineage IV isolates were further screened for the presence of inlGC2DE cluster using two additional sets of primers that amplify the most conservative regions of inlG, inlC2, inlD, and inlE, but consistent negative results (data not shown) were found for all isolates examined, indicating that the inlGC2DE gene cluster was present only in selected lineage III isolates and absent from all lineage IV isolates
Table 2.
Descriptive analysis of each gene in lineages III and IV
| Gene | No. of Isolates | Length of sequence (bp)a | Alleles | π | % polymorphismb | Syn-onymous Changes | Nonsynonymous Changes | GC % |
|---|---|---|---|---|---|---|---|---|
| Lineage III | ||||||||
| gap | 8 | 567 | 2 | 0.0004 | 0.2 | 1 | 0 | 40.0 |
| prs | 8 | 633 | 7 | 0.0118 | 3.3 | 22 | 0 | 41.0 |
| purM | 8 | 714 | 8 | 0.0149 | 4.2 | 24 | 6 | 41.9 |
| ribC | 8 | 639 | 5 | 0.0086 | 2.7 | 15 | 2 | 39.4 |
| sigB | 8 | 660 | 6 | 0.0044 | 1.4 | 9 | 0 | 38.8 |
| inlA | 8 | 2382 | 7 | 0.0193 | 5.6 | 81 | 53 | 37.2 |
| inlB | 8 | 1869 | 8 | 0.0178 | 4.7 | 59 | 29 | 36.2 |
| inlC | 6 | 885 | 6 | 0.0202 | 4.9 | 25 | 18 | 34.2 |
| inlG | 5 | 1467 | 5 | 0.05067 | 11.4 | 103 | 67 | 37.1 |
| inlC2 | 6 | 1653 | 6 | 0.0318 | 7.2 | 83 | 37 | 37.2 |
| inlD | 6 | 1713 | 6 | 0.0572 | 13.9 | 127 | 109 | 36.4 |
| inlE | 6 | 1494 | 5 | 0.0542 | 14.1 | 112 | 88 | 35.9 |
| prfA | 8 | 711 | 8 | 0.0105 | 3.1 | 18 | 5 | 33.1 |
| plcA | 8 | 951 | 8 | 0.0205 | 6.0 | 38 | 17 | 35.4 |
| hly | 8 | 1587 | 8 | 0.0128 | 3.6 | 51 | 6 | 35.5 |
| mpl | 8 | 1533 | 8 | 0.0261 | 8.1 | 97 | 29 | 38.5 |
| actA | 8 | 1812–1926c | 8 | 0.0175 | 5.2 | 45 | 52 | 39.8 |
| plcB | 8 | 870 | 8 | 0.0160 | 5.5 | 39 | 9 | 36.2 |
| noncodingd | 8 | 1197 | NCe | NC | 16.7 | NC | NC | NC |
| Full prfA cluster | 8 | 8775 | 8 | 0.0224 | 6.9 | NC | NC | 36.2 |
| Lineage IV | ||||||||
| gap | 4 | 567 | 2 | 0.0009 | 0.2 | 1 | 0 | 39.8 |
| prs | 4 | 633 | 3 | 0.0056 | 1.1 | 7 | 0 | 41.3 |
| purM | 4 | 714 | 4 | 0.0066 | 1.3 | 5 | 4 | 43.1 |
| ribC | 4 | 639 | 3 | 0.0060 | 0.9 | 6 | 0 | 39.5 |
| sigB | 4 | 660 | 2 | 0.0008 | 0.2 | 1 | 0 | 38.8 |
| inlA | 4 | 2382 | 4 | 0.0078 | 1.5 | 23 | 12 | 37.3 |
| inlB | 4 | 1869 | 4 | 0.0122 | 2.0 | 28 | 10 | 36.5 |
| inlC | 4 | 885 | 3 | 0.0155 | 2.7 | 15 | 9 | 34.2 |
| prfA | 4 | 711 | 2 | 0.0007 | 0.1 | 1 | 0 | 33.0 |
| plcA | 4 | 951 | 4 | 0.0056 | 1.1 | 8 | 2 | 35.1 |
| hly | 4 | 1587 | 3 | 0.0057 | 1.0 | 16 | 0 | 35.8 |
| mpl | 4 | 1533 | 4 | 0.0048 | 0.9 | 8 | 6 | 38.1 |
| actA | 4 | 1821–1926c | 4 | 0.0089 | 1.6 | 13 | 17 | 39.8 |
| plcB | 4 | 870 | 4 | 0.0065 | 1.3 | 8 | 3 | 35.6 |
| Noncoding | 4 | 1197d | NC | NC | 0.5 | NC | NC | NC |
| Full prfA cluster | 4 | 8775 | 4 | 0.0055 | 1.0 | NC | NC | 36.2 |
Length of sequence is in the number of base pair of protein coding region of the gene;
% polymorphism = (Number of polymorphic sites/length of sequence (bp)) × 100%;
Length variation due to a gap;
Noncoding length = length of prfA cluster minus length of 6 coding genes;
NC: not calculated;
In both lineages III and IV isolates, the GC content of housekeeping genes was higher as compared to the GC content of internalin genes and the prfA cluster (Table 2); the same trend was observed for lineage I and II isolates (data not shown). However, for each gene the average GC content was very similar for lineages III and IV (Table 2). With the exception of gap, nucleotide diversities were consistently higher among lineage III isolates as compared to lineage IV isolates (i.e., for housekeeping genes, internalin genes and the prfA cluster; see Table 2). However, when only lineage IIIA isolates are considered (by excluding FSL F2-270, a lineage IIIC isolate, from the alignment), the nucleotide diversity of lineage III isolates is lower than that in lineage IV for inlC. Each of the eight lineage III and the four lineage IV isolates had a different prfA cluster sequence. While each lineage III isolate also presented different alleles for each individual genes in the prfA cluster, some lineage IV isolates shared identical alleles for some genes in the prfA cluster (i.e., prfA, hly). Among the internalin genes, inlB, inlC, inlC2, inlD and inlG each showed different alleles for each lineage III isolates, while both inlA and inlB showed different alleles for each lineage IV isolate.
3.2. Phylogenetic analysis
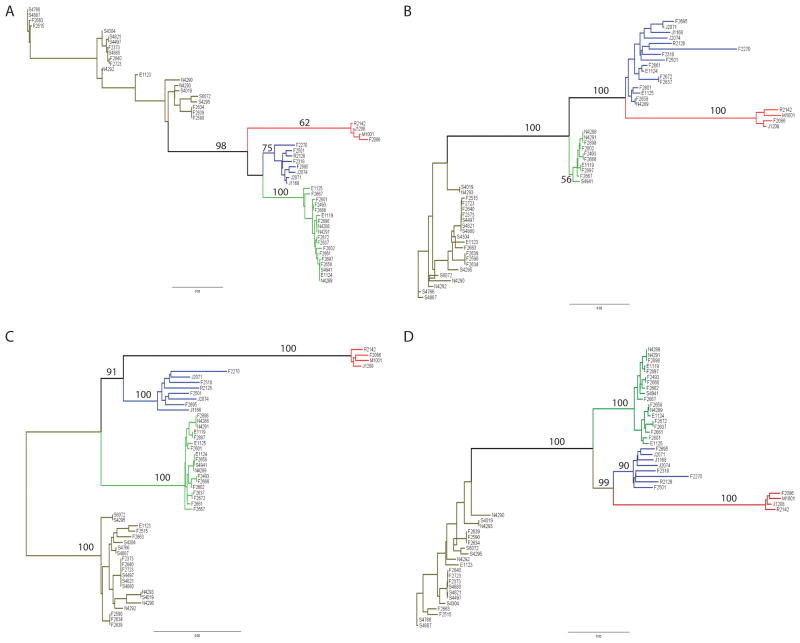
Four datasets were created to investigate the phylogenetic history of lineages III and IV, including (i) a concatenated alignment of the 5 housekeeping genes; (ii) a concatenated alignment of inlA and inlB (these genes are located in the same operon and represent the only two internalin genes present in all isolates studied); (iii) an alignment of the entire prfA cluster; and (iv) a concatenated alignment of the three previous alignments; a Neighbor-Joining (NJ) tree was generated from each dataset. In three out of the four phylogenetic trees, the four lineages formed monophyletic groups (Fig. 1), supporting their classification into different lineages. Only the NJ tree based on the inlAB alignment did not provide good bootstrap support for a monophyletic lineage III.
Figure 1.
Phylogenetic trees constructed in PAUP using the Neighbor-Joining method and substitution models selected by ModelTest. (A) Tree based on a concatenated alignment of the five housekeeping genes (gap, prs, purM, ribC and sigB); (B) Tree based on a concatenated alignment of inlA and inlB sequences; (C) Tree based on alignment of the prfA cluster; and (D) Tree based on a concatenated alignment of the three previous datasets. Numbers on major branched show the bootstrap support for the branch (%). Branches are color coded as lineage I (green), lineage II (brown), lineage III (blue) and lineage IV (red). Branch tips are labeled with the last 5 characters of the isolate designation, e.g., FSL J1-208 (Table 1) is coded as “J1208)
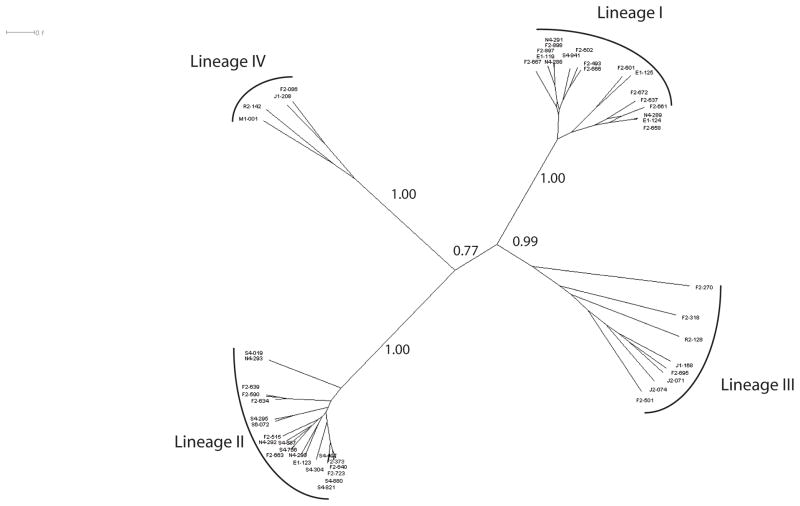
In addition, the trees based on the four datasets showed different relationships among the lineages (Fig. 1). In the housekeeping tree, lineage III isolates formed a sistergroup to lineage I isolates, while in the other three trees, lineages III and IV isolates shared a most common recent ancestor. The housekeeping alignment also provided the poorest bootstrap support (i.e., 62 %), among all datasets, for monophyletic grouping of the lineage IV isolates. The largest dataset, i.e., the concatenated alignment of housekeeping genes, inlA and inlB, and the prfA cluster, supported the monophyletic grouping of each of the four lineages with high support (i.e., bootstrap values ≥ 90%) with lineages III and IV representing divergent sister clades. As some genes in this alignment showed evidence for recombination (see below), we also constructed a phylogeny using ClonalFrame, which uses an algorithm that accounts for the presence of point mutations as well as recombination. The ClonalFrame analysis of the concatenated alignment of the prfA cluster, housekeeping genes and inlA and inlB genes also supported that lineage III and IV isolates form distinct lineages (Figure 2). The phylogenetic tree inferred by ClonalFrame also grouped lineages I and III together (as sistergroups). Overall, our analyses show that the phylogeny of L. monocytogenes lineages III and IV may be hard to define, possibly due to recurrent recombination events, but do suggest that lineages I and III may indeed be sistergroups. Interestingly, isolate FSL F2-270, which was classified as lineage IIIC by Roberts et al. (2006) was highly divergent from the other lineage III isolates in the ClonalFrame tree. This isolate also grouped separate from the other lineage III isolates in the NJ tree based on the housekeeping genes (Fig. 1a) and showed long branches, within the other lineage III isolates, in the prfA cluster and inlA-inlB tree (Fig. 1b and c). Given the relatively high divergence found in this study between isolates from lineages IIIA and IIIC, further studies should be carried to closely investigate the evolution of lineage III as more lineage IIIC isolates become available.
Figure 2.
Consensus network (33%) for the concatenated alignment with all sequence data (i.e., the five housekeeping genes, inlA and inlB, and the prfA cluster). Posterior probabilities are shown for the major branches. Analysis was run with ClonalFrame version 1.1 using default settings.
3.3. Horizontal gene transfer analysis
GENECONV was used as previously described (Nightingale et al., 2005) to identify and characterize possible recombination events. Lineage III isolates were identified as recipients in (i) six recombination events among the internalin genes (inlA, inlB, inlD, inlE and inlG [2 events]), (ii) four recombination events in the prfA cluster, and (iii) one recombination event in prs (Table 3). Only one of the recombination events identified had a lineage IV isolate as recipient (this event occurred in inlB; a lineage I isolate was identified as donor; see Table 3). While the recombination events identified among lineage III isolates involved predominantly lineage II isolates as donors (Table 3); the only recombination event identified between lineage III and IV isolates occurred in the noncoding region upstream actA and involved a lineage III isolate as recipient and a lineage IV isolate as donor). Our data indicate that both lineage III and IV isolates evolved with substantial contributions of horizontal gene transfer, which may explain at least in part the great genetic diversity observed within these two lineages (Table 2). The lineage IIIC isolate FSL F2-270, which was highly divergent from other lineage III isolates in our phylogenetic analysis, was identified as recipient of two recombinant fragments, one within inlA and one including part of mpl and its upstream noncoding region.
Table 3.
Recombination events involving lineages III or IV
| Location | Donor | Recipient |
|---|---|---|
| prs | I | III |
| purM | III | II |
| inlG | Undefineda | III |
| inlG | II | III |
| inlD | II | III |
| inlE | II | III |
| inlA | I/III | II |
| inlA | II | III |
| inlA | III | II |
| inlB | Undefined | III |
| inlB | I | IV |
| actA | II | III |
| actA | III | II |
| plcA/non-codingb | III | II |
| mpl/non-coding/actAb | III | III |
| actA/non-coding/plcBb | III | III |
| actA | Undefined | Undefined |
| actA | Undefined | Undefined |
| actA | Undefined | Undefined |
| actA | Undefined | Undefined |
| non-coding/mplb | IV | III |
”Undefined” indicates instances where the donor isolate could not be identified
as the whole prfA region was used for recombination analyses some recombinant fragments span multiple genes and non-coding (intergenic) regions
3.4. Intragenic positive selection
Three tests (M0 vs. M3, M1a vs. M2a, and M7 vs. M8) implemented in PAML were used to assess whether the different genes in the prfA cluster and the different internalin genes (i.e., inlA, inlB, inlC, inlC2, inlD, inlE, inlF, inlG) evolved by positive selection (using an alignment of the available sequences for all 51 isolates). M3 was supported over M0 for all virulence genes in this study (i.e. internalin genes and genes in the prfA cluster), which indicates that heterogeneous levels of selection occurred in different regions of these genes. In addition, we found evidence of positive selection (P < 0.05; M1a and/or M7 rejected in favor of M2a and/or M8) for hly, plcA, actA, inlA and inlC2 (Table 4). The Bayesian approach implemented in PAML identified 8, 7, 6 and 6 codon sites in plcA, actA, inlC2, and inlA as showing evidence for positive selection, respectively (ω > 1; posterior probability > 0.95; Table 4). Three of the positively selected sites in ActA (452, 464, and 479) were identified in a transmembrane domain located between sites 391 to 639 and reverted from polar to non-polar or vice versa. This may affect the ActA bacterial anchoring ability (Skoble et al., 2000). Positive selection was also found at amino acid site 117, which is one of five sites necessary for filament formation (Lasa et al., 1997). Another positively selected site was site 141, which is part of a cofilin homologous sequence (136–165) that was previously found to be important for actin nucleation and Arp2/3 stimulation (Skoble et al., 2000). The positively selected site 294 is in a region (264–390) that interacts with the host vasodilator-stimulated phosphoprotein (VASP), and, while not vital for actin tail formation, contributes to the rate of movement and the percentage of moving bacteria (Skoble et al., 2000). Site 168, which was also found to be under positive selection, falls in a region with no defined function. All the positively selected sites in actA and plcA were polymorphic in lineage III or IV isolates, with exception of sites 464 (actA) and 119 (plcA), which were conserved within each lineage, although divergent between them. None of the sites under positive selection in plcA are known to be important for its function (Moser et al., 1997). Although a recent study has shown that InlC2 elicits the humoral immune response against L. monocytogenes in guinea pigs, cattle, sheep (Yu et al., 2008) and rabbits (Yu et al., 2007), InlC2 function is not yet well enough characterized to determine whether any positively selected sites in this protein fall into functionally important sites.
Table 4.
Positive selection results for positively selected genesa.
| Gene | M1a vs M2a (p-value, ωb, pc) | M7 vs M8 (p-value, ωb, pc) | Sites identified as positively selected with posterior probabilities of >95%, and >99%(*) |
|---|---|---|---|
| actA | p-value < 0.001; ω = 3.46; p = 0.035 | p-value < 0.001; ω = 2.90; p = 0.054 | 117, 141, 168, 294, 452, 464*, 479 |
| plcA | p-value < 0.001; ω = 3.67; p = 0.038 | p-value < 0.001; ω = 3.55; p = 0.042 | 4*, 7*, 13, 17, 19, 56, 119, 211* |
| inlA | p-value = 0.030; ω = 2.99; p = 0.013 | p-value = 0.011;ω = 2.58; p = 0.021 | 3, 19, 142, 187*, 426, 594 |
| inlC2 | p-value < 0.001;ω = 7.99; p = 0.007 | p-value <0.001; ω = 7.40; p = 0.008 | 20, 39, 102*, 395*, 396*, 445* |
These analyses were performed using the available sequences for all 51 isolates; polymorphic sites among lineages III and IV isolates are detailed in the “Results” section
ω: dN/dS estimated for sites under positive selection using models M2a or M8;
p: frequency of sites under positive selection estimated using models M2a or M8
When the virulence gene alignments were further analyzed, using branch-sites models (Zhang et al., 2005), to test for positive selection along lineage III and IV branches, positive selection was only observed for actA (for both lineages III and IV; Table 5). All of the amino acid sites identified as under positive selection in lineage IV using the branch-sites model had also been identified using M8. Branch-sites model analysis of lineage III identified three sites with evidence for positive selection, which were not identified in the M8 analysis, including (i) aa site 48, which is located in the f- actin binding region (21–97) and is necessary for proper actin tail elongation (Lasa et al., 1997) and (ii) site 611, another site in the transmembrane domain that reverted from polar to non-polar.
Table 5.
Positive selection on lineage III and IV branches of the actA tree.
| Lineage | p-value | ωa | pb | Positively selected sites (P > 95%) | Amino Acidc |
|---|---|---|---|---|---|
| III | <0.001 | 5.69 | 0.043 | 48 183 611 |
T(6)/M(1)/I(1) S(3)/L(4)/P(1) P(4)/S(4) |
| IV | <0.001 | 3.57 | 0.036 | 117 141 464 |
V(4) S(4) P(4) |
ω: dN/dS ratio for positively selected sites at the tree branch of interest (i.e. lineage III or lineage IV ancestral branch)
p: Frequency of ω for positively selected sites at the branch of interest
Letters indicate the amino acid in each site, followed by their counts among lineage III or IV isolates
3.5. Intergenic positive selection
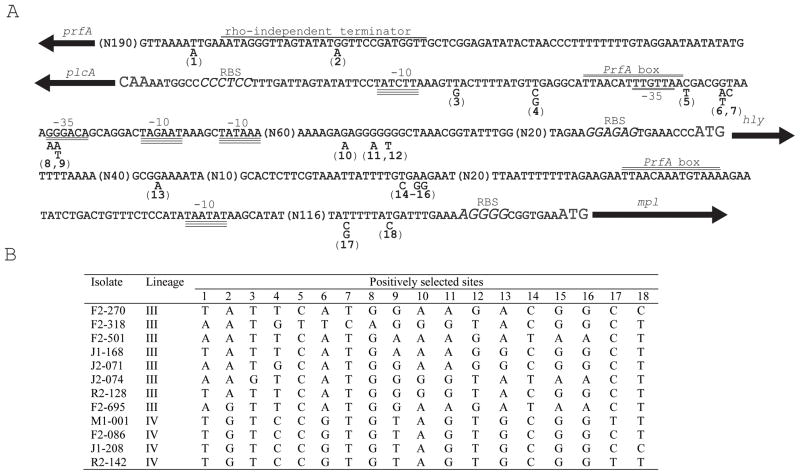
Positive selection analysis of intergenic regions, using EvoNC (Wong and Nielsen, 2004), identified three noncoding regions (prfA-plcA, plcA-hly, and hly-mpl) that may have evolved by positive selection among the 51 isolates representing all four L. monocytogenes lineages (Figure 3). In these three regions, two positively selected sites between prfA and plcA, ten sites between plcA and hly, and six sites between hly and mpl were identified. Among lineage IV isolates, 16 out of the 18 sites under positive selection were monomorphic (Figure 3B). Conversely, 17 out of the 18 sites under positive selection were polymorphic among lineage III isolates. These results suggest that positive selection may have occurred either during divergence of the lineages or during diversification of lineage III. One nucleotide located 74 bp upstream of the plcA start codon and 168 bp upstream of the hly start codon is positively selected; this nucleotide is located next to the PrfA-dependent hly promoter, and directly upstream the PrfA box. This mutation occurred in five of the fifty-one isolates, including one lineage III isolate (FSL F2-318). A SigmaA-dependent −35 region upstream of hly harbored four polymorphic sites, including two sites with evidence for positive selection. All four lineage IV isolates have the same sequence (GTGCCA) in this region while lineage III isolates presented three different sequences (GGGACA, 4 isolates; AGGACA, 1 isolate; GAGACA, 2 isolates) in the same region. All lineage I isolates were identical (GGGACA) and lineage II isolates harbored three distinct alleles (AGGATA, 3 isolates; GGGACA, 17 isolates; AGGACA, 1 isolate) in this −35 region.
Figure 3.
Positively selected sites identified in the intergenic regions between prfA and plcA, plcA and hly, and hly and mpl. (A) The figure shows all intergenic regions between prfA and mpl; coding regions are shown as arrows and selected noncoding regions that did not show evidence for positive selection are coded with N and the number of nucleotides that are not shown (e.g., N190). For all sites with evidence for positive selection, the polymorphic nucleotides are shown below the sequence, and are labeled by numbers 1 to 18. Selected features in the intragenic regions are highlighted, including (i) start codons (large font), (ii) ribosome binding sites (RBS) (large font and italics), (iii) −35 and −10 promoter regions (double and triple underline, respectively), (iv) PrfA boxes (double line above the sequence), and (v) rho-independent transcription terminators (single line above the sequence). (B) Polymorphic nucleotides among lineage III and IV isolates for sites with evidence of positive selection. Sites are labeled in Panel (A).
4. DISCUSSION
Lineage III and lineage IV isolates of L. monocytogenes are rarely associated with human clinical cases of listeriosis. They are thus usually underrepresented among isolate collections and have been understudied as compared to lineage I and lineage II isolates. Comparative analysis of selected housekeeping and internalin genes as well as the prfA cluster in lineage III and IV isolates showed that (i) L. monocytogenes lineages III and IV represent highly divergent monophyletic lineages, (ii) intragenic recombination contributed considerably to the genetic diversity of lineage III, and (iii) actA evolved by positive selection in both lineage III and IV while inlA, inlC2 and plcA showed evidence for positive selection in L. monocytogenes in general.
4.1. Lineages III and IV are divergent and monophyletic
While L. monocytogenes lineages I and II have been well established to represent two well supported divergent monophyletic lineages (see review by Orsi et al., 2010), which may show significantly different virulence characteristics (Chen et al., 2006), the phylogeny of lineages III and lineage IV isolates and their relation to lineages I and II has remained unclear, despite the fact that these lineages represent a number of phenotypic and genetic characteristics unusual for the species L. monocytogenes. While Ward et al. (2004) identified lineages I, II, and III as distinct evolutionary divisions based on prfA virulence gene cluster sequence data, their dataset did not include lineage IV isolates. Liu et al. (2006) and Roberts et al. (2006) provided phenotypic evidence that lineage III isolates might represent three distinct lineages, named IIIA, IIIB and IIIC. Roberts et al. (2006) also provided some genetic evidence based on the partial sequences of actA and sigB that these three lineages were divergent, although lineages IIIA and IIIC were more closely related to each other as compared to lineage IIIB isolates.
Recently, Ward et al. (2008) suggested that lineage IIIB should be renamed as lineage IV to make it clear that this lineage represents a completely distinct group within L. monocytogenes. Ward et al. (2008) and den Bakker et al. (2010) provided genetic evidence of the paraphyletic relationship of lineages IIIA and IIIC, which were thus grouped together and designated as lineage III. We have provided genetic evidence, based on more than 16 kb of DNA sequence data for distinct regions in the L. monocytogenes chromosome, that the four L. monocytogenes lineages are monophyletic. However, our data failed to provide definitive insights on the phylogenetic relationships between the different L. monocytogenes lineages. While the Neighbor-Joining trees based on the concatenated alignment of the prfA cluster, inlA and inlB genes and 5 housekeeping gene fragments suggest that lineage IV isolates share a most common recent ancestor with lineage III isolates, which is in disagreement with some previous studies (Orsi et al., 2008b; Roberts et al., 2006; Ward et al., 2008) and in agreement with a most recent population study (den Bakker et al., 2010), the ClonalFrame phylogenetic tree from this same concatenated alignment places lineages I and III as sister groups; however this relationship is not supported by a significant posterior probability. Our inability to reveal, with high confidence, the true phylogenetic structure of L. monocytogenes lineages is probably due to the high amount of horizontal gene transfer followed by homologous recombination observed between isolates belonging to different lineages. Future work on more lineage III and IV strains, including phylogenetic analyses utilizing full genome sequences will likely be needed to further clarify the phylogeny of these two lineages
In addition to the genetic divergence between lineages III and IV, we also observed considerable differences in internalin gene profiles among these two lineages, consistent with previous data that showed considerable diversity in the internalin gene presence/absence patterns in lineage I and II strains (Call et al., 2003; Hong et al., 2007; Jia et al., 2007; Pohl et al., 2006). Interestingly, inlC, which has been shown to be critical for cell to cell spread by L. monocytogenes (Engelbrecht et al., 1996; Rajabian et al., 2009) was missing from two lineage III isolates, consistent with previous data by Jia et al. (2007), who also found this gene absent in the lineage III strain FSL F2-695 (which represented one of the two strains without inlC in this study presented here). In addition, the full inlGC2DE cluster was missing from all lineage IV isolates and two lineage III isolates, all of these strains did contain inlC though. Using a Δ inlGC2DE strain Bergmann et al. (2002) showed that inlGC2DE might have a role in aiding InlA- and InlB-mediated endocytosis in lineages I and II strains. The absence of the gene cluster from lineage IV isolates could possibly indicate that inlGC2DE cluster does not play a critical role in bacterial adaptation or survival, and was lost in lineage IV by genetic drift. These genes may also only be required in specific hosts, which are not generally infected by lineage IV (and some lineage III) isolates. While, overall, our findings are consistent with observations that lineage III and IV isolates show considerable diversity with regard to virulence-related phenotypic characteristics (including virulence attenuation of some isolates) (Liu et al., 2006; Nightingale et al., 2008; Orsi et al., 2010; Orsi et al., 2007; Van Stelten and Nightingale, 2008), further virulence characterization of strains with well defined sequence data (such as the strain characterized here), including characterization of appropriate internalin gene null mutants in lineage III and IV strains, will be needed to clarify the role of internalin gene presence/absence in the virulence characteristics of lineage III and IV strains.
4.2. Intragenic recombination contributed significantly to the genetic diversities of lineage III virulence genes
Lineage III internalin genes and the prfA cluster are characterized by high frequencies of intragenic recombination, probably reflecting that virulence genes are under less stringent functional constraint than housekeeping genes (Nightingale et al., 2005), and are more likely to retain and incorporate the genetic fragments after acquiring them (Lan and Reeves, 2001; Lan et al., 2000; Wertz et al., 2003). Alternatively, some of these genes may experience positive selection in lineages III and IV, thus facilitating fixation of horizontally acquired virulence and internalin genes (as previously proposed by Levin and Cornejo, 2009; Orsi et al., 2007; Orsi et al., 2008b). Identification of considerable recombination among lineage III strains (including introduction of divergent sequences from other lineages) is also consistent with the observation that lineage III isolates showed higher genetic diversity in the prfA cluster, inlA, inlB, inlC, and prs and sigB, as compared to lineage IV isolates as well as compared to lineages I and II isolates (Orsi et al., 2007; Orsi et al., 2008b; Tsai et al., 2006). A similar trend was previously observed for lineage II isolates (Orsi et al., 2008a; Orsi et al., 2007; Orsi et al., 2008b), which show a much higher diversity and recombination levels than lineage I isolates. The fact that few recombination events were observed among lineage IV isolates may be due to the underrepresentation of this lineage in our dataset. However, genetic differences between lineages IV and III, such as different restriction systems that interfere with their recombination levels, cannot be ruled out. Future studies on additional lineage IV isolates, as they become available, will hopefully help clarify this.
4.3. Positive selection contributed to the evolution of virulence genes in lineages III and IV
Although positive selection in inlA, inlC2, plcA and hly was identified in the whole phylogeny (i.e., including isolates from lineages I and II), consistent with previous studies (Orsi et al., 2008a; Orsi et al., 2007; Tsai et al., 2006), positive selection in actA was also identified in the ancestral branches of lineages III and IV. Interestingly, actA is the only gene in the prfA cluster that encodes a protein attached to the bacterial cell wall (Vazquez-Boland et al., 2001). Although, unlike listeriolysin O, ActA is not readily presented to the immune system by MHC class I or class II molecules (Darji et al., 1998), its localization may require a higher degree of variation in order to avoid immune detection. As ActA may aid in host cell entry by recognizing an HSPG receptor (Alvarez-Dominguez et al., 1997), amino acid changes in ActA may also facilitate invasion of diverse host cells. Finally, as some ActA aminoacid sites under positive selection in lineages III and IV are in regions that have known functions for actin tail formation and interact with different macromolecules in the host cells, positive selection of ActA may facilitate adaptation to the intracellular environment of different host species. As additional mammalian genomes are completed, it may become possible to test this theory by comparing ActA sequences with those of their interacting proteins from various animal species.
Consistent with one previous study (Orsi et al., 2008a), which indentified evidence for positive selection in sites in the hly-plcA intergenic region in the prfA cluster, we also identified a specific nucleotide site in the −35 promoter region upstream of hly that appears to have evolved by positive selection. This mutation was found in five of the fifty-one isolates analyzed, including one lineage III isolate. Mutations in promoter sequences can affect the binding affinity of sigma factors or other regulators to the DNA, which can affect the rate of transcription initiation. The mutation identified here is of particular interest as listeriolysin, which is encoded by hly, is not only required for escape from this host cell vacuole, but is also extremely toxic to cells, and therefore its expression is strictly regulated (Shen and Higgins, 2005). While it is tempting to speculate that adaptations of hly promoters across isolates representing different lineages suggests functional importance of the positively selected site identified here (possibly reflecting different virulence associated functions), future experiments with isogenic strains with site specific mutations in this site will be required to identify the specific functions that may be associated with this change.
5. Conclusion
Consistent with two other recent studies (den Bakker et al., 2010; Ward et al., 2008), our data support that L. monocytogenes lineages III and IV form clearly distinct monophyletic groups, even though the phylogenetic relationships of these two lineages remain difficult to define, likely due to considerable horizontal gene transfer in lineage III isolates. Both horizontal gene transfer and recombination as well as positive selection of virulence genes appear to have been important forces shaping the evolution of these two lineages and seem to have contributed to their genetic and phenotypic diversity. While lineages III and IV are rarely isolated from foods, and possibly environmental sources, they have been associated with human cases and are not uncommon among isolates from animal clinical cases. While, overall, these lineages are candidates for host adapted L. monocytogenes lineages, further research on their host range using animal and tissue culture models are needed. These experiments, combined with generation and characterization of lineage III and IV null mutant strains will provide further insight into the biology and virulence of these unusual clonal groups and will help us to determine whether these groups represent unusual virulence characteristics, which could be facilitated or acquired by horizontal gene transfer and positive selection.
HIGHLIGHTS.
L. monocytogenes lineages III and IV form two distinct monophyletic groups.
Virulence genes evolved by positive selection in lineages III and IV.
Recombination contributed mainly to the evolution of lineage III isolates.
Acknowledgments
We thank Dr. Ziheng Yang and Dr. Rasmus Nielsen for their advice in PAML analysis, Dr. Henk den Bakker for helpful discussions. This work was partially supported by the National Institute of Health Award No. R01GM63259-04 and by USDA Special Research Grants 2006-34459-16952 and 2008-34459-19043.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez-Dominguez C, Vazquez-Boland JA, Carrasco-Marin E, Lopez-Mato P, Leyva-Cobian F. Host cell heparan sulfate proteoglycans mediate attachment and entry of Listeria monocytogenes, and the listerial surface protein ActA is involved in heparan sulfate receptor recognition. Infect Immun. 1997;65:78–88. doi: 10.1128/iai.65.1.78-88.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann B, Raffelsbauer D, Kuhn M, Goetz M, Hom S, Goebel W. InlA- but not InlB-mediated internalization of Listeria monocytogenes by non-phagocytic mammalian cells needs the support of other internalins. Mol Microbiol. 2002;43:557–570. doi: 10.1046/j.1365-2958.2002.02767.x. [DOI] [PubMed] [Google Scholar]
- Bierne H, Sabet C, Personnic N, Cossart P. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 2007;9:1156–1166. doi: 10.1016/j.micinf.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Call DR, Borucki MK, Besser TE. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J Clin Microbiol. 2003;41:632–639. doi: 10.1128/JCM.41.2.632-639.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ross WH, Gray MJ, Wiedmann M, Whiting RC, Scott VN. Attributing risk to Listeria monocytogenes subgroups: dose response in relation to genetic lineages. J Food Prot. 2006;69:335–344. doi: 10.4315/0362-028x-69.2.335. [DOI] [PubMed] [Google Scholar]
- Darji A, Bruder D, zur Lage S, Gerstel B, Chakraborty T, Wehland J, Weiss S. The role of the bacterial membrane protein ActA in immunity and protection against Listeria monocytogenes. J Immunol. 1998;161:2414–2420. [PubMed] [Google Scholar]
- De Cesare A, Mioni R, Manfreda G. Prevalence of Listeria monocytogenes in fresh and fermented Italian sausages and ribotyping of contaminating strains. Int J Food Microbiol. 2007;120:124–130. doi: 10.1016/j.ijfoodmicro.2007.06.009. [DOI] [PubMed] [Google Scholar]
- De Jesus AJ, Whiting RC. Thermal inactivation, growth, and survival studies of Listeria monocytogenes strains belonging to three distinct genotypic lineages. J Food Prot. 2003;66:1611–1617. doi: 10.4315/0362-028x-66.9.1611. [DOI] [PubMed] [Google Scholar]
- den Bakker HC, Bundrant BN, Fortes ED, Orsi RH, Wiedmann M. A population genetics-based and phylogenetic approach to understanding the evolution of virulence in the genus Listeria. Appl Environ Microbiol. 2010;76:6085–6100. doi: 10.1128/AEM.00447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KA, Bielawski JP, Ward TJ, Urquhart C, Gu H. Reconciling ecological and genomic divergence among lineages of Listeria under an “extended mosaic genome concept”. Mol Biol Evol. 2009;26:2605–2615. doi: 10.1093/molbev/msp176. [DOI] [PubMed] [Google Scholar]
- Engelbrecht F, Chun SK, Ochs C, Hess J, Lottspeich F, Goebel W, Sokolovic Z. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol Microbiol. 1996;21:823–837. doi: 10.1046/j.1365-2958.1996.541414.x. [DOI] [PubMed] [Google Scholar]
- Farber JM, Peterkin PI. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrer B, Candrian U, Hoefelein C, Luethy J. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J Appl Bacteriol. 1991;70:372–379. doi: 10.1111/j.1365-2672.1991.tb02951.x. [DOI] [PubMed] [Google Scholar]
- Gellin BG, Broome CV. Listeriosis. JAMA. 1989;261:1313–1320. [PubMed] [Google Scholar]
- Gray MJ, Zadoks RN, Fortes ED, Dogan B, Cai S, Chen Y, Scott VN, Gombas DE, Boor KJ, Wiedmann M. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl Environ Microbiol. 2004;70:5833–5841. doi: 10.1128/AEM.70.10.5833-5841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa S, Kimura B, Takahashi H, Koda T, Hisa K, Fujii T. Incidence of Listeria monocytogenes in raw seafood products in Japanese retail stores. J Food Prot. 2005;68:411–415. doi: 10.4315/0362-028x-68.2.411. [DOI] [PubMed] [Google Scholar]
- Hong E, Doumith M, Duperrier S, Giovannacci I, Morvan A, Glaser P, Buchrieser C, Jacquet C, Martin P. Genetic diversity of Listeria monocytogenes recovered from infected persons and pork, seafood and dairy products on retail sale in France during 2000 and 2001. Int J Food Microbiol. 2007;114:187–194. doi: 10.1016/j.ijfoodmicro.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jeffers GT, Bruce JL, McDonough PL, Scarlett J, Boor KJ, Wiedmann M. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology. 2001;147:1095–1104. doi: 10.1099/00221287-147-5-1095. [DOI] [PubMed] [Google Scholar]
- Jia Y, Nightingale KK, Boor KJ, Ho A, Wiedmann M, McGann P. Distribution of internalin gene profiles of Listeria monocytogenes isolates from different sources associated with phylogenetic lineages. Foodborne Pathog Dis. 2007;4:222–232. doi: 10.1089/fpd.2006.0081. [DOI] [PubMed] [Google Scholar]
- Jones D. Foodborne listeriosis. Lancet. 1990;336:1171–1174. doi: 10.1016/0140-6736(90)92778-g. [DOI] [PubMed] [Google Scholar]
- Kiss R, Tirczka T, Szita G, Bernath S, Csiko G. Listeria monocytogenes food monitoring data and incidence of human listeriosis in Hungary, 2004. Int J Food Microbiol. 2006;112:71–74. doi: 10.1016/j.ijfoodmicro.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Klaeboe H, Rosef O, Fortes E, Wiedmann M. Ribotype diversity of Listeria monocytogenes isolates from two salmon processing plants in Norway. Int J Environ Health Res. 2006;16:375–383. doi: 10.1080/09603120600869406. [DOI] [PubMed] [Google Scholar]
- Lan R, Reeves PR. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 2001;9:419–424. doi: 10.1016/s0966-842x(01)02133-3. [DOI] [PubMed] [Google Scholar]
- Lan Z, Fiedler F, Kathariou S. A sheep in wolf’s clothing: Listeria innocua strains with teichoic acid-associated surface antigens and genes characteristic of Listeria monocytogenes serogroup 4. J Bacteriol. 2000;182:6161–6168. doi: 10.1128/jb.182.21.6161-6168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I, Gouin E, Goethals M, Vancompernolle K, David V, Vandekerckhove J, Cossart P. Identification of two regions in the N-terminal domain of ActA involved in the actin comet tail formation by Listeria monocytogenes. Embo J. 1997;16:1531–1540. doi: 10.1093/emboj/16.7.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Cornejo OE. The population and evolutionary dynamics of homologous gene recombination in bacterial populations. PLoS Genet. 2009;5:e1000601. doi: 10.1371/journal.pgen.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Lawrence ML, Wiedmann M, Gorski L, Mandrell RE, Ainsworth AJ, Austin FW. Listeria monocytogenes subgroups IIIA, IIIB, and IIIC delineate genetically distinct populations with varied pathogenic potential. J Clin Microbiol. 2006;44:4229–4233. doi: 10.1128/JCM.01032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorber B. Listeriosis. Clin Infect Dis. 1997;24:1–9. doi: 10.1093/clinids/24.1.1. quiz 10–11. [DOI] [PubMed] [Google Scholar]
- Lukinmaa S, Aarnisalo K, Suihko ML, Siitonen A. Diversity of Listeria monocytogenes isolates of human and food origin studied by serotyping, automated ribotyping and pulsed-field gel electrophoresis. Clin Microbiol Infect. 2004;10:562–568. doi: 10.1111/j.1469-0691.2004.00876.x. [DOI] [PubMed] [Google Scholar]
- Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J, Gerstel B, Meyer JE, Chakraborty T, Wehland J, Heinz DW. Crystal structure of the phosphatidylinositol-specific phospholipase C from the human pathogen Listeria monocytogenes. J Mol Biol. 1997;273:269–282. doi: 10.1006/jmbi.1997.1290. [DOI] [PubMed] [Google Scholar]
- Nightingale K, Bovell L, Grajczyk A, Wiedmann M. Combined sigB allelic typing and multiplex PCR provide improved discriminatory power and reliability for Listeria monocytogenes molecular serotyping. J Microbiol Methods. 2007;68:52–59. doi: 10.1016/j.mimet.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Nightingale KK, Ivy RA, Ho AJ, Fortes ED, Njaa BL, Peters RM, Wiedmann M. inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl Environ Microbiol. 2008;74:6570–6583. doi: 10.1128/AEM.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale KK, Windham K, Wiedmann M. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J Bacteriol. 2005;187:5537–5551. doi: 10.1128/JB.187.16.5537-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton DM, Scarlett JM, Horton K, Sue D, Thimothe J, Boor KJ, Wiedmann M. Characterization and pathogenic potential of Listeria monocytogenes isolates from the smoked fish industry. Appl Environ Microbiol. 2001;67:646–653. doi: 10.1128/AEM.67.2.646-653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi RH, den Bakker HC, Wiedmann M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol. 2010 doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Orsi RH, Maron SB, Nightingale KK, Jerome M, Tabor H, Wiedmann M. Lineage specific recombination and positive selection in coding and intragenic regions contributed to evolution of the main Listeria monocytogenes virulence gene cluster. Infect Genet Evol. 2008a;8:566–576. doi: 10.1016/j.meegid.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi RH, Ripoll DR, Yeung M, Nightingale KK, Wiedmann M. Recombination and positive selection contribute to evolution of Listeria monocytogenes inlA. Microbiology. 2007;153:2666–2678. doi: 10.1099/mic.0.2007/007310-0. [DOI] [PubMed] [Google Scholar]
- Orsi RH, Sun Q, Wiedmann M. Genome-wide analyses reveal lineage specific contributions of positive selection and recombination to the evolution of Listeria monocytogenes. BMC Evol Biol. 2008b;8:233. doi: 10.1186/1471-2148-8-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl MA, Wiedmann M, Nightingale KK. Associations among Listeria monocytogenes genotypes and distinct clinical manifestations of listeriosis in cattle. Am J Vet Res. 2006;67:616–626. doi: 10.2460/ajvr.67.4.616. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008;4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabian T, Gavicherla B, Heisig M, Muller-Altrock S, Goebel W, Gray-Owen SD, Ireton K. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat Cell Biol. 2009;11:1212–1218. doi: 10.1038/ncb1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen OF, Skouboe P, Dons L, Rossen L, Olsen JE. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology. 1995;141 (Pt 9):2053–2061. doi: 10.1099/13500872-141-9-2053. [DOI] [PubMed] [Google Scholar]
- Roberts A, Nightingale K, Jeffers G, Fortes E, Kongo JM, Wiedmann M. Genetic and Phenotypic Characterization of Listeria monocytogenes lineage III. Microbiology. 2006;152:685–693. doi: 10.1099/mic.0.28503-0. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Sawyer S. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Sawyer SA. GENECONV: A computer package for the statistical detection of gene conversion. Distributed by the author, Department of mathematics, Washington University; St. Louis: 1999. available at http://math.wust1.edu/~sawyer. [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchat A, Swaminathan B, Broome CV. Epidemiology of human listeriosis. Clin Microbiol Rev. 1991;4:169–183. doi: 10.1128/cmr.4.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, Higgins DE. The 5′ untranslated region-mediated enhancement of intracellular listeriolysin O production is required for Listeria monocytogenes pathogenicity. Mol Microbiol. 2005;57:1460–1473. doi: 10.1111/j.1365-2958.2005.04780.x. [DOI] [PubMed] [Google Scholar]
- Skoble J, Portnoy DA, Welch MD. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J Cell Biol. 2000;150:527–538. doi: 10.1083/jcb.150.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt BG, Maiden MC. Bacterial population genetics, evolution and epidemiology. Philos Trans R Soc Lond B Biol Sci. 1999;354:701–710. doi: 10.1098/rstb.1999.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4. Sinauer Associates; 2002. [Google Scholar]
- Tsai YH, Orsi RH, Nightingale KK, Wiedmann M. Listeria monocytogenes internalins are highly diverse and evolved by recombination and positive selection. Infect Genet Evol. 2006;6:378–389. doi: 10.1016/j.meegid.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Van Stelten A, Nightingale KK. Development and implementation of a multiplex single-nucleotide polymorphism genotyping assay for detection of virulence-attenuating mutations in the Listeria monocytogenes virulence-associated gene inlA. Appl Environ Microbiol. 2008;74:7365–7375. doi: 10.1128/AEM.01138-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Boland JA, Dominguez-Bernal G, Gonzalez-Zorn B, Kreft J, Goebel W. Pathogenicity islands and virulence evolution in Listeria. Microbes and Infection. 2001;3:571–584. doi: 10.1016/s1286-4579(01)01413-7. [DOI] [PubMed] [Google Scholar]
- Ward TJ, Ducey TF, Usgaard T, Dunn KA, Bielawski JP. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl Environ Microbiol. 2008;74:7629–7642. doi: 10.1128/AEM.01127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TJ, Gorski L, Borucki MK, Mandrell RE, Hutchins J, Pupedis K. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J Bacteriol. 2004;186:4994–5002. doi: 10.1128/JB.186.15.4994-5002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz JE, Goldstone C, Gordon DM, Riley MA. A molecular phylogeny of enteric bacteria and implications for a bacterial species concept. J Evol Biol. 2003;16:1236–1248. doi: 10.1046/j.1420-9101.2003.00612.x. [DOI] [PubMed] [Google Scholar]
- Wiedmann M, Bruce JL, Keating C, Johnson AE, McDonough PL, Batt CA. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun. 1997;65:2707–2716. doi: 10.1128/iai.65.7.2707-2716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M, Mobini S, Cole JR, Jr, Watson CK, Jeffers GT, Boor KJ. Molecular investigation of a listeriosis outbreak in goats caused by an unusual strain of Listeria monocytogenes. J Am Vet Med Assoc. 1999;215:369–371. 340. [PubMed] [Google Scholar]
- Wong WS, Nielsen R. Detecting selection in noncoding regions of nucleotide sequences. Genetics. 2004;167:949–958. doi: 10.1534/genetics.102.010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yu WL, Dan H, Lin M. Novel protein targets of the humoral immune response to Listeria monocytogenes infection in rabbits. J Med Microbiol. 2007;56:888–895. doi: 10.1099/jmm.0.46977-0. [DOI] [PubMed] [Google Scholar]
- Yu WL, Dan H, Lin M. InlA and InlC2 of Listeria monocytogenes serotype 4b are two internalin proteins eliciting humoral immune responses common to listerial infection of various host species. Curr Microbiol. 2008;56:505–509. doi: 10.1007/s00284-008-9101-4. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2473–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]