Summary
There has been a recent explosion in the wealth of genomic data available to C. elegans researchers, as efforts to characterize gene expression and its regulators at a molecular level have borne significant fruit. Detailed measurement of gene expression at a variety of developmental stages, and in numerous individual tissues, has dramatically increased our understanding of cell-type-specific gene expression networks. Characterization of the targets of transcription factors, chromatin-binding proteins, and miRNAs has provided genome-wide insights into the mechanisms governing gene expression. Development of new techniques have allowed this characterization to begin to shift from whole-organism studies to tissue-, and even single-cell-level profiling, creating a first glimpse into gene regulatory circuits at the single-cell level in a living organism. Integration of these datasets has yielded novel insights into evolution, gene expression regulation, and the link between sequence and phenotype.
Introduction
Being the first multicellular animal with a full genome sequence [1], the nematode worm Caenorhabditis elegans is an ideal model system to study how a single genome can encode the blueprint to generate a fully differentiated organism with distinct cell- and tissue-types. C. elegans has been at the forefront of systems-level analysis of biological circuits, ranging from the full mapping of the full wiring diagram of all neurons [2], to the discovery of microRNA regulation of gene expression [3], to defining gene networks based on co-expression [4, 5], and to assaying the phenotype of knocking down more than 16,000 individual genes [6].
Recent technological advances in microarray and sequencing technologies have enabled high-throughput profiling of gene expression and direct identification of regulatory interactions. These high-throughput methods can show data at the level of whole organisms, at the level of individual organs, at the level of specific cell-types, and at the level of individual cells, with each step showing a finer-grained view of gene expression networks. At the individual cell level, work in C. elegans is aided by the remarkable property of an invariant cell lineage [7]. This invariance means that every nucleus in the worm can be uniquely identified at all stages of development, and once the identity of a specific nucleus is known the full developmental history of cell division leading up to that nucleus as well as its full future lineage trajectory are known. As such, C. elegans provides an ideal model system to study how alterations of single-cell gene expression or functionality can propagate throughout the organism. This review will discuss key insights that have emerged from efforts to characterize gene expression, with a focus on identification of regulatory mechanisms and networks that underlie these expression profiles. In addition, we will discuss recently developed techniques to measure single-cell gene expression in a medium-throughput manner and how this single-cell approach can yield novel insights into the basic properties underlying gene regulatory networks.
Gene expression
New tiling array techniques as well as high-throughput sequencing technologies have expanded our ability to understand the C. elegans genome. The development of C. elegans tiling microarrays has allowed array-based measurement of transcription at ~25bp-resolution, enabling similar unbiased views of transcription at the genome-wide level. In parallel, the development of high-throughput sequencing technologies has made it possible to sequence millions of short transcript reads from a cDNA library (RNA-seq), allowing the identification and quantification of any sequence that can be mapped to the C. elegans genome. Recent studies have used these two techniques to obtain a high-coverage map of mRNA transcripts at various stages of C. elegans embryonic and larval development, in the two C. elegans sexes, and in worms carrying mutations for critical steps in small RNA processing and nonsense-mediated-decay. The high-quality of this technology has enabled identification and characterization of alternative splicing, alternative promoter, and alternative 3’UTR usage [8–10].
Similar to poly-adenylated protein-coding transcripts, the exploration of the landscape of non-coding and small RNA transcripts has increased dramatically. Although dozens of expressed microRNAs have previously been identified by a combination of computational predictions and small RNA cloning techniques [11, 12], the full catalog of small RNAs was not obtained until the development of high-throughput sequencing technologies. Sequencing of small RNAs led to the discovery of a previously unseen class of small RNAs (21U-RNAs, later confirmed to be piRNAs) [13], and has also led to a full accounting of stage- and sex-specificity of microRNA expression [14]. Tiling array experiments have enabled the discovery of large non-coding RNAs, which had previously been difficult to find genome-wide [15, 16]. The most recent effort by Spencer, et al. [16] suggests that ~10% of the C. elegans genome encodes novel RNAs that were not detected by RNA-seq of poly-adenylated RNAs or small RNAs, suggesting the presence of a large fraction of previously unknown non-coding RNAs. Using this expression data in combination with conservation & secondary structure, Lu, et al. [17] predicted and characterized thousands more novel non-coding RNA transcripts, many of which show distinct stage-specific expression patterns.
In addition to analysis of gene expression from whole-worms, it is often useful to examine expression in individual tissues (i.e. muscle, skin and nervous system). Medium-to-high throughput analysis of C. elegans tissues have been performed using two approaches: 1. large-scale analysis of fluorescent reporters, and 2. transcriptome profiling of individual tissues. Due to their transparent nature, gene expression patterns can be observed simply by fusing a promoter (or other regulatory region) of interest to a fluorescent reporter, generating a transgenic strain expressing this fusion, and visualizing either manually using common microscopy techniques or automatically using profiling algorithms. The combined efforts of these approaches have obtained tissue-level descriptions of expression patterns for ~3000 genes [18–20], including more than 350 TFs in a targeted approach [18]. Although this approach requires a substantial time investment in the generation and visualization of each individual strain, it has the advantage of creating a permanent resource of strains for the field to further explore. In addition, these reporters can be used to identify subtle or unexpected expression patterns that would be missed by transcriptome profiling focusing on a specific isolated tissue.
The second approach has been to perform transcriptome profiling of specific tissues. A variety of methods have been used to isolate RNA from individual tissues:
Genetic tissue knockouts and over-expressions: certain genetic mutations can lead to either loss or over-proliferation of a specific tissue-type. Profiling of such mutant worms by DNA microarray or RNA-seq has enabled identification of germ-line-, sperm-, and oocyte-enriched transcripts [16, 21] and pharynx-enriched transcripts [22]. Although this method is easiest technically, requiring only a mutation or over-expression of interest, it is possible that some of the observed expression changes could be indirect effects of the mutation that propagate through other tissues.
Hand-dissection of individual tissues: certain tissues of the worm, including the gonad [23] and the intestine [24], can be isolated by dissection and expressed RNAs can be identified using DNA microarrays or RNA-seq. Dissection provides the most high-quality source of a specific tissue, but it remains technically challenging to cleanly isolate most of the cell-types in the worm.
FACS-sorting: A GFP marker can be used to label a specific cell-type. To purify the specific cell-type, GFP-positive embryonic cells are isolated by FACS (Fluorescence-activated cell sorting) using dissociated cells from embryos. Many embryonic tissues have been profiled using this approach, including: embryonic neurons [16, 25], touch receptor neurons [26], motor neurons [27], AWB olfactory and AFD thermosensory neurons [28], various neuronal subtypes [16], embryonic muscle [16, 29], germ-line-precursor cells [16], intestine [16, 30], and coelomyocytes [16]. Two limitations to this approach are that only embryonic tissues can be reliably dissociated and sorted, and that the cells are temporarily cultured ex vivo before RNA is isolated. However, sorting allows extremely clean purification of GFP-expressing cells, resulting in an accurate snapshot of that GFP-expressing tissue.
mRNA-tagging: To identify mRNAs expressed in a specific tissues, a tissue specific promoter is used to express an epitope-tagged protein that binds the poly-A tail of mRNAs (PolyA binding protein; PAB-1) in a tissue of interest. mRNAs expressed in the tissue are bound by the epitope-tagged protein, and can be identified by immunoprecipitating PAB-1:mRNA complexes. This approach was used to characterize genes expressed in the muscle [16, 31], neurons [16, 25], various neuronal subtypes [16, 32], hypodermis [16], excretory cells [16], coelomyocytes [16], CEP sheath cells [16], and intestine [16, 33] in various stages of C. elegans development. This approach is the most adaptable, making it possible to identify tissue-enriched transcripts in any tissue- or cell-type of interest at any stage. However, the immunoprecipitation step introduces a key limitation to this approach, as it introduces a significant amount of variability that requires numerous replicates and quality control measures. Also, as the immunoprecipitation enriches but does not quantitatively pull down PAB-1:mRNA complexes, , this technique does not measure gene expression levels per se but rather enrichment of gene expression in a given tissue.
These efforts to define tissue-specific of gene expression have contributed to a number of insights into the mechanisms of gene expression regulation. For example, using mRNA-tagging Roy, et al. [31] noticed that muscle-enriched transcripts were not randomly distributed along the chromosome; rather, they are often found in clusters of 2–5 genes. They then extended this analysis to other datasets of co-expressed genes, finding that 13 of the 15 largest co-expressed gene-sets showed a significant propensity to cluster along the chromosome. Chromosomal clustering was also observed for intestine-specific transcripts and housekeeping genes in an independent analysis, suggesting a strong role for genome organization in coordinated regulation of gene expression [33].
High-throughput gene expression studies have contributed to the identification of master regulators of tissue development in C. elegans. Identification of intestine-specific transcripts provided key insights leading to the identification of ELT-2 as the main regulator of intestine-specific gene expression. Pauli, et al. [33] used mRNA tagging to identify 1938 intestine-enriched mRNAs, McGhee et al. [24] performed expression analysis from hand-dissected intestines and identified 80 intestine-enriched transcripts in young adult worms, and McGhee et al. [30] used FACS-sorted embryonic intestine cells to identify 82 embryonic intestine-specific transcripts. These genes were enriched for those with roles in digestion, bacterial lysis, degradation of macromolecules, stress response pathways, and expression of yolk proteins. Promoter motif analyses identified a significantly-enriched TGATAA motif that was shown to direct intestine-specific expression, and further experiments confirmed ELT-2 as the major functional GATA transcription factor in intestinal development as well as adult intestinal function [24, 30, 33].
Similarly, the use of DNA microarrays to identify genes that are differentially expressed in worms lacking or over-producing pharyngeal cells led to the characterization of PHA-4 as a key regulator of pharynx development [22]. Motif analysis of pharynx-specific transcripts led to the canonical PHA-4 binding site, which was shown to be essential for pharyngeal expression patterns. The relative strength of the binding site was shown to correlate with pharyngeal expression dynamics, with stronger sites activated in embryos but lower-affinity sites activated later in development, suggesting that PHA-4 affinity is a key regulatory mechanism for regulation of gene expression in pharyngeal development. Further work identified additional secondary cis-regulatory motifs for specification of pharyngeal expression, and determined that the onset of expression of pharynx-specific genes could be predicted with greater than 85% accuracy using this regulatory code [34].
However, technical differences between experiments make it difficult to integrate these results in a rigorous way in order to understand gene regulatory networks across the entire organism. To address this issue, as well as to explore additional cell-types that had previously been uncharacterized, Spencer et al. [16] used tiling microarrays to profile 13 embryonic tissues by FACS-sorting and 12 tissues by mRNA tagging (in addition to whole-organism controls). Using this larger dataset, thousands of genes with tissue-specific expression profiles could be obtained and separated into a small subset of expression clusters. These clusters were then used to identify tissue-specific regulatory motifs, showing the effectiveness of an unbiased method to identify transcription factors that regulate expression in specific tissues [16]. These expression datasets can be combined with transcription factor binding data to screen the genome for novel transcriptional regulators acting in specific tissues ([35] & EL Van Nostrand unpublished).
The C. elegans lineage was first characterized and annotated in 1977, enabling the potential to characterize gene expression not only at the level of tissues but at the level of individual cells within each tissue. Multiple groups have begun to develop automated methods to enable high-throughput characterization of gene expression at the single-cell level [36, 37]. Using a combination of computational and manual techniques, single-cell gene expression measurements of about 100 genes are now available as a snapshot for L1 stage larvae [38], and a smaller number as time-courses for early development [39].
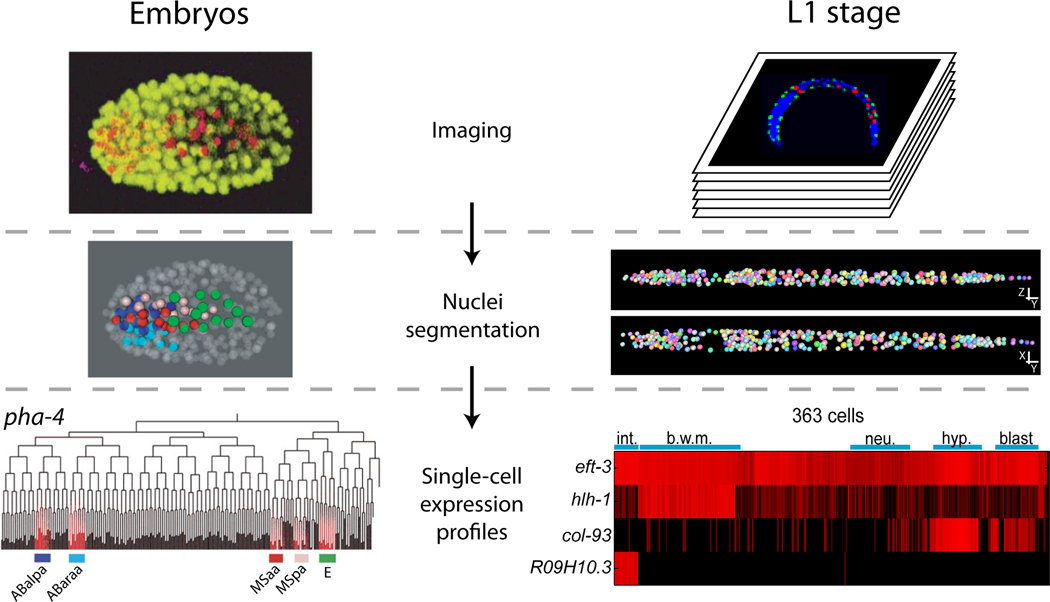
To measure single-cell gene expression during embryogenesis, Bao et al. [37] developed computational methods to analyze movies of embryos expressing two fluorescently-tagged proteins: a GFP-tagged histone protein to mark nuclei and an mCherry reporter expressed from a promoter of interest. Time-lapse confocal microscopy is used to follow expression from the initial one-cell stage through the 350-cell stage in late embryogenesis, and software (StarryNite/AceTree) is used to automatically recognize and trace nuclei across time (Figure 1) [37, 40]. Initial efforts focused on profiling expression for four well-studied developmental transcription factors (pha-4, cnd-1, hlh-1, and end-3) in gene knockout as well as wild-type worms [39].
Figure 1.
Single-cell measurement of gene expression in C. elegans. To quantify gene expression in individual cells, a promoter or regulatory region of interest is fused to a fluorescent reporter. (Left) In embryos, three-dimensional movies of fluorescence signal are tracked over time from the initial one-cell stage up until the 350 cell stage in the GFP (nuclear marker) and Cherry (desired reporter) channels (top). These images are then processed using StarryNight and AceTree [37] to segment and uniquely identify nuclei (middle). Once identified, the fluorescent reporter of interest is quantified within identified nuclei to obtain a profile of expression in each cell throughout the developmental lineage (bottom) [39]. (Right) In L1 stage larvae, three-dimensional image stacks are generated for synchronized fixed animals (top) in three channels: DAPI (nuclear stain), GFP (reference marker), and Cherry (reporter of interest). DAPI signal is used to segment nuclei, and segmented nuclei are uniquely identified using positional information in combination with a GFP reference marker (middle) [36]. Cherry signal within each nuclei is then quantified, generating a matrix of gene expression across 363 individual cells for each assayed reporter (bottom) [38].
In a parallel approach, Long et al. [36] generated automated cell lineage profiling software for L1 stage larvae. A digital atlas of nuclei positions in L1 worms was first manually annotated from confocal 3D image stacks, and software was then created to automatically identify and annotate 363 individual nuclei, including nearly all major tissue-types (intestine, muscle, neuronal, hypodermal, epithelial, and blast cells) (Figure 1) [36]. In an initial analysis, Liu et al. [38] profiled promoter fusions representing expression patterns of 93 genes at single-cell resolution. The expression level of these 93 genes in each cell was used to generate a molecular profile for that individual cell, providing the raw expression data for a number of systems-level analyses. First, they compared the roles of cell fate (i.e. muscle vs. neuron), organ identity (i.e. pharynx), cell lineage (i.e. AB.a vs. AB.b) and cell position (i.e. anterior vs posterior) on gene expression. As expected, all four were found to play roles in influencing gene expression. Unexpectedly, however, when cells were clustered purely based on expression profiles the strongest influence was organ identity. Cells of neural, epithelial, and muscle fate in the pharynx clustered more closely with other pharyngeal cells than with cells of similar fate in other organs.
Liu et al. [38] also identified an interesting and unexpected example of developmental convergence, in which two classes of nuclei that express identical sets of effector genes had different upstream regulatory circuits guiding their expression. The nuclei are generated from different parts of the cell lineage, and become united in the same hypodermal syncytium by cell fusion. Previous work had shown that both types of nuclei express similar skin-specific proteins (including collagens and adherens junction-related proteins). However, careful examination of the two nuclei showed that they expressed different upstream transcription factors but the same set of downstream skin effector molecules. Knockdown of one transcription factor (by RNAi) could re-program expression of one nucleus but not an adjacent nucleus in the same skin syncytium, indicating that two independent regulatory networks were used to direct expression of genes associated with one fate. This unique result could only be experimentally found in an animal such as C. elegans, in which the cell lineage is known.
Major challenges for the C. elegans field in the coming years will be to develop methods of manipulating gene targets at the single-cell level in order to gain insight about gene regulation with single-cell resolution.
Regulation of gene expression
While mRNA expression provides an important readout for gene expression, methods to characterize proteins that bind to DNA and regulate expression provide another critical piece to the gene regulatory puzzle. At the most basic level, accessibility of DNA for transcription is regulated by packaging of DNA into nucleosomes [41]. Thus, a genome-wide map of nucleosome positioning is a key step towards understanding why certain regions are accessible for transcription. These positions were identified in mixed-stage worms using limited mononuclease digestion followed by high-throughput sequencing, and further analysis identified motifs associated with nucleosome positioning [42, 43]. These studies show that DNA accessibility is controlled not just by association of extrinsic factors (histones and transcription factors), but also by intrinsic sequences contained within the DNA sequence itself [43].
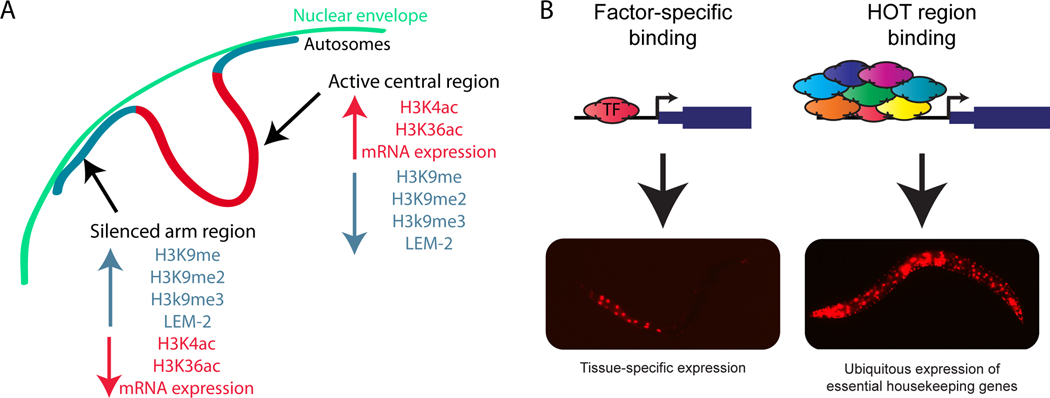
Histone tails undergo a large number of post-translational modifications such as methylation and acetylation that alter their structure and function [44]. Numerous groups have used antibodies that recognize specific histone modifications in chromatin immunoprecipitation (ChIP) followed by high-throughput sequencing (ChIP-seq) or DNA microarrays (ChIP-chip). These studies identify the DNA associated with each type of chromatin modification and thus generate a view of each specific mark throughout the genome. Some marks (such as H3K4 di- and tri-methylation and H3K27 acetylation) are associated with active transcription while others (e.g. H3K27 tri-methylation) are associated with repressed chromatin [45–49]. In the largest effort to date, the modENCODE consortium performed ChIP-chip of 19 histone modifications, one histone variant, and 8 chromatin-associated proteins in embryos and L3 larvae [49]. Using these datasets, they were able to characterize which marks are associated with activating or repressive activity, and identify differences between promoter regions and gene bodies. Unexpectedly, the 19 histone modifications and 2 histone variants revealed relatively sharp boundaries between the arms and central regions of the autosomes, with a striking correlation between these broad regions and recombination rate (Figure 3A). The chromosome center regions are gene-rich, enriched for active marks, and have less recombination, whereas the arms are gene-poor, enriched for repressive marks, and have high recombination. These arm regions also had a significant enrichment for nuclear membrane protein LEM-2, suggesting that association with the nuclear envelope may contribute to silencing of the chromosome arms [49].
Figure 3.
Unexpected findings from large-scale regulator profiling. (A) Profiling of chromatin modifications identified distinct broad chromosomal domains. Chromosome arms were enriched for silencing marks, including H3K9 methylation, and depleted for activating marks (H3K4 & H3K36 acetylation). Arms were also enriched for association with nuclear envelope protein LEM-2, suggesting localization of repressed chromosome arms with the nuclear periphery. In contrast, central regions were enriched for activating marks and depleted for repressive marks, and had enriched density of expressed genes. (B) ChIP-seq of 23 TFs led to the identification of HOT (Highly Occupied Target) regions bound by 15 or more TFs. While factor-specific targets are associated with tissue-specific expression patterns that match known roles for specific TFs, HOT region targets are characterized by high, ubiquitous expression of essential housekeeping genes. HOT regions may represent a novel mechanism to maintain expression of necessary genes by ensuring robust association with multiple TFs instead of through individual TFs.
Although chromatin state often correlates with transcription, regulation of transcription is largely controlled through nearby binding of sequence-specific transcription factors (TFs) to specific target sites in the genome. Yeast one-hybrid assays provide one approach to determine where TFs are bound to the DNA in a high-throughput manner. In this method, the DNA binding domain of a TF of interest is screened against a library of gene promoters in yeast cells to determine which show strong binding by the DNA binding domain. Hundreds of direct interactions between a TF and promoters were identified using this technique [50, 51], allowing a first glimpse of direct TF regulatory networks in the intestine [51]. However, this technique has the inherent problem of assaying interactions in yeast, which is a non-native environment. DNA targets bound by worm TFs identified using the yeast one-hybrid assay must then be validated in C. elegans.
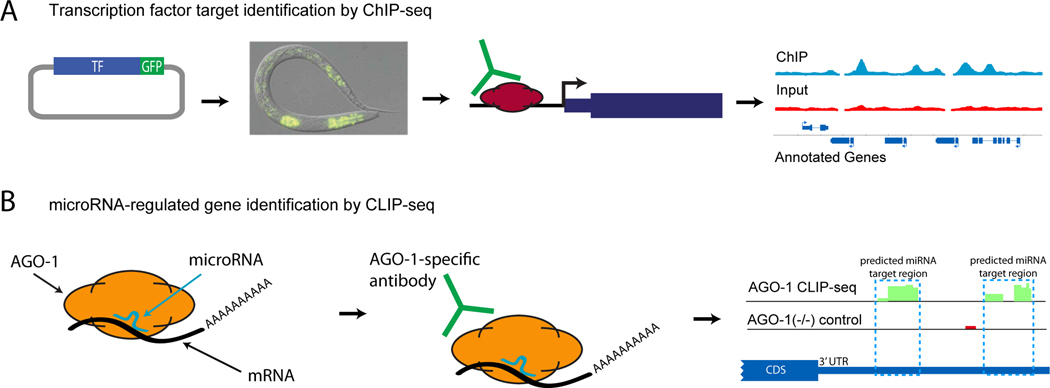
ChIP and a related DamID approach are methods to directly identify DNA bound by TFs in vivo. In ChIP, an antibody that specifically recognizes either the TF of interest, or an epitope tag that has been added to the desired TF, is used to immunoprecipitate the TF along with bound DNA (Figure 2A). This bound DNA can then be isolated and assayed by micro-array (ChIP-chip) or by high-throughput sequencing (ChIP-seq) [52] DamID involves fusing a TF of interest to a bacterial DNA adenine methyltransferase. When this fusion protein binds to DNA, adenines in GATC tetramers within ~2kb are methylated to create DpnI restriction sites. Restriction digest followed by amplification allows these methylated regions to be specifically amplified and similarly assayed by high-throughput methods [53]. Individual efforts in C. elegans have identified the targets of DAF-16 [54, 55], NFI [56], PHA-4 [52], HLH-1 [57], and DAF-12 [58], yielding insights into the roles of these TFs in core cellular processes ranging from regulation of aging and stress to development of the pharynx and muscle. Unexpected insights were gained from these genome-wide approaches: for example, Hochbaum, et al. [58] used insights from DAF-12 target identification to characterize DAF-12 as a critical regulator of robustness in regulation of seam cell divisions. daf-12 null worms show higher variability in seam cell number than wild-type in normal conditions, and the variability is greater under stress conditions. This result suggests that one role for daf-12 activity in wild-type worms is to buffer against stochastic errors in cell division under environmental stress.
Figure 2.
Identification of direct regulatory targets in C. elegans. (A) Transcription factor (TF) targets are identified by ChIP-seq. (1) A fosmid containing a TF of interest fused to a GFP marker is generated. (2) Low-copy genomic integration of this transgene is achieved by biolistic bombardment, and a stage-synchronized population of worms expressing the TF:GFP fusion is grown. (3) The TF is cross-linked to associated DNA, and the GFP:TF:DNA complex is immunoprecipitated using an anti-GFP antibody. (4) TF-bound DNA is isolated and subjected to high-throughput sequencing to identify TF binding regions genome-wide. (B) microRNA-regulated genes are identified by CLIP-seq of AGO-1. (1) microRNA association with mRNA target genes is mediated by the RISC complex, including Argonaute protein AGO-1. In live worms, AGO-1:microRNA:mRNA complexes are stabilized by crosslinking. (2) Anti-AGO-1 antibody is used to immunoprecipitate complexes. (3) Complex-bound mRNA regions are isolated and subjected to high-throughput sequencing to identify sites of microRNA/AGO-1 association genome-wide. An AGO-1 deletion strain is used as a negative control.
A more global picture of TF regulatory networks emerged from ChIP-seq analysis of 22 TFs in a variety of developmental stages by the C. elegans modENCODE consortium [59]. This dataset provides the first large-scale look at direct TF regulation in C. elegans, as well as an initial view of overlaps between individual TF binding networks. Individual TF ChIP-seq datasets also yielded interesting insights: for example, GEI-11 was found to specifically associate with small RNA promoters, suggesting that it may play a previously unknown role in regulation of transcription of small RNAs. This effort has continued since publication, and 109 different ChIP-seq datasets for 60 TFs are now publicly available (http://intermine.modencode.org/), allowing researchers to easily determine whether a gene or gene set of interest shows direct binding by specific TFs.
This limited dataset also enabled the discovery of a novel mechanism for regulation of housekeeping genes (Figure 3B) [35]. HOT (Highly Occupied Target) regions are genomic loci bound significantly by 15 or more of the 22 TFs assayed by ChIP-seq. These regions appear to signify a previously uncharacterized mechanism of gene regulation in which hundreds of TFs are associated with the same DNA region in order to direct expression of house-keeping genes. These regions are located proximal to highly expressed, ubiquitous, essential transcripts, and are characterized by a unique mix of chromatin features associated with highly active regions.
The discovery of HOT regions has led to the further delineation of TF target sites as “factor-specific” or “non-factor-specific”, a distinction with significant relevance regarding whether a TF target site marks a gene with a specific function related to that of the TF or whether it is a general target for many TFs with no specific connection to any particular TF. Knowledge about whether TF targets are specific or general is essential when constructing integrated regulatory networks, and dramatically improves inference of TF biological function from ChIP-seq targets ([35] and EL Van Nostrand unpublished). This distinction between factor-specific and non-factor-specific targets may also help to explain the common finding that genes that are direct targets identified by ChIP often do not show changes in expression when the TF is knocked down. These unchanged targets may simply represent that expression of that gene is regulated by a large number of TFs as opposed to one or a few specific regulators ([35]; EL Van Nostrand unpublished).
Combining information from ChIP seq studies with expression data from TF knockdown or overexpression mutants shows which genes are not only bound, but are also regulated by the transcription factor of interest. High-throughput sequencing of cDNAs derived from ELT-2 knockout worms identified genes that showed decreased expression, and identified a significant enrichment for the ELT-2 TGATAA binding site in their promoters [30]. These genes had strong overlap with intestine-enriched datasets, suggesting a role for ELT-2 in intestine-specific gene expression. Identification of targets that have increased expression in HLH-1 over-expression mutants revealed enrichment for muscle-expressed genes [60]. By focusing on TFs, unc-120 and hnd-1 were identified as additional key regulators of muscle development, and further experiments determined that the combination of these three TFs is both necessary and sufficient to drive bodywall muscle differentiation in C. elegans development.
The mapping of regulatory targets for factors that operate at the post-transcriptional level has only just begun, but will provide an additional layer of information for understanding regulatory circuits in C. elegans. Initial efforts to computationally identify miRNA targets involved identifying mRNAs that contained a sequence matching the reverse complement of the miRNA, and then used additional sequence properties including cross-species conservation and local nucleotide frequencies to enrich for true regulatory targets [61]. Predicted targets allowed inference of biological roles for certain miRNAs whose targets were enriched for specific functional categories, but individual predicted interactions still require experimental validation. A direct approach to identify miRNA-regulated mRNAs is to immunoprecipitate ALG-1 (the C. elegans Argonaute protein that is directed to target mRNA by guiding miRNAs) and then identify the associated miRNAs by high-throughput sequencing (CLIP-seq) (Figure 2B) [62]. This approach has helped to identify the subtle sequence properties that enable miRNA regulation, which remains difficult to predict de novo. In addition, a significant enrichment was observed for miRNA regulation of mRNAs encoding proteins involved in the miRNA pathway (including ALG-1 itself), suggesting that auto-regulation of the miRNA pathway may be critical to robustness of miRNA regulation.
An additional layer of regulation at the RNA level is provided by regulation of alternatively splicing events. Barberan-Soler et al. [63] profiled 352 alternatively spliced exons in 12 splicing factor mutants to obtain a general view of splicing regulation in staged C. elegans embryos. Clustering of relative exon inclusion or exclusion rates in these mutants yielded examples of splicing events regulated only by single factors and splicing that is co-regulated by multiple splicing factors, and identified numerous examples of stage-specific splicing regulation. Further work to identify direct targets of RNA binding proteins using CLIP-seq should elucidate the network for regulation of splicing.
Data integration / network building
The ultimate goal of systems biology in C. elegans is to combine detailed datasets to obtain a molecular understanding of the circuits that underlie development of complex tissues and systems. This goal has been a focus since early genomic efforts in C. elegans, when a co-expression matrix generated from large datasets of microarray studies was used to identify coordinately regulated gene sets de novo from unrelated studies [4, 5]. More recent efforts to generate genetic networks have shown that integration of phenotypic as well as interaction information yields a significant benefit [64], supporting the potential of the use of C. elegans as a model for network building. With direct regulator-target interactions obtained from ChIP-seq (and related technologies), these networks can be re-interpreted to delineate between direct and indirect effects on gene expression.
The integration of genomics data in C. elegans has also provided interesting insights into basic properties of the evolution of regulatory networks. Characterization of global transcription patterns in natural isolates as well as mutation-accumulation lines provided a unique perspective on evolution. Denver, et al. [65] showed that selective forces played a strong role in shaping the mutational landscape to avoid dramatic changes in gene expression networks. They also identified a significant increase in mutation rates on chromosome arms over central regions in natural isolated, but not mutation-accumulation, lines. Although this can be explained by selection effects due to higher gene density in core regions, the distinct chromatin states and nuclear envelope associations described by Liu et al. [49] may give hints as to the mechanisms driving mutational effects on regulatory networks. In addition, the in-depth knowledge of certain developmental circuits has begun to enable detailed studies of the subtle molecular features of gene regulatory networks. Signal transduction pathways were over-represented for stronger selection whereas carbohydrate, amino acid and lipid metabolism were shifted towards weaker selection, showing that different functional categories are subjected to significantly different selective pressures [65].
In the intestine, Raj et al. [66] used a previously characterized intestinal transcription factor network to study the correlation between genetic mutations and intestinal phenotypes. Focusing on mutations in this network with variable phenotypes, they showed that this variability could be traced back to the variability of specific nodes within the network, and was in fact a feature of the redundancy present in the network. To assay expression within individual embryos, mRNA molecules were fluorescently labeled and individually counted to obtain a molecule-level measure of gene expression. They determined that expression of key intestine developmental factor elt-2 in early embryonic development (in skn-1 mutants) was dependent not on absolute expression levels of upstream regulator end-1, but on whether expression exceeded a specific threshold value. Once end-1 expression reaches this level, elt-2 activity appears to trigger a feed-forward loop to achieve final high levels of stable elt-2 expression. In wild-type worms, even though end-1 and end-3 are genetically redundant in elt-2 activation, variability of end-3 had significant effects on the timing of elt-2 activation [66]. Overall, these results suggest that the basic view of regulatory networks provide a first insight into regulation of a complex phenotype, it should not be ignored that more subtle variation in specific nodes of the network can be equally important.
Decades of work have identified and characterized subtle phenotypes in various systems in C. elegans. The availability of high-throughput screening of gene knockouts in C. elegans (using RNAi libraries) provides a unique position to use phenotypes in addition to molecular studies to formulate genetic networks. Using the C. elegans gonad as a model tissue, Green et al. [67] profiled over 500 essential genes for 94 different phenotypic effects in the germline, visualized simply by looking for alteration of histone and plasma membrane fluorescence markers. The resulting phenotypes were sufficient to generate robust genetic networks for the first two cell divisions in embryos, and yielded functional predictions for 106 out of 116 uncharacterized sterile genes. This work shows the remarkable potential for combining high-throughput identification of subtle tissue-level phenotypes with genome-wide understanding of regulatory networks.
Conclusions
The past few years have seen an explosion in the ability to quantify and identify gene expression, and to understand this expression in the context of regulatory networks. We now have a detailed map of gene expression at various stages at the whole-organism level and of dozens of individual tissues at specific developmental time-points. Dozens of regulatory chromatin marks and transcription factors have similarly been characterized at specific stages in development. The integration of these datasets has just begun, but has already yielded insights into novel regulatory mechanisms of ubiquitous housekeeping genes, previously unknown connections between recombination and chromosome-level chromatin domains, natural selection and its role in shaping the mutational landscape, and the complex circuitry underlying tissue differentiation.
As many of the whole-organism characterizations described above have been used in various species ranging from yeast to human, it is important to note the advantages of C. elegans in this area. Although whole-organism and specific tissues can be isolated from these organisms, the invariant cell lineage and transparency of the worm makes it possible to measure gene expression at the resolution of uniquely identifiable single cells in intact animals. This remarkable property of C. elegans enables a finer-grained view of regulatory circuits, which can provide important insights into how intra-cellular regulatory networks lead to inter-cellular phenotypes. Going forward, the unique position of C. elegans as an organism that is tractable for genome-wide, tissue-specific, and even in some cases cell-specific genomic and phenotypic experiments places it at the forefront of using systems biology to diagram the links between genome and function.
Acknowledgements
The authors would like to acknowledge NHGRI, NIA, the Glenn Foundation, the Smith Fellowship, and the Stanford Genome Training Program for funding. We are also grateful to members of the Kim lab for fruitful discussions and comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 2.White JG, et al. The structure of the ventral nerve cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275(938):327–348. doi: 10.1098/rstb.1976.0086. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Kim SK, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293(5537):2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 5.Stuart JM, et al. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302(5643):249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 6.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421(6920):231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 7.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56(1):110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 8.Ramani AK, et al. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res. 2011;21(2):342–348. doi: 10.1101/gr.114645.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillier LW, et al. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res. 2009;19(4):657–666. doi: 10.1101/gr.088112.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangone M, et al. The Landscape of C. elegans 3 ' UTRs. Science. 2010;329(5990):432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambros V, et al. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13(10):807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 12.Lim LP, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127(6):1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 14.Kato M, et al. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009;10(5):R54. doi: 10.1186/gb-2009-10-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He H, et al. Mapping the C. elegans noncoding transcriptome with a whole-genome tiling microarray. Genome Res. 2007;17(10):1471–1477. doi: 10.1101/gr.6611807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spencer WC, et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21(2):325–341. doi: 10.1101/gr.114595.110. The authors profiled gene expression from 13 embryonic tissues purified by FACS-sorting and 12 larval tissues by mRNA tagging using tiling microarrays. Bioinformatic analysis identified tissue-specific gene clusters, providing a valuable resource for the worm community. These sets of tissue-specific genes were also used to identify regulatory motifs controlling tissue-specific gene expression.
- 17.Lu ZJ, et al. Prediction and characterization of noncoding RNAs in C. elegans by integrating conservation, secondary structure, and high-throughput sequencing and array data. Genome Res. 2011;21(2):276–285. doi: 10.1101/gr.110189.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reece-Hoyes JS, et al. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt-Newbury R, et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5(9):e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupuy D, et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol. 2007;25(6):663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- 21.Reinke V, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6(3):605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 22.Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295(5556):821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, et al. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics. 2009;10:213. doi: 10.1186/1471-2164-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGhee JD, et al. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev Biol. 2007;302(2):627–645. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 25.Von Stetina SE, et al. Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol. 2007;8(7):R135. doi: 10.1186/gb-2007-8-7-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, et al. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418(6895):331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- 27.Fox RM, et al. A gene expression fingerprint of C. elegans embryonic motor neurons. BMC Genomics. 2005;6:42. doi: 10.1186/1471-2164-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colosimo ME, et al. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr Biol. 2004;14(24):2245–2251. doi: 10.1016/j.cub.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 29.Meissner B, et al. An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet. 2009;5(6):e1000537. doi: 10.1371/journal.pgen.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGhee JD, et al. ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev Biol. 2009;327(2):551–565. doi: 10.1016/j.ydbio.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy PJ, et al. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418(6901):975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- 32.Kunitomo H, et al. Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails. Genome Biol. 2005;6(2):R17. doi: 10.1186/gb-2005-6-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pauli F, et al. Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development. 2006;133(2):287–295. doi: 10.1242/dev.02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaudet J, et al. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol. 2004;2(11):e352. doi: 10.1371/journal.pbio.0020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330(6012):1775–1787. doi: 10.1126/science.1196914. This paper integrated multiple types of modENCODE data and yielded a number of novel insights into gene expression regulation. ChIP-seq of 23 transcription factors identified HOT regions, providing a first glimpse to a novel mechanism for regulation of highly expressed, ubiquitous, essential genes. ChIP-chip of 19 histone modifications and various other chromatin factors identified distinct broad chromatin domains on the autosomes. Gene-dense central regions were characterized by enrichment of active marks and low recombination rates, whereas gene-poor arm regions were enriched for repressive marks, high recombination rates, and association with the nuclear envelope. The paper also includes analysis of gene regulatory networks built from ChIP-seq data, datasets of tissue- and stage- specific gene expression and alternative splicing, and predictive algorithms for chromatin and transcription factor binding.
- 36. Long F, et al. A 3D digital atlas of C. elegans and its application to single-cell analyses. Nat Methods. 2009;6(9):667–672. doi: 10.1038/nmeth.1366. The authors describe software to automatically straighten, segment, and annotate single nuclei in the first larval stage worm. This software enables automated single-cell gene expression profiling in L1 worms.
- 37. Bao Z, et al. Automated cell lineage tracing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2006;103(8):2707–2712. doi: 10.1073/pnas.0511111103. The authors develop STARRYNITE software to enable automatic segmentation, identification, and tracking of individual nuclei in movies of worm embryonic development.
- 38. Liu X, et al. Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell. 2009;139(3):623–633. doi: 10.1016/j.cell.2009.08.044. Expression of 93 genes in 353 individual cells was profiled in the first larval stage worm. This matrix of single-cell expression provided the basis for a variety of analyses studying the role of gene regulation at the single-cell level. The relative roles of organ type, cell type, cell lineage and position within the body on gene expression are compared. Nuclei within the same syncytium were observed to have expression profiles that differ depending upon lineage, suggesting convergence of two independent regulatory networks to one final cell fate.
- 39. Murray JI, et al. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods. 2008;5(8):703–709. doi: 10.1038/nmeth.1228. Four developmental transcription factors were profiled at single-cell resolution from the one-cell through the 350-cell stage in C. elegans embryogenesis. This detailed profiling allowed for exact mapping of the timing of activation of these regulatory factors, confirming previous experimental approaches. The authors showed that this approach was successful in mutant strains as well, enabling single-cell profiling of gene expression in response to various genetic alternations.
- 40.Murray JI, et al. The lineaging of fluorescently-labeled Caenorhabditis elegans embryos with StarryNite and AceTree. Nat Protoc. 2006;1(3):1468–1476. doi: 10.1038/nprot.2006.222. [DOI] [PubMed] [Google Scholar]
- 41.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98(3):285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 42.Valouev A, et al. A high-resolution nucleosome, position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18(7):1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458(7236):362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41(3):376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ooi SL, Priess JR, Henikoff S. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2006;2(6):e97. doi: 10.1371/journal.pgen.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu SG, Fire A. Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma. 2010;119(1):73–87. doi: 10.1007/s00412-009-0235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whittle CM, et al. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 2008;4(9):e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu T, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21(2):227–236. doi: 10.1101/gr.115519.110. As part of the modENCODE project, genome-wide profiling was performed in C. elegans embryos and L3 larvae for 19 histone modifications, one histone variant, and eight chromatin-associated proteins. Clustering of chromatin marks yielded groups of coordinately regulated marks that could be associated with active or repressed genes. Other marks specifically mark the X chromosome, giving insight into the mechanisms of X chromosome dosage compensation in C. elegans. Autosomes were identified to have distinct arm and central domains characterized by distinct chromatin features.
- 50.Barrasa MI, et al. EDGEdb: a transcription factor-DNA interaction database for the analysis of C. elegans differential gene expression. BMC Genomics. 2007;8:21. doi: 10.1186/1471-2164-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deplancke B, et al. A gene-centered C. elegans protein-DNA interaction network. Cell. 2006;125(6):1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 52.Zhong M, et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 2010;6(2):e1000848. doi: 10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18(4):424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 54.Schuster E, et al. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol. 2010;6:399. doi: 10.1038/msb.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oh SW, et al. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38(2):251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 56.Whittle CM, et al. DNA-binding specificity and in vivo targets of Caenorhabditis elegans nuclear factor I. Proc Natl Acad Sci U S A. 2009;106(29):12049–12054. doi: 10.1073/pnas.0812894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lei H, et al. A widespread distribution of genomic CeMyoD binding sites revealed and cross validated by ChIP-Chip and ChIP-Seq techniques. PLoS One. 2010;5(12):e15898. doi: 10.1371/journal.pone.0015898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hochbaum D, et al. DAF-12 Regulates a Connected Network of Genes to Ensure Robust Developmental Decisions. PLoS Genet. 2011;7(7) doi: 10.1371/journal.pgen.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Niu W, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2011;21(2):245–254. doi: 10.1101/gr.114587.110. As part of the modENCODE project, ChIP-seq was performed for 22 transcription factors spanning multiple stages, developmental roles, and tissue-specificities. Target sites were identified and used to build an inter-transcription factor regulatory network. GEI-11 was found to significantly associate with small RNA promoters, suggesting a specific role in regulation of small RNA transcription.
- 60.Fukushige T, et al. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev. 2006;20(24):3395–3406. doi: 10.1101/gad.1481706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lall S, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol. 2006;16(5):460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 62. Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17(2):173–179. doi: 10.1038/nsmb.1745. This study describes an approach to directly identify microRNA targets in vivo. Using the knowledge that ALG-1 (an Argonaute protein) is guided by microRNAs to complementary sites in specific mRNAs, the authors performed CLIP-seq to immunoprecipitate ALG-1:microRNA:mRNA complexes, and profile the bound mRNAs by high-throughput sequencing. This genome-wide list of microRNA binding sites led to the discovery of significant targeting of genes involved in microRNA biogenesis, suggesting strong autoregulation in the microRNA pathway.
- 63.Barberan-Soler S, et al. Co-regulation of alternative splicing by diverse splicing factors in Caenorhabditis elegans. Nucleic Acids Res. 2011;39(2):666–674. doi: 10.1093/nar/gkq767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simonis N, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6(1):47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Denver DR, et al. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 2005;37(5):544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- 66.Raj A, et al. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463(7283):913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Green RA, et al. A High-Resolution C. elegans Essential Gene Network Based on Phenotypic Profiling of a Complex Tissue. Cell. 2011;145(3):470–482. doi: 10.1016/j.cell.2011.03.037. This study provides an interesting approach to building regulatory networks, starting from phenotype instead of gene expression. 500 genes were profiled for 94 subtle phenotypes on germ-line development and function, yielding various phenotypic classes. These phenotypes could be used to generate and visualize genetic networks of germ-line function at varying depths of co-functionality.