Abstract
Using DNA microarrays to generate transcriptional profiles of the aging process is a powerful tool for identifying biomarkers of aging. In C. elegans, a number of whole-genome profiling studies identified genes that change expression levels with age. High-throughput RNAi screens in worms determined a number of genes that modulate lifespan when silenced. Transcriptional profiling of the fly head identified a molecular pathway, the ‘response to light’ gene set, that increases expression with age and could be directly related to the tendency for a reduction in light levels to extend fly lifespan. In mouse, comparing the gene expression profiles of several drugs to the gene expression profile of caloric restriction identified metformin as a drug whose action could potentially mimic caloric restriction in vivo. Finally, genes in the mitochondrial electron transport chain group decrease expression with age in the human, mouse, fly, and worm.
Introduction
Aging is a process in which there is a progressive loss of function in multiple tissues resulting in an increasing probability of death. There are a large variety of genetic and environmental ways in which lifespan can be extended in model organisms. These include mutation of genes in the insulin-like signaling pathway [1], caloric restriction [2], overexpression of sirtuins such as Sir2 [3], or even by inducing mild heat shock in worms [4]. Since lifespan can be extended in many ways, it seems that aging is a complex process and can be influenced independently by a relatively large number of age-associated pathways [5]. Rather than focusing on one or even several pathways in studying the molecular processes of aging, an increasingly powerful approach has been to screen the entire genome for age-associated genes and genetic pathways. DNA microarrays can be used to screen the genomes of several organisms for genes and pathways that either increase or decrease expression levels with age. In the last several years, DNA microarray studies of aging gene expression have been vigorously pursued in C. elegans, D. melanogaster, mice and humans.
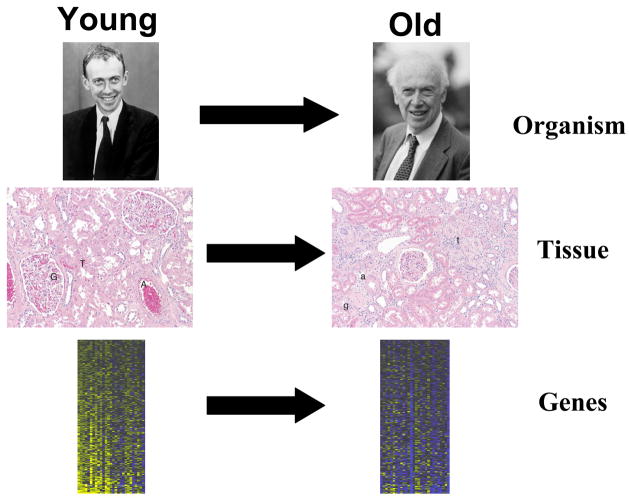
A major aim of these studies is the identification of novel biomarkers of aging. For several decades, the aging field has searched for measurable biological parameters that can accurately predict the physiological decline of an aging organism [6]. Specific aging biomarkers are apparent at the level of the whole organism, and also at the level of tissues. Given the complex nature of aging, it is hoped that transcriptional profiling studies will be able to identify molecular biomarkers of aging that are more sensitive and more specific than cellular or tissue biomarkers (Figure 1). In this review, we will examine the results of studies using transcriptional profiling and other systems biology techniques to study the effects of aging in worms, flies, mice and humans. Here, we mainly restrict our focus to studies completed within the last 2 years. For a more complete review of systems biology of aging, see [7].
Figure 1. Biomarkers of aging.
Examples of biomarkers of aging in young (left) and old (right) individuals. Top: At the level of the whole organism, biomarkers such as wrinkled skin and whitened hair are apparent in old individuals. Center: Examples of histology in human kidney cortex. A, G, and T refer to arterioles, glomeruli and tubules respectively. Each of these histological features displays very different morphology in old vs. young individuals. Bottom: Heat map of genes that decrease expression level in elderly human kidneys. These genes are molecular biomarkers of aging.
Transcriptional profiles of aging in C. elegans
The roundworm C. elegans is a model system particularly suited to studies of aging, exhibiting a short lifespan of approximately two weeks and offering the availability of a robust genetic toolset. Previously, whole-genome transcriptional profiles of worm populations of varying ages were generated using DNA microarrays [8,9]. Lund et al. used cDNA microarrays to profile worms at six time points after reaching adulthood and discovered a transcriptional profile consisting of 164 genes that either increased or decreased expression with age. Transcriptional profiles have also been generated comparing mutant worms with altered longevity (such as sir-2.1, daf-2, daf-16 and age-1) to wild-type worms in order to identify downstream genes involved in lifespan [10•–13]. For instance, Viswanathan et al. tested the effects of resveratrol, a compound known to extend lifespan in worms, in several mutant backgrounds [10•]. They found that resveratrol failed to extend lifespan in sir-2.1 mutant worms, suggesting that resveratrol acts through the sir-2.1 pathway. Viswanathan et al. also generated a transcriptional profile of resveratrol-treated worms, and observed that genes involved in the endoplasmic reticulum (ER) stress response displayed increased gene expression after resveratrol treatment. Silencing of abu-11, an ER stress response gene, by RNAi abolished lifespan extension by reveratrol treatment.
A concept of interest is to use biomarkers of aging to predict the lifespan of individuals within a population with an identical age and sharing the same environment. Levels of lipofuscin, a pigment thought to correlate with physiological age, were found to predict the remaining lifespan of worms [14]. Golden et al. performed DNA microarray analyses on individual worms, with the aim of discovering genes that vary stochastically amongst individuals within an age-matched population [15,16].
RNAi libraries may be used to systematically silence essentially every gene within the C. elegans genome. RNAi-silenced worms can then be examined for longevity phenotypes to uncover genes and pathways involved in aging. A number of genome-wide RNAi screens for longevity have been recently completed in C. elegans [17•–20]. These studies found that a large number of genes expressing components of the electron transport chain promoted longevity when silenced, and these genes typically produced the largest increases to lifespan [17•–20]. This supports the importance of the oxidative phosphorylation system in the determination of lifespan. Genetic epistasis experiments suggest that some of the genes could potentially act in the daf-2 insulin receptor pathway whereas other genes might act in the caloric restriction pathway [17•]. Although these studies use epistasis to assign genes to pathways, further work remains to be accomplished at the molecular and biochemical levels in order to definitively show the functional relationships between these longevity genes and mechanisms with which they interact. Notably, the results of these studies did not show a high degree of overlap, suggesting that future RNAi screens will continue to find additional longevity genes and that the total number of genes that can affect longevity is quite high.
Gene expression studies of aging in D. melanogaster
The fruit fly D. melanogaster is relatively short-lived and aging in the fly has been studied using gene expression profiling. A benefit to working in the fly is that it can easily be dissected into component body parts and organs, allowing for the study of aging in specific tissues. Pletcher et al. used DNA microarrays to measure transcriptional changes in young and old flies, under normal and calorically-restricted dietary conditions, and discovered 885 age-regulated and 827 calorically-restricted transcripts [21]. The rate of age-related change in these genes was ameliorated under calorically-restricted conditions, suggesting that these genes are physiological biomarkers of aging in the fly. Landis et al. examined the transcriptional profiles of flies that were raised in the presence of 100% oxygen and compared it to a transcriptional profile of aging in flies, discovering that several pathways were similarly regulated between the two conditions [22]. This result supports the oxidative damage theory of aging in the fly.
Girardot et al. recently compared the transcriptional profiles of 3-day old flies to 40-day old flies at the level of the whole organism, but also on dissected heads and thoraxes, using Affymetrix DNA chips [23•]. Another group also profiled the effects of aging on the fly head using cDNA microarrays [24]. Both groups agreed substantively on which genes are age-regulated in the fly head and found that energy pathways and particularly proton transporters significantly decrease expression with age. Interestingly, Girardot et al. found that this expression decrease with age occurred less in the thorax than in the head, arguing that the dynamics of age-regulation vary from tissue to tissue in the fly. Both groups found that the genes involved in the response to light decreased with age in the fly head. It has been known for some time that flies grown in darkness have increased median lifespans [25,26] when compared to flies grown in dim or bright light. The effect of reduced light on lifespan may be related to reduced expression of the genes involved in the response to light in old age.
DNA microarray studies of aging in mice
The common mouse M. musculus is a popular model for aging, despite its relatively long lifespan when compared to simple organisms such as worms or flies. The first transcriptional profiles of aging were generated in a limited number of mouse tissues such as muscle [27] and brain [28]. Recently, transcriptional profiles of such diverse organs as adipose tissue [29], heart [30], liver [31••], and macrophages [32] have been studied. A recent study by Edwards et al. used an empirical Bayes approach to identify 712 transcripts that are age-regulated in mouse muscle that were enriched for genes active in the p53 pro-apoptotic pathway in addition to genes encoding mitochondrial proteins involved in energy generation [33]. The p53-related genes increase expression level as age progresses, suggesting that age-associated DNA damage may lead to a p53 response in the muscle of older mice. The p53-related genes also exhibited a reduced expression level in mice undergoing caloric restriction, suggesting that they are physiological biomarkers of aging in the mouse.
There is widespread interest in discovering biomarkers of aging for their ability to act as surrogate endpoints in screening for pharmaceuticals that extend longevity. Dhahbi et al. used the molecular profile of caloric restriction generated by DNA microarrays in mouse liver as such a surrogate endpoint. Drugs that mimic the transcriptional response to caloric restriction may also act to delay or ameliorate the effects of aging [31••]. Dhahbi et al. compared the transcriptional profile of caloric restriction to profiles of the livers from mice given anti-diabetic medications metformin, glipizide, and rosiglitazone. They found that the effects of metformin on liver expression showed a highly significant overlap with the molecular profile for caloric restriction.
A novel experiment by Bahar et al. assayed the robustness of the transcriptional network with age in mouse cardiomyocytes [34••]. Bahar et al. used RT-PCR to measure the variance in expression of a number of genes in cardiomyocytes sampled from young and old mice, and found that cardiomyocytes from old mice displayed a larger variance in gene expression than those of cardiomyocytes from younger mice. This suggests a mechanism of aging in which the entire transcriptome degrades with age, leading to noisier gene expression and a general loss of transcriptional control. Small increases in levels of gene dysregulation in old mice could be catastrophic to the entire genetic network when amplified across all genes expressed in a cell, and result in cellular dysfunction and senescence.
Expression profiles of aging in humans
Despite its great relevance, the study of aging in humans is difficult due to the wide variability in genetic backgrounds and environmental influences in different individuals, in addition to the scarcity of human tissue samples for study. Nonetheless, transcriptional profiles of many human tissues, including brain [35,36], blood [37], eye [38], kidney [39,40], muscle [41–43••], and skin [44] have been generated. Several studies have used large sample sizes to overcome the genetic and environmental variability inherent in human populations, permitting these studies to achieve a higher level of statistical significance [35,39,43]. In recent years, there has been a trend towards comparing the transcriptional profiles of several different tissues to each other, aiming to determine genes and genetic pathways that show common patterns of age-regulation in different tissues. In a study of aging in the kidney, Rodwell et al. [39] compared the transcriptional profile of the medulla to that of the cortex, and found that two sections of kidney composed of very different cell types shared similar patterns of age-regulated gene expression. Fraser et al. [36] compared areas of the frontal cortex with the cerebellum and failed to find any significant overlap in age-regulation. Finally, in a comparison of the molecular profiles of aging muscle, kidney and brain, Zahn et al. discovered a common signature of aging in these three tissues consisting of six genetic pathways, including the mitochondrial electron transport chain, which decreases expression with age [43••]. These results suggest that while certain tissues may age very similarly to one another, such as the kidney cortex and medulla, for other tissues, particularly tissues with widely divergent function, there exists only small numbers of common aging pathways.
To what degree do different species display similar patterns of age-regulation? To date, only a few studies have compared transcriptional profiles of aging to detect the genes and pathways that are publicly age-regulated between multiple species and those that are privately age-regulated within a single species. Recently, Zahn et al. have compared transcriptional profiles of aging in worms, flies, mice and humans to search for genetic pathways that show similar age-regulation in all four species. They found that one genetic pathway, the electron transport chain, that decreases expression with age in all organisms investigated [43••]. The electron transport chain may prove to be a particularly useful biomarker for aging studies because it shows an overall two-fold decrease in expression over the course of life, regardless of the average lifespan of the organism.
Conclusion
A large number of transcriptional profiling studies of aging have been completed and the gene expression data have been made publicly available. These molecular profiles explore the effects of aging in a multitude of experimental conditions, among many tissues and many relevant species. In nearly all studies, only those results generated by that study are analyzed, and results from other studies of gene expression profiles of aging are not examined. In the future, it will become important to integrate the results from each of these studies in order to determine whether age-related effects are reproducible. Data integration could be used to classify tissues into different aging pathways according to similarities in their expression profiles. Data integration could also show which genes and pathways are age-regulated in a tissues-specific fashion or organism wide. Finally, data integration between species will continue to classify age-related changes as either private to one species or public across different species. Results from meta-analysis of existing data from several publications will help uncover significant, novel insights about the aging process, as well as identify key biomarkers of physiological aging. Beyond biomarkers, identifying the genes that specify and control, rather than merely correlate with lifespan will broaden our understanding of what aging is and how aging occurs in various organisms. To this end, the identification of promoter motifs in age-regulated genes as well as the transcription factors that bind them will aid in elucidating the molecular mechanisms of aging. In the future, as systems biology techniques progress, it may become possible to study aging at a multitude of levels within the cell, from transcriptional changes to changes in protein quality or quantity, to changes in metabolic flux through genetic pathways. Collating this information will provide a global fingerprint of aging, enabling one to use systems level analysis to dissect apart complex changes involved in aging.
Acknowledgments
We would like to thank the National Institutes of Health and the Ellison Medical Foundation for funding support. Jacob Zahn is supported under a National Science Foundation fellowship and the Stanford Genome Training Grant. We are grateful to Stuart Kim’s laboratory for continuing discussion on issues referenced in this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 2.Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Olsen A, Vantipalli MC, Lithgow GJ. Lifespan extension of Caenorhabditis elegans following repeated mild hormetic heat treatments. Biogerontology. 2006;7:221–230. doi: 10.1007/s10522-006-9018-x. [DOI] [PubMed] [Google Scholar]
- 5.Kirkwood TB, Austad SN. Why do we age? Nature. 2000;408:233–238. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- 6.Butler RN, Sprott R, Warner H, Bland J, Feuers R, Forster M, Fillit H, Harman SM, Hewitt M, Hyman M, et al. Biomarkers of aging: from primitive organisms to humans. J Gerontol A Biol Sci Med Sci. 2004;59:B560–567. doi: 10.1093/gerona/59.6.b560. [DOI] [PubMed] [Google Scholar]
- 7.Kim SK. Genome-wide Views of Aging Gene Networks. In: Guarente L, Partridge L, editors. Molecular Biology of Aging. Cold Spring Harbor Laboratory Press; 2007. In press. [Google Scholar]
- 8.McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 9.Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE. Transcriptional profile of aging in C. elegans Curr Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 10•.Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 11.McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 12.McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- 13.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 14.Gerstbrein B, Stamatas G, Kollias N, Driscoll M. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans. Aging Cell. 2005;4:127–137. doi: 10.1111/j.1474-9726.2005.00153.x. [DOI] [PubMed] [Google Scholar]
- 15.Golden TR, Hubbard A, Melov S. Microarray analysis of variation in individual aging C. elegans: approaches and challenges. Exp Gerontol. 2006;41:1040–1045. doi: 10.1016/j.exger.2006.06.034. [DOI] [PubMed] [Google Scholar]
- 16.Golden TR, Melov S. Microarray analysis of gene expression with age in individual nematodes. Aging Cell. 2004;3:111–124. doi: 10.1111/j.1474-9728.2004.00095.x. [DOI] [PubMed] [Google Scholar]
- 17•.Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 20.Curran SP, Ruvkun G. Lifespan Regulation by Evolutionarily Conserved Genes Essential for Viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol. 2002;12:712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- 22.Landis GN, Abdueva D, Skvortsov D, Yang J, Rabin BE, Carrick J, Tavare S, Tower J. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101:7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23•.Girardot F, Lasbleiz C, Monnier V, Tricoire H. Specific age-related signatures in Drosophila body parts transcriptome. BMC Genomics. 2006;7:69. doi: 10.1186/1471-2164-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SN, Rhee JH, Song YH, Park DY, Hwang M, Lee SL, Kim JE, Gim BS, Yoon JH, Kim YJ, et al. Age-dependent changes of gene expression in the Drosophila head. Neurobiol Aging. 2005;26:1083–1091. doi: 10.1016/j.neurobiolaging.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 25.Allemand R, Cohet Y, David J. Increase in the longevity of adult Drosophila melanogaster kept in permanent darkness. Exp Gerontol. 1973;8:279–283. doi: 10.1016/0531-5565(73)90040-5. [DOI] [PubMed] [Google Scholar]
- 26.Massie HR, Whitney SJ. Preliminary evidence for photochemical ageing in Drosophila. Mech Ageing Dev. 1991;58:37–48. doi: 10.1016/0047-6374(91)90118-j. [DOI] [PubMed] [Google Scholar]
- 27.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 28.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 29.Higami Y, Barger JL, Page GP, Allison DB, Smith SR, Prolla TA, Weindruch R. Energy restriction lowers the expression of genes linked to inflammation, the cytoskeleton, the extracellular matrix, and angiogenesis in mouse adipose tissue. J Nutr. 2006;136:343–352. doi: 10.1093/jn/136.2.343. [DOI] [PubMed] [Google Scholar]
- 30.Dhahbi JM, Tsuchiya T, Kim HJ, Mote PL, Spindler SR. Gene expression and physiologic responses of the heart to the initiation and withdrawal of caloric restriction. J Gerontol A Biol Sci Med Sci. 2006;61:218–231. doi: 10.1093/gerona/61.3.218. [DOI] [PubMed] [Google Scholar]
- 31••.Dhahbi JM, Mote PL, Fahy GM, Spindler SR. Identification of potential caloric restriction mimetics by microarray profiling. Physiol Genomics. 2005;23:343–350. doi: 10.1152/physiolgenomics.00069.2005. [DOI] [PubMed] [Google Scholar]
- 32.Chelvarajan RL, Liu Y, Popa D, Getchell ML, Getchell TV, Stromberg AJ, Bondada S. Molecular basis of age-associated cytokine dysregulation in LPS-stimulated macrophages. J Leukoc Biol. 2006;79:1314–1327. doi: 10.1189/jlb.0106024. [DOI] [PubMed] [Google Scholar]
- 33.Edwards MG, Anderson RM, Yuan M, Kendziorski CM, Weindruch R, Prolla TA. Gene expression profiling of aging reveals activation of a p53-mediated transcriptional program. BMC Genomics. 2007;8:80. doi: 10.1186/1471-2164-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 35.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 36.Fraser HB, Khaitovich P, Plotkin JB, Paabo S, Eisen MB. Aging and gene expression in the primate brain. PLoS Biol. 2005;3:e274. doi: 10.1371/journal.pbio.0030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan Q, Christensen K, Christiansen L, Frederiksen H, Bathum L, Dahlgaard J, Kruse TA. Genetic dissection of gene expression observed in whole blood samples of elderly Danish twins. Hum Genet. 2005;117:267–274. doi: 10.1007/s00439-005-1308-x. [DOI] [PubMed] [Google Scholar]
- 38.Segev F, Mor O, Segev A, Belkin M, Assia EI. Downregulation of gene expression in the ageing lens: a possible contributory factor in senile cataract. Eye. 2005;19:80–85. doi: 10.1038/sj.eye.6701423. [DOI] [PubMed] [Google Scholar]
- 39.Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, Xiao W, Mindrinos M, Crane E, Segal E, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melk A, Mansfield ES, Hsieh SC, Hernandez-Boussard T, Grimm P, Rayner DC, Halloran PF, Sarwal MM. Transcriptional analysis of the molecular basis of human kidney aging using cDNA microarray profiling. Kidney Int. 2005;68:2667–2679. doi: 10.1111/j.1523-1755.2005.00738.x. [DOI] [PubMed] [Google Scholar]
- 41.Welle S, Brooks AI, Delehanty JM, Needler N, Bhatt K, Shah B, Thornton CA. Skeletal muscle gene expression profiles in 20–29 year old and 65–71 year old women. Exp Gerontol. 2004;39:369–377. doi: 10.1016/j.exger.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 42.Giresi PG, Stevenson EJ, Theilhaber J, Koncarevic A, Parkington J, Fielding RA, Kandarian SC. Identification of a molecular signature of sarcopenia. Physiol Genomics. 2005;21:253–263. doi: 10.1152/physiolgenomics.00249.2004. [DOI] [PubMed] [Google Scholar]
- 43••.Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lener T, Moll PR, Rinnerthaler M, Bauer J, Aberger F, Richter K. Expression profiling of aging in the human skin. Exp Gerontol. 2006;41:387–397. doi: 10.1016/j.exger.2006.01.012. [DOI] [PubMed] [Google Scholar]