Abstract
Background
FtsZ, the major cytoskeletal protein in bacterial cytokinesis, assembles in vitro into protofilaments, which can further associate into sheets, bundles or tubes. We have constructed 16 site-directed mutants of E. coli ftsZ, and tested them for GTP hydrolysis and assembly in vitro, and for their ability to complement the temperature sensitive ftsZ84 mutation in E. coli.
Results
The mutants were grouped into three classes. Benign mutants, which mapped mostly to the front and back surface of the protofilament, were able to complement ftsZ84 in vivo and showed normal assembly in vitro. GTP contact mutations had less than 10% of wild type GTPase activity. They could all assemble in vitro, and several of these mutants could complement ftsZ84. A third, and newly discovered, class of mutations mapped to the sides of the protofilaments. These lateral mutants had mostly normal GTPase and assembly in vitro, but none of them complemented ftsZ84. The non-complementing mutants showed greatly reduced expression from the pBS58 vector, suggesting possible dominant negative effects.
Conclusions
Several mutants with greatly reduced GTPase could still complement ftsZ84, suggesting that the high level of GTPase observed in vitro is not essential for in vivo function. All of the lateral mutants failed to complement ftsZ84, which suggests that these surfaces of the protofilaments are important for function in cell division. These lateral surfaces may mediate association of FtsZ protofilaments into pairs or small sheets, although their structure is apparently different from the sheets assembled in DEAE dextran or calcium.
Background
FtsZ assembles into protofilaments in vitro [1, 2, 3, 4], and these are thought to form the cytoskeletal framework of the bacterial cell division machine of prokaryotes [5, 6, 7]. The FtsZ protein is homologous to tubulin, and the orientation of FtsZ subunits in the protofilament can be deduced by comparing the atomic structures of tubulin and FtsZ [8, 9, 10]. If the tubulin subunit is thought of as a cube, it would have six faces. The top and bottom faces of the subunit, which we call longitudinal, contact subunits above and below it in the protofilament; the right and left faces, which we call lateral, contact subunits in adjacent protofilaments; and the front and back faces correspond to the outside and inside of the microtubule. Since the FtsZ protofilament appears to be a homolog of the tubulin protofilament [2, 11] we will use these same designations for FtsZ.
To identify amino acids critical for FtsZ function, we constructed 16 mutations, mostly changing conserved aspartate and glutamate residues to alanine. Initially we selected amino acids for mutation based on conservation across different species, but after the atomic structure of FtsZ was determined [8] we designed other mutations to target specific structural questions. The main focus of the present study was to characterize the in vitro assembly and GTPase of the mutant proteins, but we also did a preliminary survey of their in vivo function by testing their ability to complement the temperature sensitive mutant ftsZ84.
More than a dozen mutations of FtsZ have been characterized in previous studies. We include these previously characterized mutations in our analysis of the structurally important side chains of FtsZ.
Results
Expression of mutant proteins
We made 16 site directed mutants of E. coli FtsZ. The amino acids mutated were all exposed on the surface, and their positions are mapped on the atomic structure of Methanococcus FtsZ in Fig. 1. Their locations are also indicated in Table 1. The mutant proteins were expressed in the pET expression system, where all gave medium or high levels of expression. Some proteins were partly or largely insoluble when expressed in BL21(DE3), but all were soluble when expressed in the mutant BL21 strain C41 [12].
Figure 1.
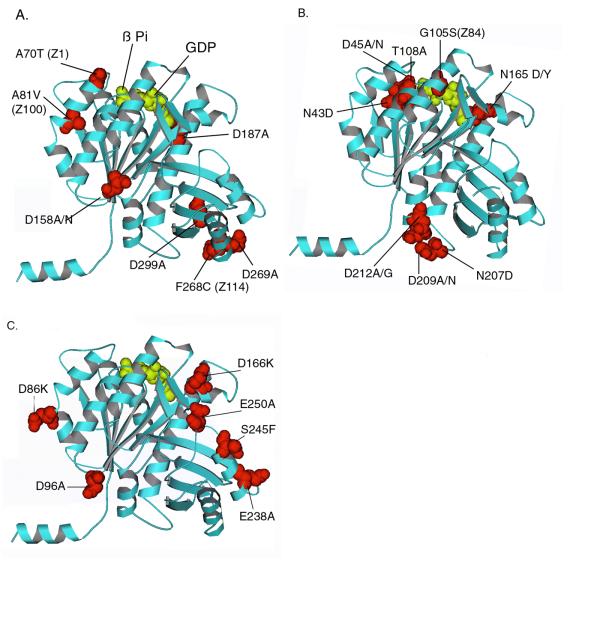
Mapping mutants on the atomic structure of Methanococcus FtsZ [8]. (A) Benign mutations; (B) GTP-contact mutations; (C) Lateral mutations. The amino acid numbers refer to the E. coli sequence, which were mapped to the corresponding amino acid in the Methanococcus sequence. Note that all of these amino acids are conserved in these two sequences as well as in most other FtsZ. Mutants identified in genetic screens are also indicated by their previous name (Z1, Z84). The subunits are oriented as they should be in a protofilament, based on the structure of tubulin viewed from outside the microtubule [9].
Table 1.
Summary of FtsZ mutations
| Mutation | aa no. | Location | Comple | GTPase | Asmb. | Asmb. | Asmb. | Asmb. | Asmb. | |||
| aa.no. | MjFtsZ | on FtsZ | ment? | %WT | GTP | GTP | GTP, | GTP, | GDP, | Ref | ||
| EcFtsZ | (1) | 42° (2) | Mg | Ca | 0.06 Dd | 0.6 Dd | 0.6 Dd | s | ||||
| Wild type | + | 100 | PF (3) | + | S (3) | T (3) | T (3) | (4) | ||||
| Benign mutations | ||||||||||||
| A70T(Z1) | A97 | Top-G | + (c) | 14 (a); | PF | S | T | T | a/c/ | |||
| <10 (d) | d | |||||||||||
| A81V/F268C | A108 | Top | + | 15 (d) | ||||||||
| (Z100) | (Buried) | |||||||||||
| D158A | D185 | Front | + | 120 | PF | S(-) | T(-) | T(-) | a | |||
| D158A | + | 155 (b) | b | |||||||||
| D158N | + | b | ||||||||||
| D187A | D212 | Back-G | + | PF | S | T | T | a | ||||
| F268C(Z114) | E293 | BtmRtBk | + (c) | 70 (d) | c/d | |||||||
| D269A | D294 | BtmRtBk | + | 10 | PF | S | T | T | a | |||
| D299A | D324 | Back | +/? | 200 | PF | S | T | T | a | |||
| GTP contact mutations | ||||||||||||
| N43D | N70 | Buried-Gγ | - | 31 (b) | b | |||||||
| D45A | D72 | Top-Gγ | - | 5 | NONE | + | T | T | T | a | ||
| D45N | - | 5 (b) | b | |||||||||
| G105S(Z84) | G132 | Top-G | +TS(f) | ~ 10 (g) | S | S | T | f/g/ | ||||
| a | ||||||||||||
| T108A(Z3) | T135 | Buried-G | - | ~ 0 (c) | NONE | c/e | ||||||
| N165D/Y | E192 | Buried-G | - | 17 (b) | b | |||||||
| N207D | N233 | Btm-Gsyn | - | 5 (b) | b | |||||||
| D209A | D235 | Btm-Gsyn | - | 7 | PF | + | T | T | T | a | ||
| D209N | - | b | ||||||||||
| D212G(Z2) | D238 | Btm-Gsyn | +TS2(c) | 0.5 (h) | T | c/e/h | ||||||
| D212A | - | 7 | NONE | + | S+T | T | T | a | ||||
| D212N | 35 | S (i) | i | |||||||||
| D212C | 17 | S (i) | i | |||||||||
| D212E | 17 | i | ||||||||||
| Lateral mutations | ||||||||||||
| D86K | E113 | Left | - | 49 | twPF+T | + | S+T | S+T | T | a | ||
| D96A | D123 | Left | - | PF | + | S | NONE | a | ||||
| D166K/F268V | E193/E293 | Right | - | 15 | PF | + | S (-) | S (-) | NONE | a | ||
| E238A | E264 | Right | - | 145 | PF | + | S | T | T | a | ||
| S245F | N271 | Right | - | 75 | PF+T | + | S | T | T | a | ||
| E250A | D276 | Right | - | 67 | PF | + | S (-) | S (-) | T (-) | a | ||
| E250K/D253K | D276/D278 | Right | - | 23 | PF | + | S (-) | S (-) | NONE | a | ||
1. Mutated amino acids are all surface residues, and their locations on the atomic structure are shown in Fig. 1. Top = the top surface, forming the interface in the protofilament; most "Top" amino acids tested also contact the GTP, as indicated by Top-G; N43 and D45 probably contact the gamma phosphate, indicated by -Gγ. Btm = the bottom surface, the other interface in the protofilament. N207, D209 and D212 form the "synergy" loop and probably contact the GTP of the subunit below; these are indicated Btm-Gsyn. Front = the front surface (corresponding to the outside of the microtubule). Back = the back surface (inside of the microtubule). Right = the right lateral surface. Left = the left lateral surface. N165 is largely buried, and makes contact with the GDP (buried-G). 2. Complementation tests in the present study (ref. a) were done with ftsZ84 (Ts) mutant cells. The mutant FtsZ was on the pBS58 plasmid. + indicates that the mutant plasmid supported cell growth and division in liquid culture overnight at 42°C. - indicates that the mutant gave only filamentous cells with limited growth. Complementations in ref. b were done with both ftsZ84 and a genomic FtsZ null, with identical results. A blank indicates that this was not tested; TS = temperature sensitive. 3. Assembly was in MEMK 6.5, with 1 mg/ml FtsZ and 2 mM GTP or GDP, and monitored by electron microscopy. A blank in any assembly condition means this was not tested, and NONE means no polymers were found by electron microscopy. Assembly in GTP (without Ca or DEAE dextran) produced single protofilaments (PF) in wild type and most mutants. D86K produced twined protofilaments (twPF). Assembly in 20 mM Ca produced protofilament bundles when indicated by a +. Assembly in DEAE dextran normally produced sheets of protofilaments (S) at 0.06 mg/ml, and tubes (T, protofilaments in the curved conformation) at 0.6 mg/ml. 4. References: a, the present work; b, Table 2 of Wang et al., [13]; c, Bi and Lutkenhaus, [24, 38]; d, Fig.5B of Dai et al.[25]; e, Mukherjee et al, [1]; f, Phoenix and Drapeau [20] and Powell and Court [21]; g, RayChaudhuri and Park, [19]; and de Boer et al., [17]; h, Trusca et al., [23]; i, Scheffers et al., [39].
Since we were mutating only surface-exposed charged groups, we expected that the mutant proteins would fold normally. One mutant protein, E274A was completely inactive in assembly, GTPase activity and in vivo function. Gel filtration and sedimentation showed that this protein was aggregated, and the loss of activity may be a consequence of this aggregation, rather than the specific location of the mutation. E274A is not included in Table 1 or Fig. 1. The best evidence that the other mutant proteins are properly folded is that every one of them assembled in the presence of DEAE dextran into specific polymers recognizable by electron microscopy.
In vivo complementation tests
A complementation system consisting of E. coli strain JFL101 (ftsZ84(ts)) and plasmid pBS58 [13] was used to test the ability of our mutant FtsZs to support cell division. JFL101 carries the temperature sensitive ftsZ84 mutation at the genomic locus, and is unable to divide at 42°C, although it can grow and divide normally at 30°C. pBS58 is a very low copy plasmid carrying a genomic fragment containing ftsQ, ftsA and ftsZ, and all of the relevant promoters. This supplies a 2-3 fold excess of FtsZ (and also FtsQ and FtsA) and complements ftsZ84, permitting cell growth and division at 42°C when carried in JFL101 [13].
Each of our mutations was transferred from pET into the ftsZ gene of pBS58. These mutants in pBS58 were tested for the ability to complement the ftsZ84(ts) mutation of JFL101, supporting growth and division when grown in liquid LB containing 1% NaCl with 100 μg/mL spectinomycin. Cells that showed prominent filamentation and inability to generate a turbid culture after overnight growth at 42°C were designated non-complementing. The results of the complementation tests are given in Table 1.
We used quantitative western blotting to check the level of expression from the pBS58 plasmid (Fig. 2). We first found that the level of FtsZ expression was approximately the same in three strains of E. coli at both 30 and 42°C. pBS58 with wild type ftsZ caused a three- to fivefold increase in total FtsZ levels when transformed into MC1000 or JFL101, consistent with the previous report [13]. pBS58 carrying four of the benign mutations caused a similar 3-5-fold increase in total FtsZ. These plasmids all complemented the temperature sensitive ftsZ84 mutation in JFL101. However, seven mutants that failed to complement showed total FtsZ levels approximately equal to that from cells without pBS58, implying that expression from the plasmid was severely reduced.
Figure 2.
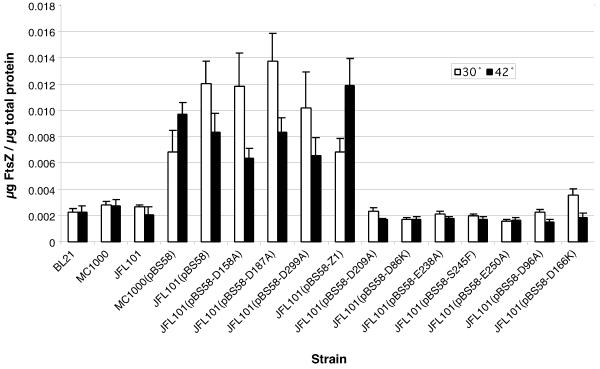
FtsZ expression levels in cells containing ftsZ mutants on pBS58 FtsZ levels were determined by quantitative western blotting and normalized to total protein levels to control for cells of varying length or cell density.
The failure to complement ftsZ84 at the non-restrictive temperature can therefore be attributed to the low expression, but the important question is why the expression has been so reduced. A likely explanation is that these mutant FtsZs are toxic at 30°C, so that the only clones obtained are ones that have altered the pBS58 plasmid to reduce expression. Consistent with this, clones of pBS58 carrying D96A and D166K/F268V showed aberrant Z-ring formation at 30°C, and E250K/D253K showed fragility and cell lysis. This suggests that the mutant protein is expressed at a low level, and it has a toxic effect.
That wild type ftsZ and four benign mutants gave substantial expression from pBS58, while expression was repressed in seven non-complementing mutants, suggests that many or all of the latter have dominant negative effects. These results also point to the limitations of the JFL101 (pBS58) system to characterize phenotypes of mutant ftsZ. This plasmid uses the endogenous ftsZ promoters, which are constitutively activated and may be subject to repression by second site mutations. To fully characterize the in vivo phenotypes, we are turning to a system where expression of the mutant protein can be repressed for cloning, and induced at controlled levels for phenotypic examination. We will also test whether the mutant ftsZs can support division in an ftsZ null. These studies are in progress to resolve the present uncertainty of the phenotypes in the JFL101(pBS58) system.
In vitro GTPase and assembly
Table 1 summarizes the GTPase and assembly properties of the mutant proteins. Data are also included for mutants characterized in previous studies.
GTPase activity was characterized for the purified proteins in MEMK6.5, where wild type FtsZ had a GTPase activity of 6.5 mol GDP per mol FtsZ per min [14]. The activity of mutant proteins is expressed as a percent of the wild type activity. Entries followed by a letter in parentheses are data from other studies (the letter indicates the reference, see footnote to the table).
FtsZ assembly was tested in MEMK6.5, 1 mg/ml FtsZ, 2 mM GTP or GDP, and either no polycation, 20 mM calcium, or 0.06 or 0.6 mg/ml DEAE dextran. Assembly was assayed by negative stain electron microscopy, and the type of polymers assembled is indicated for each condition [15].
We need to recall that the polymers formed by wild type FtsZ with DEAE dextran and GTP depend on the relative concentration of FtsZ and polycation [14, 15]. At 1 mg/ml FtsZ, 0.06 mg/ml DEAE dextran gives sheets of straight protofilaments, whereas with 0.6 mg/ml DEAE dextran the polymers were mostly helical tubes. We have speculated that at 0.6 mg/ml DEAE dextran, hydrolysis of GTP may occur before the polymers are fully formed and rigidified, allowing the GDP-FtsZ to adopt the curved conformation. At 0.06 mg/ml DEAE dextran on the other hand, straight protofilaments may be associated into sheets faster than hydrolysis, and lateral contacts in the sheet may prevent the protofilaments from adopting the curved conformation. Assembly in GDP plus 0.6 mg/ml DEAE dextran is very reproducible, giving only helical tubes [15].
The mutations are grouped into three classes, based on their ability to complement ftsZ84 and their location on the 3-D structure of the protein.
Benign mutations
The first box in Table 1 lists benign mutations, which complement ftsZ84 and are therefore functional in cell division. These mutant proteins also showed normal assembly in our in vitro assay. However, D269A had only 10% of the wild type GTPase activity. Two previously characterized mutation proteins, A70T (FtsZ1), A81V/F268C (FtsZ100), have comparably reduced GTPase in vitro, but appear to be fully functional for cell division.
Most of the benign mutations are located on the front or back of the FtsZ subunit, i.e., the faces corresponding to the outside or inside of the microtubule (Fig. 1a). This suggests that even highly conserved amino acids on these surfaces may not be crucial for function in cell division.
Mutations of amino acids that contact GTP
Several of our mutants had substantially reduced GTPase activity in vitro, failed to complement ftsZ84, and mapped on the 3-D structure in direct contact with the GTP. Table 1 also lists several mutation with similar characteristics characterized in other studies. G105S (FtsZ84) and D212G (FtsZ2) were able to function in cell division but had temperature sensitive and other phenotypes, so were not listed with benign mutations.
ftsZ85 is temperature sensitive for Z-ring assembly and division in vivo [16], but neither its GTPase nor assembly in vitro showed any temperature sensitivity. Thus de Boer et al. [17] reported that the GTPase activity was essentially the same at 30, 37 and 44°C. Our in vitro assembly showed the same products assembled at 30°C and 42°C (data not shown). FtsZ84 produced normal sheets of straight protofilaments in 0.06 mg/ml DEAE dextran, but they were abnormal in also producing sheets at 0.6 mg/ml DEAE dextran (with GTP). In fact, the sheets formed by FtsZ84 were exceptionally large and well ordered (Fig. 3a). Tubes could be obtained by assembly in GDP, but it seems that when assembled in GTP the impaired hydrolysis favors the straight conformation. We have not yet examined FtsZ84 for assembly of protofilaments in GTP and protofilament sheets in calcium.
Figure 3.
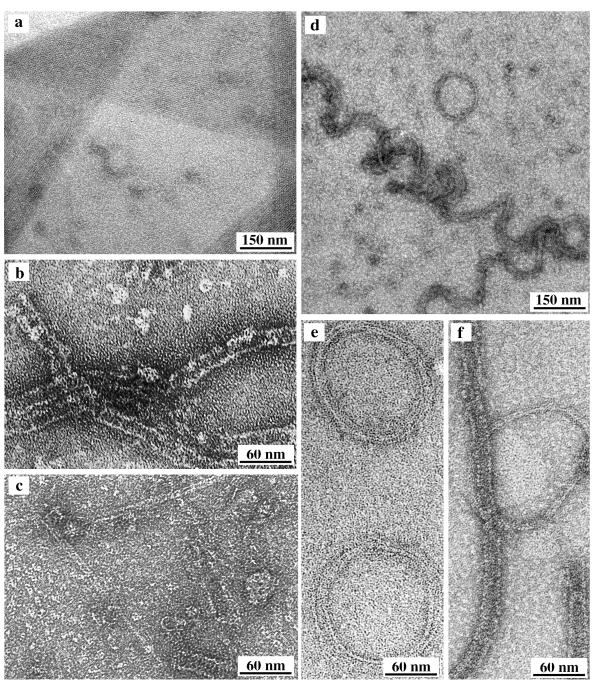
Assembly of three FtsZ mutants. (a) FtsZ84 assembled with GTP and 0.6 mg/ml DEAE dextran formed protofilament sheets of exceptional size and regularity (although the quality of the sheets varied for different preparations). Note that wild type FtsZ would have formed exclusively tubes under these conditions. (b) D86K assembled into tubes in MEMK6.5. No DEAE dextran and no GTP or GDP were added. (c) D86K at 0.5 mg/ml in MEMK6.5 plus 2 mM GTP (no DEAE dextran) formed twin protofilaments and short tubes. (d,e) D209A plus 0.6 mg/ml DEAE dextran and 10 mM GTP formed long spirals of protofilament bundles, and some circular hoops. (f) D209A in 2 mM GMPCPP also formed hoops, and these are mixed with tubes typical of assembly in GDP.
A cluster of mutations (N207, D209, D212) strongly affecting GTPase activity are located on the opposite side of the subunit from the GTP (Fig. 1b). This short strand of polypeptide, which we call the "synergy" loop, apparently contacts the GTP molecule of the subunit beneath it in the protofilament and contributes essential residues to the hydrolysis mechanism. (See [5, 18] for an expanded discussion of this hypothesis and references to mutants of tubulin that map to this loop.)
D209A showed a novel assembly form. In GTP plus 10 mM EDTA, or in 10 mM GTP (which also chelates the Mg), the D209A protein assembled into protofilament bundles that formed long spirals or closed circular hoops (Fig. 3d,e). Both the spirals and the hoops were about 120 to 150 nm in diameter. These structures seem to be closely related to each other, with the hoops being formed when the protofilament ends join after one turn. In 2 mM GMPCPP, a slowly hydrolyzable analog of GTP, D209A assembled a mixture of hoops and tubes (Fig. 3f).
D212A was also tested in 2 mM GMPCPP, where it made only tubes in most circumstances. However, in 0.06 mg/ml DEAE dextran it made some twin protofilaments, which were quite straight. D209A could also make straight, single protofilaments in the absence of DEAE dextran, while D212A could not. Both mutants assembled bundles of straight protofilaments in 20 mM calcium. This demonstrates that the synergy loop mutants can make straight protofilaments under certain conditions. However, the synergy loop mutants and D45A seem highly shifted toward the curved conformation, even when assembled with GMPCPP.
D45 is close to and probably contacts the γ-phosphate of the GTP, and was the most severe of this group of mutations. The alanine substitution reduced the in vitro GTPase activity to 5% of wild type and appeared to eliminate the ability to form straight protofilaments, either alone or with 0.06 mg/ml DEAE dextran. This mutant protein could still assemble into curved protofilaments and form tubes.
Mutations on the protofilament sides (lateral mutations)
A second group of mutants mapped to the sides of the subunits, which we originally believed to be the surfaces that bond protofilaments to each other in the protofilament sheets (see Discussion). Three of these were part of our original group of mutations, changing conserved asp or glu residues to ala. Three others were designed to make very disruptive changes in the presumed lateral contacts, changing D166 to K, S245 to F, and constructing a double mutant E250K/D253K. E250 and D253 are adjacent to a hydrophobic pocket formed by the highly conserved sequence PLL(D/E).
The lateral mutants had mostly normal GTPase activity, which was expected since the GTPase is thought to be triggered by assembly into protofilaments, which does not involve the lateral surfaces. There were two major surprises with these lateral mutants. First, they gave mostly normal in vitro assembly of both sheets and tubes, in contrast to our design goal to disrupt lateral interactions of protofilaments. Second, all of the lateral mutants were incapable of complementing the ftsZ84 mutation. These points are addressed in the Discussion.
D86K had an interesting assembly property, in that it showed a strong tendency to assemble into tubes. In the complete absence of DEAE dextran, D86K assembled into tubes in GDP or without added nucleotide (Fig. 3b). In fact, D86K was largely assembled into tubes when released from the expressing bacteria. In the presence of GTP, D86K formed a mixture of tubes and straight twin protofilament pairs (Fig. 3c). Wild type FtsZ also formed twin protofilaments, but their occurrence was exaggerated with D86K. D86K could also form protofilament sheets when assembled in DEAE dextran, as could all of the other lateral mutants (Table 1). D166K had a lower GTPase and did not assemble as well, but this may have been due to problems in the protein purification.
Discussion
High GTPase is not essential for cell division
Several mutations reduce the GTPase activity to 5-15% of wild type as measured in vitro, yet can still support cell division. Best studied is ftsZ84 (G105S), which has only 10% of the GTPase activity of wild type FtsZ over the entire range from 30 to 44°C [17, 19]. In spite of this greatly reduced GTPase in vitro, FtsZ84 can function for cell division at 30°C, and also at 42°C when over-expressed 2-3 fold [20, 21], or when ZipA is overexpressed 2-fold [22]. FtsZ2 (D212G) has 200-fold lower GTPase than wild type [23], yet it can support division when overexpressed (however, this division occurs with a mini-cell phenotype at 30°C, and cell lysis occurs at 42°C) [24]. FtsZ1 (A70T) and FtsZ100 (A81V/F268C - it is apparently the A81V substitution that reduces the GTPase, since F268C has near wild type activity [25]) have about 15% wild type GTPase activity and can support cell division [25]. Our present study has identified one additional mutant, D269A, that is capable of supporting cell division, but has lost 90% of its GTPase activity. Most of these mutant proteins seem to have normal assembly in vitro.
The ability of these five mutants to function in cell division is surprising, and raises two possibilities. One is that GTP hydrolysis may be substantially moderated in vivo. For example, the polymer structure in vivo (still unknown) might impose a new rate-limiting step in hydrolysis, which is lower even than the hydrolysis rate of the mutants in vitro. Alternatively, the GTPase activity in vitro may be ten times higher than actually needed for division. In vivo experiments will be needed to address this question further.
Curved and straight protofilaments, and mutations contacting GTP
In a recent study [15] we found that FtsZ protofilaments could adopt either a straight or curved conformation. The straight conformation was seen in single protofilaments assembled from purified FtsZ with GTP or the slowly hydrolyzable analog GMPCPP, and in protofilament sheets assembled with DEAE dextran. The curved conformation generated tubular polymers, which were assembled with DEAE dextran and GDP. We concluded that the straight and curved conformations were favored by GTP and GDP, respectively.
We therefore expected that mutations that severely blocked GTP hydrolysis would assemble tubes in GDP, but that assembly in GTP would give only straight protofilaments. All of these mutants formed tubes in the presence of DEAE dextran and GDP, as expected. The straight conformation was preferred for G105S (FtsZ84) in GTP and DEAE dextran, also as expected. However, the other three GTP-contact mutants (D45A, D209A, D212A) preferred to make tubes even in GTP. This did not agree with our initial expectations that reduced GTP hydrolysis would lock the protofilaments into the straight conformation.
A recent paper has proposed that hydrolysis of GTP results in a substantial movement of the T3 loop, and suggested that this movement might be involved in the transition to the curved conformation [26]. D45 is adjacent to the T3 loop, so removal of its side chain might permit movements similar to those that occur when the γ-phosphate is removed. The positions of D209 and D212 are less certain, but they are likely close to or in contact with the T3 loop of the adjacent subunit. We suggest the hypothesis that removal of any of the side chains in this region may permit movement of the T3 loop into the GDP conformation, similar to removal of the γ-phosphate.
The mystery of lateral contacts
Our initial goal in making the lateral mutants was to target amino acids making lateral contacts between protofilaments. We expected some of these mutants to disrupt assembly of sheets and/or tubes, and we would then be able to test whether they also disrupted cell division. Protofilament sheets have not been demonstrated in vivo, so this could be an important test of whether the lateral interactions are actually used in cell division. However, none of these lateral mutations seemed to affect the assembly of sheets in vitro - sheets assembled normally in both DEAE dextran and calcium, and tube assembly was also mostly normal.
A closer examination of the atomic structure may explain why our lateral mutants failed to prevent assembly of DEAE dextran sheets in vitro. The protofilaments in both sheets and tubes assembled in DEAE dextran are spaced 5.3-5.4 nm apart [15, 27], but the subunit is not that wide. The widest aspect of the FtsZ subunit is seen in the front view, i.e., as presented in Fig. 1. Most of the amino acids that we mutated could only make contact if the subunits were spaced 4 - 4.5 nm apart. The only structure that could bridge a 5.3 nm spacing is the N-terminal segment, which is highly extended in Methanococcus FtsZ and perhaps also in E. coli. It seems clear from the dimensional analysis that the lateral residues we mutated could not form the bridges between protofilaments in the DEAE dextran sheets. The N-terminal helix is a likely candidate for lateral contacts in these sheets.
There is, however, a second type of lateral contact that can assemble protofilament sheets or pairs with a much closer spacing. We observed a second form of protofilament sheet when assembled on cationic lipid monolayers, in which the protofilaments were only 3.7 nm apart [2]. Löwe and Amos [11] showed pairs of Methanococcus FtsZ protofilaments with a ~ 3.9 nm spacing. We believe that these sheets and pairs with the 3.7 - 3.9 nm protofilament spacing may assemble by contacts of the lateral surfaces that we mutated, and may be representative of an essential polymer in vivo.
In view of the mostly normal in vitro GTPase and assembly, it was remarkable that all lateral mutants failed to complement ftsZ84. So far this failure to complement has only been tested in the JFL101(pBS58) system, but the reduced expression of the mutant proteins suggests a possible dominant negative effect. These lateral surfaces may therefore be important for function in cell division. One possibility is that they could provide the binding sites for accessory proteins. However, FtsA [13, 28, 29, 30] and ZipA [31,32, 33] are the only two proteins known to bind directly to FtsZ, and they bind to the highly conserved C-terminal peptide [34]. An alternative is that these lateral surfaces mediate association of protofilaments with each other, to form protofilament pairs or small sheets. These would likely correspond to the in vitro structures with 3.7-3.9 nm spacing.
Wang et al., have recently described 13 site directed mutants of ftsZ in Caulobacter crescentus [35], and tested them in a strain in which the genomic wild type ftsZ could be fully expressed or completely suppressed. They targeted clusters of adjacent residues containing two or more charged residues, which were changed to alanine. Six of their mutants were fully able to support cell division, and were listed as wild type in phenotype. Four were not able to support cell division and were listed as recessive-lethal. Each of these mutations did, however, cause significant disruptions in division when expressed with wild type ftsZ, so they had a partially dominant effect. Three mutations were dominant-lethal, as was a G109S mutant designed to duplicate the E. coli ftsZ84.
Two of these mutants are particularly relevant to our study. Their DE254AA includes our E250A. They listed it as lethal-recessive, although noting that it produced filamentous cells with constrictions when expressed along with wild type ftsZ. Their ability to obtain transformants expressing the mutant protein contrasts with our results that the only transformants we obtained had significantly reduced expression levels of the mutant protein. It may be that the aberrations induced by this mutant protein were sufficient select for down-regulated expression from the pGS58 vector, or the aberrations may be more severe in E. coli. A second mutation of Wang et al., demonstrate mutations can have very different phenotypes in different species. In Caulobacter G109S was dominant-lethal, while in E. coli the corresponding G104S(ftsZ84) has a wild type phenotype at 30°C, and can support cell division at 42°C when overexpressed.
Conclusions
We have tested 16 site-directed mutants of E. coli FtsZ for assembly and GTPase activity in vitro, and for whether they can function in vivo to complement ftsZ84. Mutations on the front and back of the FtsZ protofilament were mostly benign: they had normal in vitro assembly, and could complement ftsZ84. Several of these, however, had significantly reduced in vitro GTPase, demonstrating that the high level of GTPase is not essential for assembly or function in vivo. Another class of mutations altered residues contacting the GTP. These could all assemble in DEAE dextran, although with some abnormalities, and they had < 10% of the wild type GTPase. These mutants failed to complement ftsZ84 when expressed on pBS58. A third class of mutations mapped to the sides of protofilaments. These lateral mutations did not interfere with GTPase nor with assembly of protofilaments, and surprisingly showed mostly normal assembly in DEAE dextran. We conclude that these lateral surfaces are not involved in assembly of the DEAE dextran polymers. However, four mutations on the right side and two on the left failed to give clones that complement ftsZ84, suggesting that these lateral surfaces are important for the function of FtsZ in cell division. These lateral surfaces may mediate association of FtsZ protofilaments into pairs or small sheets, although with a structure different from the in vitro polymers stabilized by DEAE dextran.
Materials and Methods
Site-directed mutagenesis of FtsZ
All FtsZ mutants with amino acid substitutions were constructed by an overlap extension PCR method [36]. We used genomic DNA for a template, and for each mutant transcribed the complete ftsZ sequence (adding NdeI and BamHI cloning sites at the ends) using error-correcting Pfu polymerase [37] and outward-facing primers containing the desired mutations. Mutated ftsZ genes were cloned into the pET11 plasmid for expression of FtsZ, permitting purification of the mutant protein.
The FtsZ mutants were all created originally in the pET expression vector. In order to test whether they could function in vivo, we transferred the mutations to the plasmid pBS58 (provided by Dr. Joe Lutkenhaus, University of Kansas Medical Center, Kansas). To construct a pBS58 ftsZ mutant plasmid (pBS58ftsZmut), the EcoRI and BstEII fragment of ftsZ in pBS58 was replaced with the same part of ftsZ from pET-ftsZmut. Because EcoRI and BstEII sites are not unique in pBS58 and pET, additional cloning steps were needed. A 2.4 kb HindIII fragment from pBS58 that contains the whole ftsZ gene was first subcloned into vector Litmus38 (New England Biolabs) whose EcoRI site has been deleted, and created a plasmid Litmus38(RI)+H3. The EcoRI and BstEII fragment with a desired mutation from pET-ftsZmut was put into Litmus38(RI)+H3 precut by the same enzymes. Finally, the HindIII fragment from Litmus38(RI)+H3mut was ligated into pBS58 whose HindIII segment had been removed. The orientation of gene in the resulting plasmid pBS58ftsZmut was examined by EcoRI digestion.
Ten mutants were sequenced from the pET vector: D45A, A70T, D86K, D158A, D212A, E238A, E250A, D269A, E274A, and D299A. Nine mutants were sequenced from pBS58: A70T, D96A, D158A, D166K/F268V, D187A, D209A, S245F, D269A and E274A. In each case we sequenced approximately 600-700 bp around the mutation. Only one showed a change other than the desired mutation. D166K also contained the mutation F268V; since this is a conservative substitution, and since the similar F268C(Z114) is benign [38], we do not think the extra mutation is important.
Quantitation of FtsZ expression
Bacterial cultures, including strains transformed with pBS58 carrying various alleles of ftsZ, were either grown to A600 = 0.5-0.7 at 30°, or grown to A600 ~ 0.15 at 30°, then grown for 90 minutes at 42°. All cultures were grown in LB (+ 100 μg/mL spectinomycin if necessary). Harvesting and extraction of the cultures and quantitation of FtsZ levels in these extracts was carried out as described [14], except that FtsZ levels were compared to total protein levels instead of cell numbers. Total protein levels in the SDS extracts were determined by BCA assay (Pierce) after removing the reducing reagent from SDS buffer by precipitating protein from the extract with 10% trichloroacetic acid, washing in 10% trichloroacetic acid and resuspending the pellet in 0.1 N NaOH. This procedure recovered >95% of control protein from SDS sample buffer over the range of protein concentrations recommended for use in the BCA assay (data not shown).
Purification of FtsZ
FtsZ mutant proteins were over-expressed in BL21(DE3) cells and purified by two different approaches. Initially, we purified wild type and mutated FtsZ by two-step ammonium sulfate precipitation [14]. Briefly, cells were suspended in 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 1 mM PMSF, treated with 0.4 mg/mL lysozyme and lysed by freeze-thaw and sonication. We have subsequently found that lysis in a French pressure cell gives a much more reproducible recovery of soluble proteins, and have used this for later preparations. We also found that inducing at an A600 of 1.0 rather than 0.5 gave more soluble protein. FtsZ was purified from the bacterial supernatant by discarding a precipitate at 20% saturated ammonium sulfate, then precipitating the remaining FtsZ by raising the ammonium sulfate level to 25% saturation [14]. This procedure gives an excellent yield of pure, active wild type FtsZ [14], and was used for most of the studies described below with no additional steps.
We later found that some mutant FtsZs were not efficiently precipitated by 25% ammonium sulfate, so we adopted a method using higher ammonium sulfate for the initial precipitation, followed by DEAE chromatography [3, 19]. FtsZ was precipitated by adding ammonium sulfate to 35% saturation. The resulting protein pellet was resuspended in 10 ml of buffer A (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 10 % glycerol and 1 mM PMSF) and dialyzed against 500 ml of buffer A overnight. The sample was loaded onto a DEAE-Sephacel column at 4°C and eluted with a gradient of 0.05-1 M KCl in buffer A. FtsZ eluted at 0.4 M KCl, separated from a later peak containing nucleic acid. The FtsZ was desalted in MEMK6.5 (100 mM MES, pH 6.5 adjusted with KOH, 1 mM EGTA, 5 mM Mg acetate) and stored at -80°C. SDS-PAGE showed several high molecular weight contaminants in the FtsZ precipitated by 35% ammonium sulfate, and these were not removed by the DEAE column. The 20/25% ammonium sulfate protocol seems to give a better purification of proteins that can be precipitated at 25%, but the alternative protocol is important for some mutants that do not precipitate completely at 25% ammonium sulfate. The following proteins were purified by the DEAE-Sephacel procedure for GTPase assays: A70T(FtsZ1), D158A, D269A and G105S (FtsZ84).
Wild type FtsZ was usually about 70% soluble when expressed in BL21(DE3), and the bacteria were lysed with the French press. Some mutant proteins seemed less soluble, giving from 5% to 40% soluble protein (D45A, D96A, D86K, D209A, E238A, D166K, listed in increasing order of solubility). Remarkably, when the pET plasmids were expressed in E. coli C41, a modified BL21(DE3) [12], D45A was expressed at a lower level but was 50% soluble, and all other proteins were expressed at a high level and were >90% soluble.
In vitro assembly, electron microscopy and GTPase assay
All in vitro assays were done in MEMK6.5 buffer. Assembly reactions contained 1 mg/ml FtsZ (determined by our calibrated BCA assay [14]), 2 mM GTP or GDP and variable concentrations of DEAE-dextran or calcium. The reaction mixture was incubated on ice for 5-10 min, then at 37°C for 5 min, and negatively stained electron microscope specimens were prepared.
Assembly in calcium was tested in the crude lysed bacterial supernatants, by adding GTP to 1 mM and calcium chloride to 20 mM. The sample was brought to room temperature for five min. Assembly of FtsZ polymers was initially indicated by formation of a gel, and negative stain electron microscopy confirmed a network of protofilament sheets in most of these gelled samples (in some the background obscured any structures). The most decisive indication of assembly was obtained by centrifugation at 100,000 × g for 30 min. This pelleted a clean band of FtsZ in the gelled samples, whereas without calcium nothing was pelleted.
GTPase activity was assayed as described previously [14] with some modifications. Briefly, the reaction was started by adding 3 μl of 10 mM GTP (Sigma) and 5 μCi {α-32P} GTP (3000 Ci/mmol, Amersham) to 27 μl of FtsZ in MEMK6.5 and incubated immediately at 37°C. To terminate the reaction, 3 μl of the reaction mixture was withdrawn every three minutes and mixed with 7 μl of 10 mM EDTA in 50 mM Tris-HCl, pH 8.0 (this is an important modification of our previous protocol, as the EDTA stops GTP hydrolysis). One microliter of this mixture was applied to a polyethyleneimine cellulose thin layer chromatography plate and developed in 0.75 M KH2PO4. GDP produced was determined and the GTPase activity was expressed as mols GDP per mol FtsZ per min. GTPase of mutant proteins were scaled as a percent of wild type, which was 6.5 mols GTP per mol FtsZ per min.
Acknowledgments
Acknowledgements
We thank Gina Briscoe for technical assistance; Dr. Joe Lutkenhaus, University of Kansas for the pBS58 plasmid and JFL101; Dr. David Bramhill, Merck for the FtsZ84 protein. Supported by NIH grant GM28553.
Contributor Information
Chunlin Lu, Email: h.erickson@cellbio.duke.edu.
Jesse Stricker, Email: jesse_stricker@hotmail.com.
Harold P Erickson, Email: h.erickson@cellbio.duke.edu.
References
- Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg L, Simon M, Erickson HP. Polymerization of FtsZ, a bacterial homolog of tubulin. is assembly cooperative? J Biol Chem. 2001;276:11743–1153. doi: 10.1074/jbc.M009033200. [DOI] [PubMed] [Google Scholar]
- Erickson HP. FtsZ, a tubulin homolog, in prokaryote cell division. Trends Cell Biol. 1997;7:362–367. doi: 10.1016/S0962-8924(97)01108-2. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J, Addinall SG. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- Bramhill D. Bacterial cell division. Annual Review of Cell & Developmental Biology. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Erickson HP. Atomic structures of tubulin and FtsZ. Trends in Cell Biology. 1998;8:133–137. doi: 10.1016/s0962-8924(98)01237-9. [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 1999;18:2364–2371. doi: 10.1093/emboj/18.9.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli : mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Wang XD, Huang JA, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Stricker J, Erickson HP. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima- quantitation, GTP hydrolysis, and assembly. Cell Motility & the Cytoskeleton. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Lu CL, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–170. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall SG, Cao C, Lutkenhaus J. Temperature shift experiments with ftsZ84 (Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J Bacteriol. 1997;179:4277–4284. doi: 10.1128/jb.179.13.4277-4284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P, Crossley R, Rothfield L. The essential bacterial cell division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- Nogales E, Downing KH, Amos LA, Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- RayChaudhuri D, Park JT. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- Phoenix P, Drapeau GR. Cell division control in Escherichia coli K-12: some properties of the ftsZ84 mutation and suppression of this mutation by the product of a newly identified gene. J Bacteriol. 1988;170:4338–4342. doi: 10.1128/jb.170.9.4338-4342.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell BS, Court DL. Control of ftsZ expression, cell division, and glutamine metabolism in Luria-Bertani medium by the alarmone ppGpp in Escherichia coli. J Bacteriol. 1998;180:1053–1062. doi: 10.1128/jb.180.5.1053-1062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J Bacteriol. 1998;180:3946–3953. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. Isolation and characterization of ftsZ alleles that affect septal morphology. J Bacteriology. 1992;174:5414–5423. doi: 10.1128/jb.174.16.5414-5423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Mukherjee A, Xu Y, Lutkenhaus J. Mutations in ftsZ that confer resistance to SulA affect the interaction of FtsZ with GTP. J Bacteriol. 1994;176:130–136. doi: 10.1128/jb.176.1.130-136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz JF, Kralicek A, Mingorance J, Palacios JM, Vicente M, Andreu JM. Activation of Cell Division Protein FtsZ. control of switch loop T3 conformation by the nucleotide γ-phosphate. J Biol Chem. 2001;276:17307–17315. doi: 10.1074/jbc.M010920200. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Stoffler D. Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to α/β and gamma tubulin. J Cell Biol. 1996;135:5–8. doi: 10.1083/jcb.135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din N, Quardokus EM, Sackett MJ, Brun YV. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol Microbiol. 1998;27:1051–1063. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- Ma X, Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of escherichia coli ftsZ [In Process Citation]. J Bacteriol. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Mukherjee A, Lutkenhaus J. Recruitment of ZipA to the division site by interaction with FtsZ. Mol Microbiol. 1999;31:1853–1861. doi: 10.1046/j.1365-2958.1999.01322.x. [DOI] [PubMed] [Google Scholar]
- Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, Seehra J, Somers W. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19:3179–3191. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, Rhee ZC, de Boer PA. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J Bacteriol. 2000;182:5153–5166. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. The FtsZ protofilament and attachment of ZipA - structural constraints on the FtsZ power stroke. Curr Opin Cell Biol. 2001;13:55–60. doi: 10.1016/s0955-0674(00)00174-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jones BD, Brun YV. A set of ftsZ mutants blocked at different stages of cell division in Caulobacter. Mol Microbiol. 2001;40:347–360. doi: 10.1046/j.1365-2958.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Erickson HP. Expression in Escherichia coli of the thermostable DNA polymerase from Pyrococcus furiosus. Protein Expr Purif. 1997;11:179–184. doi: 10.1006/prep.1997.0775. [DOI] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA). J Bacteriology. 1990;172:5602–5609. doi: 10.1128/jb.172.10.5602-5609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers D, de Wit JG, Den Blaauwen T, Driessen AJ. Substitution of a conserved aspartate allows cation-induced polymerization of FtsZ. FEBS Lett. 2001;494:34–37. doi: 10.1016/s0014-5793(01)02310-9. [DOI] [PubMed] [Google Scholar]


