Abstract
Background
Pathogenic Leptospira species cause leptospirosis, a zoonotic disease of global importance. The spirochete displays active rotative mobility which may contribute to invasion and diffusion of the pathogen in hosts. FliY is a flagellar motor switch protein that controls flagellar motor direction in other microbes, but its role in Leptospira, and paricularly in pathogenicity remains unknown.
Results
A suicide plasmid for the fliY gene of Leptospira interrogans serogroup Icterohaemorrhagiae serovar Lai strain Lai that was disrupted by inserting the ampicillin resistance gene (bla) was constructed, and the inactivation of fliY gene in a mutant (fliY-) was confirmed by PCR and Western Blot analysis. The inactivation resulted in the mRNA absence of fliP and fliQ genes which are located downstream of the fliY gene in the same operon. The mutant displayed visibly weakened rotative motion in liquid medium and its migration on semisolid medium was also markedly attenuated compared to the wild-type strain. Compared to the wild-type strain, the mutant showed much lower levels of adhesion to murine macrophages and apoptosis-inducing ability, and its lethality to guinea pigs was also significantly decreased.
Conclusion
Inactivation of fliY, by the method used in this paper, clearly had polar effects on downstream genes. The phentotypes observed, including lower pathogenicity, could be a consequence of fliY inactivation, but also a consequence of the polar effects.
Background
The genus Leptospira is composed of both saprophytic and pathogenic species [1]. Pathogenic Leptospira spp., such as L. interrogans, L. borgpetersenii, L. weilii and L. kirschner, are the causative agents of leptospirosis, a serious world-wide disease in humans and animals [2,3]. The disease in humans occurs mostly after contact, often through skin wounds, with soil or water contaminated by urine of infected animals. Its severity varies from mild to rapidly fatal. Severe symptoms are characterized by visible jaundice involving hepatic injury, acute renal failure, carditis and hemorrhage, and case fatality varies from a few percent to 25% [3-6]. However, the mechanisms of disease caused by pathogenic Leptospira spp. remain largely unknown.
Both pathogenic and saprophytic leptospires express two endoflagella (periplasmic flagella). One of the endoflagella is attached at one end of the cell and is located between the protoplasmic cylinder and the outer membrane sheath [7-9]. The endoflagella, rotating within the periplasmic space, are responsible for spirochete motility. In pathogenic Leptospira species, this motility is considered to contribute to invasion into hosts and diffusion within the hosts during infection [9,10]. In previous studies, we found that pathogenic leptospires can adhere to host cells with one or two termini of the microbial bodies, while non-pathogenic leptospiral strains lacked this ability [11,12]. The adhering positions were located at the terminal knobs in which flagellar basal bodies are found [1,7]. At the bottom of the flagellar structure, there is a basal body composed of MS and C rings [13,14]. In flagellated bacteria, some proteins in the Fli family form the C ring, which functions as the flagellar rotor and contains the directional switching capability of the flagellar motor [15-18]. However, a possible role for the leptospiral endoflagella in pathogenicity has never been explored.
A complete set of flagella-associated genes were found in the genomic sequences of L. interrogans serovar Lai strain Lai and serovar Copenhageni strain Fiocruz L1-130, including four genes that encode flagellar motor switch proteins (FliG, FliM, FliN and FliY) [19,20]. In bacteria, the flagellar motor switch proteins play a critical role in control of flagellar motor direction [14,17,18]. Thus far FliY has been found in some spirochetes and a few bacteria but does not exist in most bacteria [21,22]. Particularly, FliY of Bacillus subtilis was shown to be a CheY-P-hydrolyzing protein in the chemotactic signaling cascade [22]. In addition, leptospiral FliY carries a carboxy-terminal domain of 60 amino acid residues that is homologous to a domain of YscQ in Yersinia pestis [19,20]. The YscQ protein was identified as a member of the flagellar associated type III secretion system (T3SS), with multiple functions such as controlling the directional rotation of flagella and the export of virulence factors including Yop proteins [23,24]. The C ring of Escherichia coli does not have FliY, but its FliN has a high sequence homology with FliY of L. interrogans strain Lai [19] and FliN is an essential agent for motility and virulence protein export [25]. These data suggest that FliY of pathogenic Leptospira species may have important functions in motility and virulence.
In the present study, we constructed a fliY gene knock-out (fliY-) mutant of L. interrogans serovar Lai strain Lai based on homologous recombination using a suicide plasmid. To examine the possible role of FliY in pathogenesis, the mutant and wild-type strain were compared in assays of motility in liquid medium and migration on semisolid agar, adhesion to macrophages, stimulation of apoptosis in infected host cells, and lethality to guinea pigs.
Results
Products of fliY gene amplification and rFliY expression
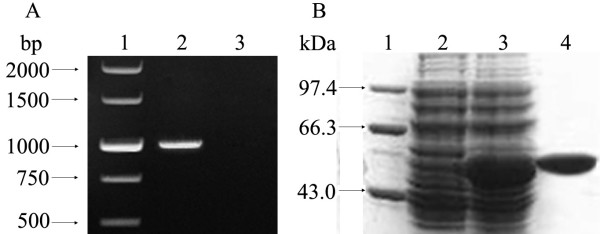
The amplification segments with expected size of the entire fliY gene (1065 bp) from L. interrogans serovar Lai strain Lai were obtained by PCR (Fig 1A). The cloned fliY gene had 100% nucleotide sequence identity with the reported sequences in GenBank (Accession No.: NC_004343, NC_005823) [10,11]. The recombinant plasmid, E. coli BL21DE3pET32a-fliY, expressed rFliY under inducement of isopropyl-β-D-thiogalactopyranoside (IPTG), and the purified rFliY by Ni-NTA affinity chromatography showed a single band on a polyacrylamide gel after electrophoresis (Fig 1B). The rabbits immunized with rFliY could produce rFliY-specific serum antibody and the immunodiffusion titer of antiserum against rFliY was 1:4.
Figure 1.
Amplification and expression of the fliY gene and purification of the rFliY protein. Panel A, showing PCR analysis. Lane 1: DNA marker (TaKaRa, China); lane 2: the amplification segment of the entire fliY gene; lane 3: blank control. Panel B, showing SDS-PAGE analysis. Lane 1: protein marker (TaKaRa); lane 2: pET32a with no insertion of the fliY gene; lane 3: the expressed recombinant protein, rFliY; lane 4: the purified rFliY protein.
Characterization of the fliY- mutant
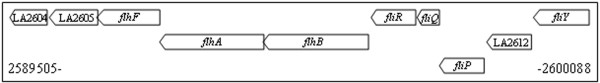
To create a fliY- mutant of L. interrogans, we cloned the fliY gene into p2NIL and inserted an ampicillin gene at the Bgl II site near the 5' end. This plasmid was then introduced into L. interrogans followed by selection for ampicillin resistance, to create a fliY bla mutant. Sequencing data indicated that the fliY gene and ampicillin resistance gene (bla) segments in suicide plasmid p2NILfliY-bla had the same orientation, and the nucleotide sequences were the same as in the original cloned fliY and bla genes. The fliY- mutant could grow in 100 μg/ml ampicillin-contained Korthof liquid medium for at least 3 months in our laboratory. The generation time of the mutant (about 10 d) was the same as that of the wild-type strain. Subsequent PCR analysis confirmed that the mutant maintained a modified fliY gene that was larger (2019 bp) than the wild-type gene (1065 bp), into which inserted the ampicillin resistance gene (954 bp) had been inserted (Fig 2A). The Western Blot analysis also revealed the absence of expression of FliY in the mutant (Fig 2B). Furthermore, the absence of mRNAs for the fliP and fliQ genes, downstream of fliY gene, indicated that the transcription of the two genes were inhibited (data not shown). In fact, ten genes (fliY, LA2612, fliP, fliQ, fliR, flhB2, flhA, flhF, LA2605 and LA2604) should be transcribed by the same operon, based on the genome structure predicted by the software, MicrobesOnline Operon Predictions (Fig 3).
Figure 2.
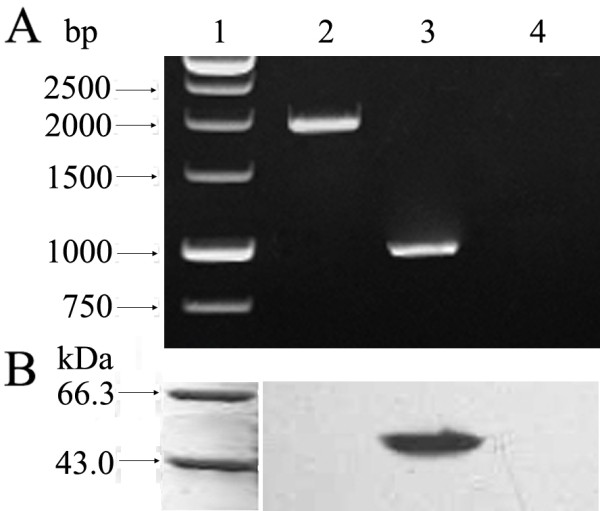
Confirmation for insertion mutantion of fliY gene in the fliY- mutant. Panel A, showing PCR analysis. Lane 1: DNA marker (TaKaRa); lane 2: the amplification segment (2019 bp) of mutated fliY gene from the fliY- mutant; lane 3: the amplification segment (1065 bp) of the fliY gene from the wild-type strain; lane 4: blank control for PCR. Panel B, showing Western Blot analysis. Lane 1: protein marker (TaKaRa); lane 2: the fliY- mutant lacking the FliY protein; lane 3: the wild-type strain expressing the FliY protein; lane 4: blank control for Western Blot assay. rFliY antiserum was used as the primary antibody.
Figure 3.
Genes present with the fliY gene within the same predicted operon. Annotation of the genes (gene name/product): fliY/flagellar motor switch protein; LA2612/flagellar protein required for flagellar formation; fliP/flagellar biosynthesis protein; fliQ and fliR/flagellar biosynthetic proteins and type III secretion apparatus proteins; flhB/flagellar protein; flhA/flagellar biosynthesis protein; flhF/flagellar GTP-binding protein; LA2605/ParA protein; and LA2604/hypothetical protein.
Persistently lower motility of the fliY- mutant
Normally, leptospires have a typical motive manner with rotation. However, all microbes of the fliY- mutant in liquid Korthof medium by dark-field microscopy only had 40% of rotative motion frequency per minute of the wild-type strain, but presented a similar shape to the wild-type strain (data not shown). On semisolid Korthof agar plates, the colonies of the fliY- mutant were noticeably smaller (2-3 mm in diameter) than that of the wild-type strain (6-8 mm in diameter) (Fig 4), consistent with attenuated motility of the mutant.
Figure 4.
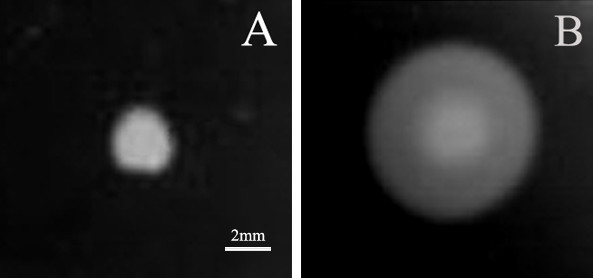
Colony sizes of the fliY-mutant and wild-type strain on semisolid Korthof agar. The colonies with different sizes formed by the fliY- mutant (A) and wild-type strain (B) on semisolid Korthof agar. The leptospires were cultured on 8% RS semisolid Korthof plate for three weeks. This experiment was repeated three times.
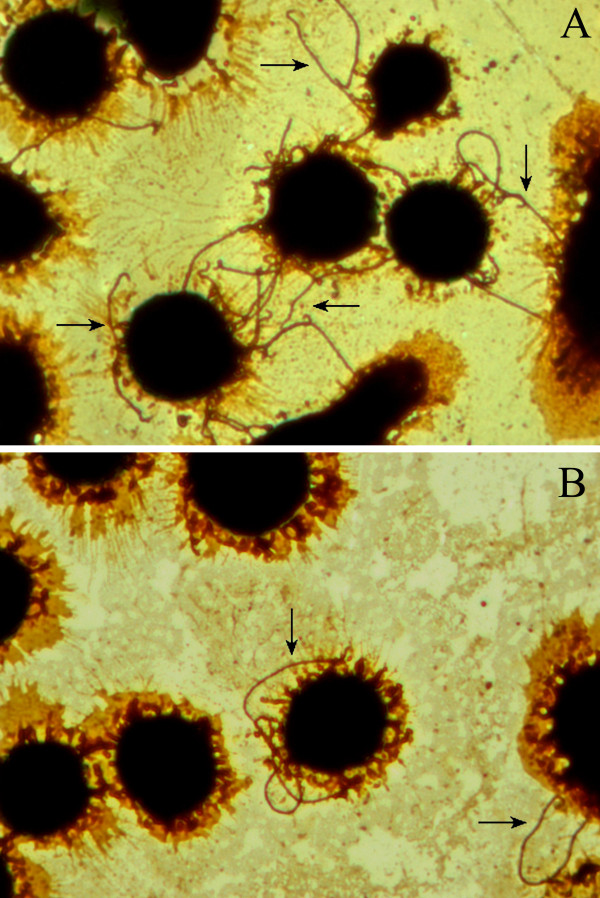
Altered adhesion of the fliY- mutant
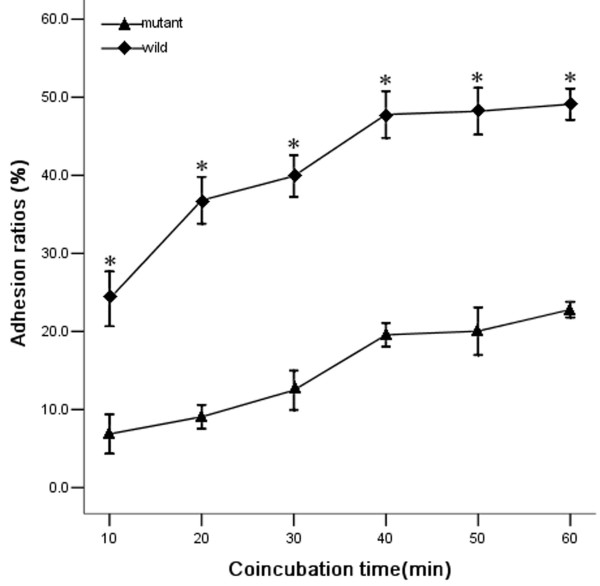
The wild-type L. interrogans strain Lai could adhere to the surface of J774A.1 cells with one or both bacterial ends (Fig 5A). The attached wild-type leptospires were visible on the cell surface after 10 min post inoculation (p.i.) and the adhesion ratios approached a plateau after 40 to 60 min p.i. (Fig 6). However, the fliY- mutant was significantly impaired in its ability to adhere to the macrophages, compared to the wild-type strain (P < 0.05) (Fig 5B and Fig. 6).
Figure 5.
Adhesion of the fliY- mutant and wild-type strain to J774A.1 cells. Adhesion of the wild-type strain (A) and fliY- mutant (B). The arrow indicates the adhering leptospires on J774A.1 cells. This experiment was repeated three times. Magnification × 400.
Figure 6.
Adhesion ratios of the fliY- mutant and wild-type strain to J774A.1 cells after different incubation times. Adhesion was quantified as described in Methods. *: P < 0.05, wild-type strain compared with the mutant.
Host-cell apoptosis induced by the wild-type and the fliY- mutant strains
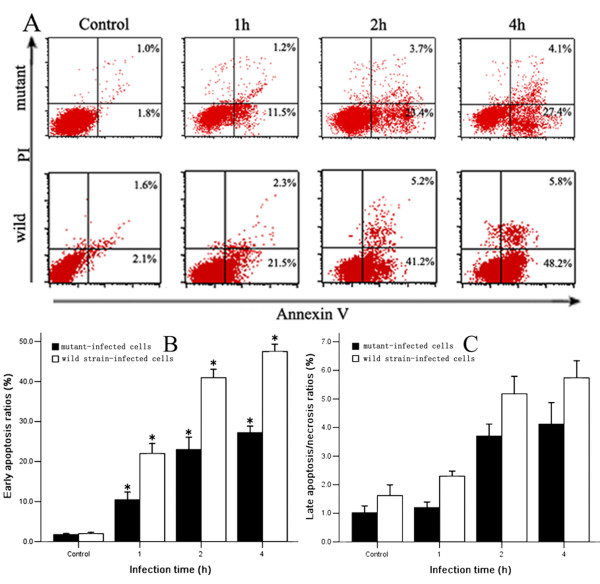
As shown in Fig 6, the wild-type L. interrogans strain Lai induced apoptosis of J774A.1 cells, and the maximal apoptotic ratio (48.2 ± 2.9%) appeared after 4 h coincubation, as detected by flow cytometry (Fig 7A). However, the ability of the fliY- mutant to cause apoptosis was markedly decreased, and the levels of apoptosis and late apoptosis/necrosis at all the different incubation times were significantly lower than those induced by the wild-type strain (P < 0.05) (Fig 7B and 7C).
Figure 7.
Apoptosis ratios of J774A.1 cells induced by the fliY- mutant and wild-type strain. Panel A: lower left quadrants indicate unstained normal cells; lower right quadrants, the early apoptotic cells binding Annexin-V; upper left quadrants, the necrotic cells binding PI; and upper right quadrants, the late apoptotic/necrotic cells binding both Annexin-V and PI. Cell death was measured by flow cytometry as described in Methods. Panel B: proportion of early apoptotic cells (annexin-V+/PI-) after infection for different times. The data are expressed as mean ± SD for three independent experiments. Panel C: proportion of late apoptotic/necrotic cells (annexin-V+/PI+) after infection for different times. The data are expressed as mean ± SD for three independent experiments. *:P < 0.05, wild-type strain compared with the mutant.
Attenuated lethality of the fliY- mutant strain in guinea pigs
The lethality to guinea pigs of the wild-type L. interrogans strain Lai was significantly larger than of the fliY- mutant during a 10d post-challenge period (Table 1). No animals infected by the fliY- mutant strain died comparing with 100% death, which were infected by wild-type strain with the same dosage. When the challenge dosage for the fliY- mutant was increased to ten times the dosage used for the wild-type strain, only 60% of the animals infected with the fliY- mutant died.
Table 1.
Lethality of the fliY- mutant and the wild-type strain in infected guinea pigs.
| Strain | Challenge dosage (×108 per animal) |
Animal (n) |
Dead/surviving (n/n) |
Death rate (%) |
|---|---|---|---|---|
| Wild-type Mutant | 6 | 10 | 10/0 | 100 |
| 6 | 10 | 0/10 | 0 | |
| 12 | 10 | 0/10 | 0 | |
| 30 | 10 | 0/10 | 0 | |
| 60 | 10 | 6/4 | 60 |
Discussion
Recent reports have shown that flagellin and other flagella-associated proteins from many bacteria participate in adhesion to host cells and colonization of hosts [26-28]. In vitro studies have suggested that the role of flagella could be to increase invasion into host cells and survival within macrophages [29,30]. However, the correlation between flagella and pathogenicity of pathogenic Leptospira spp. had not been investigated until now. L. interrogans serogroup Icterohaemorrhagiae serovar Lai strain Lai is the most prevalent pathogenic leptospiral strain, which is responsible for over 70% of human leptospirosis cases in China [31]. We therefore inactivated the fliY gene in L. interrogans strain Lai using a suicide plasmid, which is a frequently adopted strategy for determining the function of a target gene. Recently, Croda and his colleagues used plasmid pB2SK to successfully construct a suicide plasmid with spectinomycin resistance for inactivating the ligB gene of L. interrogans serovar Copenhageni strain Fiocruz L1-130 [32]. In the present study we first used another plasmid, p2NIL, with an ampicillin resistance gene (bla) to construct a fliY gene knock out (fliY-) mutant. A fliY- mutant has been constructed, but that fliY inactivation by ampicillin cassette insertion also negatively affected downstream genes; therefore, care has to be taken when interpreting the phenotypes observed for this mutant.
The inactivation of the fliY gene has shown different effects on formation of flagella in different bacteria. In Bacillus subtilis, the deletion of fliY resulted in the loss of flagella [33]. However, the flagella were still produced in the fliY-deleted strain of Bacillus cereus [34]. Although the leptospiral fliY- mutant generated in this study displayed remarkably attenuated motility compared to the wild-type strain, it maintained the typical spiral shape and propeller movement which is caused by the periplasmic endoflagella [1,7]. As mentioned previously, the major function of flagellar motor switch proteins is to control flagellar motor direction [16,19-22]. Thus, we infer that the fliY gene inactivation should not affect the formation of the endoflagella.
It is well known that adhesion to host cells is a primary and critical step for bacterial infection [35,36]. Recently, the importance of cell adhesion for pathogenic Leptospira spp. has been demonstrated [11,12,37,38]. Adhesion to host cells also acts as an essential role for pathogenicity of other spirochetes [39,40]. Mononuclear macrophages are the most important phagocytes in the human innate and acquired immnune systems. However, many pathogenic bacteria can evade host immunity by inducing apoptosis of macrophages [41-43]. Similarly, pathogenic Leptospira spp. can escape from the host immune system by promoting macrophage apoptosis [11,44-46]. In the present study, we provide evidence that the ability of the fliY- mutant to adhere to J774A.1 cells, to induce apoptosis in the cells, and to cause death in guinea pigs is much lower than for the wild-type strain. All the phentotypes observed, including lower pathogenicity, could be a consequence of fliY inactivation, or a consequence of the polar effects, or of both.
T3SS is one of protein export systems used by most Gram-negative bacteria [47]. Morphologically, as a transmembrane channel, T3SS is composed of multiple protein complexes called an injectisome, responsible for transporting virulence factors into host cells, some of which cause cell metabolic disorder and death [47-49]. However, the flagellar export apparatus can also function as a bacterial virulence protein secretion system [50]. For example, FliF of Pseudomonas aeruginosa, a flagellar associated protein component in the MS ring, is involved in adhesion by controlling secretion of bacterial adhesins [51]. Although the T3SS and flagellar export apparatus are two relatively separate systems in many pathogenic bacteria [52], the T3SS and flagellar export apparatus in Yersinia enterocolitica play a common role in secretion of bacterial phospholipases during infection [53]. Taken together, these observations suggest that inactivation of the leptospiral fliY gene (or of the downstream located fliPQ genes) may decrease the export of some unknown adhesion- and cytotoxicity-associated virulence proteins.
Conclusion
Inactivation of fliY clearly had polar effects on downstream genes. The phentotypes observed, including decreasing motility, adhesion to macrophages and host-cell apoptosis, and attenuating lethality in infected guinea pigs, could be a consequence of fliY inactivation, but also a consequence of the polar effects.
Methods
Bacterial strains and cell lines
L. interrogans serogroup Icterohaemorrhagiae serovar Lai strain Lai was offered by the National Institute for the Control of Pharmaceutical and Biological Products in Beijing, China. The leptospires were cultured in Korthof liquid medium containing 8% heat-inactivated rabbit serum (RS) at 28°C. To maintain virulence, the strain was passaged intraperitoneally in specific pathogen-free Dunkin-Hartley ICO:DH (Poc) guinea pigs (2 weeks old, each weighing about 120 g) before use, according to the description by Merien et al. and Viriyakosol et al. [44,54]. Animal protocols were approved by the Animal Ethics Review Committee of Zhejiang University.
Cell line and culture
The murine mononuclear-macrophage-like cell line (J774A.1) was obtained from the American Type Culture Collection (Rockville, MD, USA). The cells were cultured in RPMI 1640 medium (GIBCO, USA), supplemented with 10% heat-inactivated fetal calf serum (FCS) (GIBCO), 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma, USA) at 37°C in an atmosphere of 5% CO2.
PCR and sequencing
Genomic DNA of L. interrogans strain Lai was extracted using Bacterial Genomic DNA Extraction Kit (BioColor, China). Plasmid pUC19, which has an ampicillin resistant gene (bla) cassette including promotor in E. coli DH5a, was prepared by Mini-plasmid Rapid Isolation Kit (BioDev, China). Primers for amplifications of the fliY and bla genes are shown in Table 2. A commercial PCR Kit (TaKaRa, China) was used to amplify the fliY and bla genes. The products were detected on 1.5% ethidium bromide pre-stained agarose gel by electrophoresis, purified using PCR Product Purification Kit (BioColor), and ligated into plasmid pUCm-T using T-A Cloning Kit (BioColor) to form recombinant plasmids pUCm-TfliY. pUCm-Tbla sequencing was performed by Invitrogen Co. Ltd in China.
Table 2.
Primer information for amplification of the fliY and bla genes.
| Gene | Primer sequence (5'-3') | Product size |
|---|---|---|
| fliY | F: GCC GGA TCC (BamH I) ATG GGT GAA GGT TCC CTA TCA CAG | 1065 bp |
| R: GCC AAG CTT (Hind III) TCA CTT ACC CTC CGG CTT AAT CCG | ||
| bla | F: GCC AGA TCT (Bgl II) TCT AAA TAC ATT CAA ATA TGT | 954 bp |
| R: GCC AGA TCT (Bgl II) CTT GGT CTG ACA GTT ACC AAT | ||
| fliP | F: ATG AAA ATG AGA CAT AAA | 804 bp |
| R: TCA TTT ATA ACT CCT TAC | ||
| fliQ | F: ATG ACG GAA TTA GAC GTT ATG | 264 bp |
| R: CTA AAA TTT TTC GAT CAT CAA |
F: forward primer, R: reverse primer.
Expression, purification and immunization of recombinant FliY
pUCm-TfliY and expression vector pET32a (Novagen, USA) were digested with BamH I and Hind III, respectively. The recovered fliY segment was ligated into linearized pET32a using T4 DNA ligase (TaKaRa), and then transformed into E. coli BL21DE3 (Novagen) to form E. coli BL21DE3pET32a-fliY. Recombinant FliY (rFliY) was expressed under inducement of 0.5 mM IPTG for 4 h at 37°C. The expressed rFliY was extracted by Ni-NTA affinity chromatography and the purity of rFliY was determined by SDS-PAGE. New Zealand rabbits, provided by the Laboratory Animal Center of Zhejiang University, were immunized intradermally four times at an interval of once a week with the purified rFliY that was pre-mixed with Freund's adjuvant. On the 15th day after the last immunization, the rabbit serum was collected and the immunodiffusion test was used to examine the titer of antiserum.
Generation and characterization of the fliY- mutant
Plasmid p2NIL used in this study was kindly offered by Dr. Tanya Parish and Dr. Amanda C. Brown. The fliY segment from pUCm-TfliY was inserted into p2NIL at the BamH I/Hind III sites to form p2NILfliY. The plasmid has an origin of replication for E. coli (oriE), a kanamycin resistance gene (kan), and a multiple cloning site [55]. Since there is a unique Bgl II site within the fliY gene sequence (942th-947th bp at the 5' end), p2NILfliY was cut with Bgl II, dephosphorylated and ligated with ampicillin amplification segment (bla) including the promotor (10th-16th bp at 5' end) flanked by a Bgl II site to form a suicide plasmid, p2NILfliY-bla. The suicide plasmid was transformed into E. coli DH5a for amplification in Luria-Bertani (LB) medium supplemented with both 100 μg/ml ampicillin and 50 μg/ml kanamycin, and then recovered for sequencing. The p2NILfliY-bla plasmid was then denatured by alkali treatment as previously described [56,57], and electrocompetent leptospires were prepared according to Saint Girons' protocol [58]. The competent leptospiral cells were mixed with 2 μg p2NILfliY-amp DNA, and then bathed on ice for 10 min for electrotransformation. Finally, the mixture was transferred to 1 ml of 8% RS Korthof liquid medium for a 48 h incubation at 28°C. The fliY- mutant was selected on 8% RS Korthof plates containing 100 μg/ml ampicillin. Individual ampicillin-resistant colonies were inoculated in 8% RS Korthof liquid medium supplemented with 100 μg/ml ampicillin. The steps to construct the suicide plasmid and to generate fliY- mutant are summarized in Fig 8.
Figure 8.
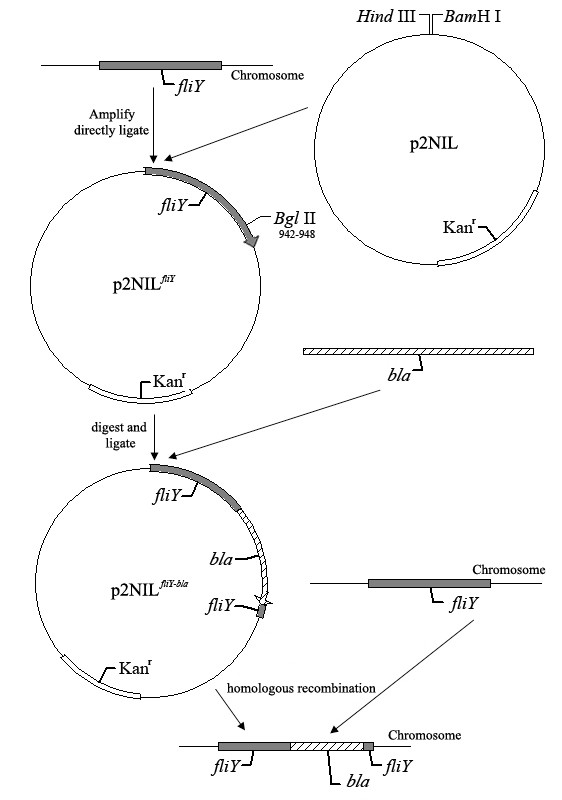
Strategy for preparing the fliY- mutant using the suicide plasmid p2NILfliY-bla.
Confirmation of the fliY gene inactivation in mutants
The fliY- mutant was cultured at 28°C in 8% RS Korthof liquid medium containing 100 μg/ml ampicillin. Genomic DNA of the mutant was extracted using Bacterial Genomic DNA Extraction Kit (BioColor), and the disrupted fliY gene in the mutant was identified by PCR and the Western Blot assay. The product of the fliY-bla gene is larger in the mutant (2019 bp) than the fliY gene in the wild-type strain (1065 bp). By using 1:2500 diluted anti-rFliY serum as the primary antibody and 1:3000 diluted HRP-labeling goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, USA) as the secondary antibody, a Western Blot assay was performed to detect the expression of FliY protein in the mutant. In the genomic sequence of L. interrogans serovar Lai strain Lai, the fliP and fliQ genes are located downstream from the fliY gene. In order to further confirm the inactivation of the fliY gene, two separate RT-PCRs were performed to detect mRNAs of the fliP and fliQ genes, with primers shown in table 2. In addition, an operon predictor tool http://www.microbesonline.org/ was used for analysis of the operon structure.
Motility assay
The motility and shapes of the fliY- mutant and wild-type strain in 8% RS Korthof liquid medium were observed under dark-field microscope after incubation at 28°C for 10 d (the primary generation), 50 d (the 5th generation) and 100 d (the 10th generation). The colony sizes of the mutant and wild-type strain on 8% RS semisolid Korthof plate (0.25% agar) that had been incubated at 28°C for three weeks were measured for three times as described above.
Fontana silver staining
J774A.1 cells (5 × 104 cells/ml) were seeded on coverslips in 12-well tissue culture plates (Corning, USA) and pre-incubated for 24 h at 37°C in an atmosphere of 5% CO2. The freshly cultured leptospires of the fliY- mutant and wild-type strain were harvested by centrifugation (12,000 × g, 15min, 15°C) and washed twice with autoclaved PBS. The pellets were suspended in pre-warmed antibiotics-free 10% FCS RPM1640 to a final concentration of 108 leptospires/ml by dark-field microscopy with a Petroff-Hausser counting chamber (Fisher Scientifics, PA). The cell monolayers were washed three times with autoclaved PBS and then infected with each of the suspensions at an MOI of 100 (100 leptospires per cell) for 10, 20, 30, 40, 50 and 60 min, respectively. After infection, the coverslips were washed three times with PBS to remove non-adherent leptospires, fixed in 5% formaldehyde, stained with silver nitrate, and observed under a light microscope [59]. The adhesion ratio was defined as the number of adhering leptospires per 100 infected host-cells × 100% [11].
Assessment of cell death by flow cytometry
Apoptosis was measured by flow cytometry using annexin-V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) labeling as previously published [11,60]. The J774A.1 cell monolayers were infected with either the fliY- mutant or wild-type strain with an MOI of 100 at 37°C for 1, 2, or 4 h [46]. After infection, the cells were washed three times with PBS, collected with a cell scratcher, and centrifuged at 1,000 × g for 5 min. The pellets were washed three times with PBS, resuspended in annexin-V binding buffer with FITC-conjugated annexin-V, and incubated for 15 min at room temperature in the dark, following the manufacturer's instructions (Caltag Laboratories, USA). After PI was added, the cell suspension was immediately analyzed by FACSCalibur flow cytometry and CellQuest Pro software (Beckman Coulter, USA). Cells in the early apoptotic phase bind annexin-V but exclude PI, and those in the late apoptotic/necrotic phase stain with both annexin-V and PI, while necrotic cells stain with PI alone [60].
Animals and challenge infections
The Dunkin-Hartley guinea pigs (150 ± 5 g, 3 weeks old) used in this study were provided by the Laboratory Animal Center of Zhejiang University. The animals were challenged intraperitoneally with different dosages of either wild-type L. interrogans serovar Lai strain Lai or the fliY- mutant, and then observed for 10 d [1]. The animal experiments were approved by the Animal Ethics Review Committee of Zhejiang University.
Statistical analysis
Data from a minimum of three experiments were averaged and presented as mean ± SD (standard deviation). One-way analysis of variance (ANOVA) followed by the Dunnett's multiple comparisons test were used to determine significant differences. Statistical significance was defined as P value ≤ 0.05.
Authors' contributions
SL carried out the molecular genetic studies, immunoassays and drafted the manuscript. AS cultured the leptospires and participated in immunoassays. DMO participated in study design and revised the manuscript. SW and JZ carried out analysis and interpretation of data. JY conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript, and agreed to having it published.
Contributor Information
Sumei Liao, Email: lsmplum@yahoo.com.cn.
Aihua Sun, Email: sunah@zjmc.net.cn.
David M Ojcius, Email: dojcius@ucmerced.edu.
Senlin Wu, Email: keyanchu1999@126.com.
Jinfang Zhao, Email: 3021851002@zju.edu.cn.
Jie Yan, Email: med_bp@zju.edu.cn.
Acknowledgements
This work was supported by a Grant (30370072) from the National Natural Science Foundation of China and a grant (2007XZA02) from the Natural Scientific National Foundation of Zhejiang Medical College of China. We are grateful to Dr. Tanya Parish and Dr. Amanda C. Brown (Center for Infectious Disease, Institute for Cell and Molecular Science, Queen Mary's School of Medicine and Dentistry, UK) for having graciously provided the plasmid p2NIL used in this study.
References
- Faine S, Adher B, Bloin C, Perolat P. Leptospira and leptospirosis. 2. Australia: MedSci; 1999. [Google Scholar]
- Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–771. doi: 10.1016/S1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- McBride AJ, Athanazio DA, Reis MG, Ko AI. Leptospirosis. Curr Opin Infect Dis. 2005;18:376–386. doi: 10.1097/01.qco.0000178824.05715.2c. [DOI] [PubMed] [Google Scholar]
- Lomar AV, Diament D, Torres JR. Leptospirosis in Latin America. Infect Dis Clin N Am. 2000;14:23–39. doi: 10.1016/S0891-5520(05)70216-6. vii-viii. [DOI] [PubMed] [Google Scholar]
- Levett PN. Leptospirosis. Clin Microbio Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslin FX. Global aspects of emerging and potential zoonoses: a WHO perspective. Emerg Infect Dis. 1997;3:223–228. doi: 10.3201/eid0302.970220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks GF, Butel JS, Morse SA. Medical Microbiology. 22. U.S.A.: McGraw-Hill; 2001. pp. 291–293. [Google Scholar]
- Wolgemuth CW, Charon NW, Goldstein SF, Goldstein RE. The flagellar cytoskeleton of the spirochetes. J Mol Microbiol Biotechnol. 2006;11:221–227. doi: 10.1159/000094056. [DOI] [PubMed] [Google Scholar]
- Li C, Motaleb A, Sal M, Goldstein SF, Charon NW. Spirochete periplasmic flagella and motility. Mol Microbiol Biotechnol. 2000;2:345–354. [PubMed] [Google Scholar]
- Charon NW, Goldstein SF. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu Rev Genet. 2002;36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- Li LW, Ojcius DM, Yan J. Comparison of invasion of fibroblasts and macrophages by high- and low-virulence Leptospira strains: colonization of the host-cell nucleus and induction of necrosis by the virulent strain. Arch Microbiol. 2007;188:591–598. doi: 10.1007/s00203-007-0280-3. [DOI] [PubMed] [Google Scholar]
- Dong HY, Hu Y, Xue F, Sun D, Ojcius DM, Mao YF, Yan J. Characterization of the ompL1 gene of pathogenic Leptospira species in China and cross-immunogenicity of the OmpL1 protein. BMC Microbiol. 2008;8:223–235. doi: 10.1186/1471-2180-8-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM. How bacteria assemble flagella. Ann Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- McCarter LL. Regulation of flagella. Curr Opin Microbiol. 2006;9:180–186. doi: 10.1016/j.mib.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Francis NR, Irikura VM, Yamaguchi S, DeRosier DJ, Macnab RM. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci USA. 1992;89:6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao RPN, Jaffe H, Reese TS, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- Thomas DR, Morgan DG, DeRosier DJ. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc Natl Acad Sci USA. 1999;96:10134–10139. doi: 10.1073/pnas.96.18.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Blair DF. The bacterial flagellar motor: structure and function of a complex molecular machine. Inter Rev Cytol. 2004;233:93–134. doi: 10.1016/S0074-7696(04)33003-2. full_text. [DOI] [PubMed] [Google Scholar]
- Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, Zhang YX, Xiong H, Lu G, Lu LF, Jiang HQ, Jia J, Tu YF, Jiang JX, Gu WY, Zhang YQ, Cai Z, Sheng HH, Yin HF, Zhang Y, Zhu GF, Wan M, Huang HL, Qian Z, Wang SY, Ma W, Yao ZJ, Shen Y, Qiang BQ, Xia QC, Guo XK, Danchin A, Saint Girons I, Somerville RL, Wen YM, Shi MH, Chen Z, Xu JG, Zhao GP. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, Haake DA, Verjovski-Almeida S, Hartskeerl RA, Marques MV, Oliveira MC, Menck CF, Leite LC, Carrer H, Coutinho LL, Degrave WM, Dellagostin OA, El-Dorry H, Ferro ES, Ferro MI, Furlan LR, Gamberini M, Giglioti EA, Góes-Neto A, Goldman GH, Goldman MH, Harakava R, Jerônimo SM, Junqueira-de-Azevedo IL, Kimura ET, Kuramae EE, Lemos EG, Lemos MV, Marino CL, Nunes LR, de Oliveira RC, Pereira GG, Reis MS, Schriefer A, Siqueira WJ, Sommer P, Tsai SM, Simpson AJ, Ferro JA, Camargo LE, Kitajima JP, Setubal JC, van Sluys MA. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol. 2004;186:2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Muff TJ, Ordal GW. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J Biol Chem. 2004;279:21787–21792. doi: 10.1074/jbc.M311497200. [DOI] [PubMed] [Google Scholar]
- Straley SC, Skrzypek E, Plano GV, Bliska JB. Yops of Yersinia spp. pathogenic for humans. Infect Immun. 1993;61:3105–3110. doi: 10.1128/iai.61.8.3105-3110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields KA, Plano GV, Straley SC. A low-Ca2+ response (LCR) secretion (ysc) locus lies within the lcrB region of the LCR plasmid in Yersinia pestis. J Bacteriol. 1994;176:569–579. doi: 10.1128/jb.176.3.569-579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Billings S, Wang X, Sharp L, Blair DF. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J Bacteriol. 1995;177:3496–3503. doi: 10.1128/jb.177.12.3496-3503.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect Immun. 1998;66:1000–1007. doi: 10.1128/iai.66.3.1000-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottemann KM, Lowenthal AC. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun. 2002;70:1984–1990. doi: 10.1128/IAI.70.4.1984-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Currie CG, Mackie S, Tree J, McAteer S, McKendrick I, McNeilly TN, Roe A, La Ragione RM, Woodward MJ, Gally DL, Smith DG. An investigation of the expression and adhesin function of H7 flagella in the interaction of Escherichia coli O157:H7 with bovine intestinal epithelium. Cell Microbiol. 2009;11:121–137. doi: 10.1111/j.1462-5822.2008.01244.x. [DOI] [PubMed] [Google Scholar]
- Weinstein DL, Carsiotis M, Lissner CR, O'Brien AD. Flagella help Salmonella typhimurium survive within murine macrophages. Infect Immun. 1984;46:819–825. doi: 10.1128/iai.46.3.819-825.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Ezaki T, Miura H, Matsui K, Yabuuchi E. Intact motility as a Salmonella typhi invasion-related factor. Infect Immun. 1988;56:1967–1973. doi: 10.1128/iai.56.8.1967-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B. Advances in research on leptospira and human leptospirosis in China. Chin Med Sci J. 1992;7:239–243. [PubMed] [Google Scholar]
- Croda J, Figueira CP, Wunder EA Jr, Santos CS, Reis MG, Ko AI, Picardeau M. Targeted mutagenesis in pathogenic Leptospira species: disruption of the LigB gene does not affect virulence in animal models of leptospirosis. Infect Immun. 2008;76:5826–5833. doi: 10.1128/IAI.00989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff DS, Ordal GW. Identification and characterization of FliY, a novel component of the Bacillus subtilis flagellar switch complex. Mol Microbiol. 1992;6:2715–2723. doi: 10.1111/j.1365-2958.1992.tb01448.x. [DOI] [PubMed] [Google Scholar]
- Senesi S, Celandroni F, Salvetti S, Beecher DJ, Wong AC, Ghelardi E. Swarming motility in Bacillus cereus and characterization of a fliY mutant impaired in swarm cell differentiation. Microbiology. 2002;148:1785–1794. doi: 10.1099/00221287-148-6-1785. [DOI] [PubMed] [Google Scholar]
- Nougayre JP, Fernandes PJ, Donnenberg MS. Adhesion of enteropathogenic Escherichia coli to host cells. Cell Microbiol. 2003;5:359–372. doi: 10.1046/j.1462-5822.2003.00281.x. [DOI] [PubMed] [Google Scholar]
- Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Cinco M, Domenis R, Perticarari S, Presani G, Marangoni A, Blasi E. Interaction of leptospires with murine microglial cells. New Microbiol. 2006;29:193–199. [PubMed] [Google Scholar]
- Choy HA, Kelley MM, Chen TL, Moller AK, Matsunaga J, Haake DA. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect Immun. 2007;75:2441–2450. doi: 10.1128/IAI.01635-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn J, Fischer JR, Leong JM. Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol Microbiol. 2005;57:1182–1195. doi: 10.1111/j.1365-2958.2005.04759.x. [DOI] [PubMed] [Google Scholar]
- Edwards AM, Jenkinson HF, Woodward MJ, Dymock D. Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect Immun. 2005;73:2891–2898. doi: 10.1128/IAI.73.5.2891-2898.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack DM, Raupach B, Hromockyj AE, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills SD, Boland A, Sory MP, Smissen P van der, Kerbourch C, Finlay BB, Cornelis GR. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc Natl Acad Sci USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albee L, Shi B, Perlman H. Aspartic protease and caspase 3/7 activation are central for macrophage apoptosis following infection with Escherichia coli. J Leukoc Biol. 2007;81:229–237. doi: 10.1189/jlb.0506358. [DOI] [PubMed] [Google Scholar]
- Merien F, Baranton G, Perolat P. Invasion of Vero cells and induction of apoptosis in macrophages by pathogenic Leptospira interrogans are correlated with virulence. Infect Immun. 1997;65:729–738. doi: 10.1128/iai.65.2.729-738.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Zheng W, Li LW, Mao Y, Yan J. Pathogenesis of leptospirosis: interaction of Leptospira interrogans with in vitro cultured mammalian cells. Med Microbiolo Immunol. 2007;196:233–239. doi: 10.1007/s00430-007-0047-0. [DOI] [PubMed] [Google Scholar]
- Jin DD, Ojcius DM, Sun D, Dong HY, Luo YH, Mao YF, Yan J. Leptospira interrogans induces apoptosis in macrophages via caspase-8- and caspase-3-dependent pathways. Infect Immun. 2009;77:799–809. doi: 10.1128/IAI.00914-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Bio Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber K, Frey J, Burnens AP, Kuhnert P. Detection of type III secretion genes as a general indicator of bacterial virulence. Mole Cell Probes. 2003;17:25–32. doi: 10.1016/S0890-8508(02)00108-1. [DOI] [PubMed] [Google Scholar]
- Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán JE, Aizawa SI. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- Young GM, Schmiel DH, Miller VL. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. Cloning and characterization of Pseudomonas aeruginosa fliF, necessary for flagellar assembly and bacterial adherence to mucin. Infect Immun. 1996;64:2130–2136. doi: 10.1128/iai.64.6.2130-2136.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecsas JJ, Strauss EJ. Molecular mechanisms of bacterial virulence: type III secretion and pathogenicity islands. Emerg Infect Dis. 1996;2(4):270–288. doi: 10.3201/eid0204.960403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren SM, Young GM. An amino-terminal secretion signal is required for YplA export by the Ysa, Ysc, and flagellar type III secretion systems of Yersinia enterocolitica biovar 1B. J Bacteriol. 2005;187:1430–40. doi: 10.1128/JB.187.17.6075-6083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viriyakosol S, Matthias MA, Swancutt MA, Kirkland TN, Vinetz JM. Toll-like receptor 4 protects against lethal Leptospira interrogans serovar Icterohaemorrhagiae infection and contributes to in vivo control of leptospiral burden. Infec Immun. 2006;74:887–895. doi: 10.1128/IAI.74.2.887-895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish T, Stoker NG. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- Hinds J, Mahenthiralingam E, Kempsell KE, Duncan K, Stokes RW, Parish T, Stoker NG. Enhanced gene replacement in mycobacteria. Microbiology. 1999;145:519–527. doi: 10.1099/13500872-145-3-519. [DOI] [PubMed] [Google Scholar]
- Picardeau M, Brenot A, Saint Girons I. First evidence for gene replacement in Leptospira spp. inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol Microbiol. 2001;40:189–199. doi: 10.1046/j.1365-2958.2001.02374.x. [DOI] [PubMed] [Google Scholar]
- Saint Girons I, Bourhy P, Ottone C, Picardeau M, Yelton D, Hendrix RW, Glaser P, Charon N. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa: construction of an L. biflexa-Escherichia coli shuttle vector. J Bacteriol. 2000;182:5700–5705. doi: 10.1128/JB.182.20.5700-5705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanan R, Rajendran P, Thyagarajan SP, Smythe LD, Norris MA, Symonds ML, Dohnt MF. Leptospira autumnalis isolated from a human case from Avadi, India, and the serovar's predominance in local rat and bandicoot populations. Ann Trop Med Parasitol. 2000;94:503–506. doi: 10.1080/00034983.2000.11813569. [DOI] [PubMed] [Google Scholar]
- Perfettini JL, Gissot M, Souque P, Ojcius DM. Modulation of apoptosis during infection with Chlamydia. Methods Enzymol. 2002;358:334–344. doi: 10.1016/s0076-6879(02)58099-x. full_text. [DOI] [PubMed] [Google Scholar]