Abstract
Purpose
FERM domain containing protein 7 (FRMD7) mutations are associated with X-linked idiopathic congenital nystagmus (ICN). The purpose of this study is to identify a novel splice variant of FRMD7 (FRMD7-S) in both humans and mice with a shortened exon 4 relative to the original form of FRMD7 (FRMD7-FL),and to detect the role of FRMD7-FL and FRMD7-S in the process of neuronal development.
Methods
The splice variant of FRMD7 was identified by PCR. Expression levels of hFRMD7-FL and hFRMD7-S transcripts in developing human fetal brain were tested by RT–PCR, and expression levels in the human pluripotent embryonic carcinoma NTera 2/cl.D1 (NTERA-2; NT2) cell line with all-trans retinoic acid (ATRA) or bone morphogenetic protein-2 (BMP-2) treatment were tested by real-time qPCR. hemaglutinin (HA)-tagged recombinant plasmids DNA encoding hFRMD7-FL and Myc-tagged recombinant plasmids DNA encoding hFRMD7-S were used to transiently transfect the human NT2 cells. Further, immunofluorescence experiments were performed to determine the co-localization of the two fusion proteins. Finally, using co-immunoprecipitation analyses, we demonstrated that FRMD7-FL and FRMD7-S interacted with each other.
Results
A novel splice variant of FRMD7 (FRMD7-S) with a shortened exon 4 relative to the original form of FRMD7 (FRMD7-FL) was identified from the cDNA of the human NT2 cell line and mouse fetal brain. The FRMD7 transcripts showed similar tissue distributions and were upregulated following all trans retinoic acid (ATRA)-induced differentiation of NT2 cells. FRMD7-FL and FRMD7-S co-localized and co-immunoprecipitated with each other. Further, overexpression of FRMD7-FL in NT2 cells resulted in altered neurite development and upregulation of FRMD7-S.
Conclusions
Although the significance of the 45 bp deletion remains unknown, our observations suggest that the FRMD7 isoforms may play a significant role during neuronal differentiation and development.
Introduction
Idiopathic congenital nystagmus (ICN) is an oculomotor disorder characterized by involuntary horizontal oscillations of the eyes that presents at birth or appears in the first months of life, but does not usually worsen over time. ICN is distinct from other ocular disorders in which nystagmus is acquired later in life (e.g., cataracts, glaucoma, albinism) or is accompanied by eye, brain, or other health abnormalities [1]. The prevalence of ICN is estimated to be 24 per 10,000 [2], and although some techniques can improve vision (e.g., glasses, contact lenses, eye muscle surgery), nystagmus is usually permanent and cannot be corrected or cured [3].
Previous studies have speculated that ICN represents a primary defect in the brain regions involved in ocular motor control [4], although the precise pathogenic mechanisms underlying ICN are currently unknown. Mutations in the human FERM domain containing protein 7 (FRMD7) gene (NM_194277), which encodes the FERM domain containing protein 7 that is a member of the FERM family, are associated with X-linked ICN [5]. Approximately 50% of X-linked pedigrees and 5% of sporadic ICN cases have been linked to FRMD7 mutations, and more than 35 FRMD7 mutations have been reported worldwide in families with X-linked ICN from various ethnic backgrounds [1,6,7].
The 298 amino acid FERM domain was originally identified in protein 4.1 [8], and subsequent studies reported the structural, transport, and membrane-localizing functions of this domain [9-11]. Notably, the hFRMD7-FL protein is highly homologous to FARP1 (FERM, RhoGEF, and pleckstrin domain protein 1) and FARP2 proteins, which are known to play significant roles in neuronal development. FARP1 is necessary and sufficient for promoting lateral motor column dendritic growth, and FARP2 is a key molecule involved in the response of neuronal growth cones to class-3 semaphorins [12,13]. Moreover, recent studies have demonstrated that inhibition of FRMD7 expression in mouse neuroblastoma cell line (NEURO2A) during neuronal differentiation is associated with significant delays in neurite growth and disrupted F-actin/G-actin dynamics [14]. In a mouse model, FRMD7 expression levels were low in all adult tissue samples, whereas FRMD7 expression levels were higher in embryos and underwent a sharp increase at embryonic day 18 in brain tissue [15]. These findings provide evidence that FRMD7 plays a critical role during neuronal morphogenesis, in synapse function, and in neurite growth, but further studies will be required to uncover the precise mechanisms associated with FRMD7 function.
The NT2 cell line, which is a human embryonic carcinoma cell line, differentiates into post-mitotic neuron-like cells following treatment with all trans retinoic acid (ATRA) [16,17] and into non-neural epithelial cell lineages following exposure to bone morphogenetic protein 2 (BMP-2) [18,19]. Therefore, NT2 cells provide an ideal model system that mimics normal neuronal differentiation in the brain according to many established criteria.
Previous studies have revealed that alternative splicing occurs much more frequently in the brain relative to other tissues [20,21], and alternative splicing is increasingly recognized for its role in neurologic disease [22,23]. In the current study, we have identified and characterized a novel FRMD7 splice variant in humans and mice, hFRMD7-S (GenBank FJ717411) and mFRMD7-S, respectively; the splice variant contains a 45 bp truncation in the fourth exon and produces an altered form of the FERM domain. Herein, we investigate the expression patterns, role in neurite development, subcellular localization, and interactions associated with the FRMD7-S splice variant relative to the original FRMD7-FL isoform in NT2 cells.
Methods
Cell culture and ATRA/BMP-2-induced differentiation
Human NT2 cells (Cell Culture Center of Peking Union Medical College, Peking, China) were maintained in DMEM/F12 medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Prior to the induction of differentiation, the NT2 cells were incubated in 25 cm2 cell culture flasks in 4 ml of medium in a 37 °C, high humidity, 5% CO2 incubator. The cells were trypsinized and sub-cultured at a ratio of 1:3 twice weekly. For the time course analyses, the NT2 cells were seeded on a 3.0×106/10 cm2 plate and treated the following day with 10 μM all-trans retinoic acid (ATRA) dissolved in DMSO [16,17] or with 50 ng/ml bone morphogenetic protein-2 (BMP-2) dissolved in normal saline [18,19]. The cells were collected at 12, 24, 36, 48, and 72 h after treatment for the early time points. For the later time points (5, 8, and 12 days), the cells were reseeded every three days in the continued presence of ATRA or BMP-2 and collected on the corresponding days.
Collection of human and mouse fetal tissue
Freshly isolated 16-week post-conception (wpc) aborted human fetal tissues were collected from the Women's Hospital School Of Medicine Zhejiang University. The experimental procedures were approved by the Human Ethical Committee of the Zhejiang University. Mouse fetal brain tissue was isolated from 18-day mouse fetuses. A scissors was used to produce 1–3 mm3 pieces of tissue, which were snap frozen in cryogenic vials in liquid nitrogen and stored at −80 °C. Total RNA from each sample was prepared for subsequent RT–PCR experiments.
RNA isolation, reverse transcription PCR, and quantitative real-time PCR analysis
Total RNA was isolated from human and mouse fetal tissues and NT2 cells using TRIZOL reagent (Invitrogen, San Diego, CA), according to the manufacturer’s instructions. Real-time quantitative PCR (RT-qPCR) assays were performed using a Light CyclerTM real-time PCR thermocycler (Roche, Shanghai, China). Briefly, 1.5 μg of total RNA was reverse transcribed using oligo (dT) 12–18 with M-MLV Reverse Transcriptase (Promega, Madison, WI) according to the manufacturer's instructions. Relative levels of gene expression were analyzed using the SYBR® Premix Ex Taq™ (TaKaRa, Dalian, China) using the recommended PCR conditions. The sequences of the RT-qPCR primers are shown in Table 1. The relative mRNA levels of target genes were calculated relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the (=2-△△CT) method [24].
Table 1. Primer sequences.
| ID | Name | Sequence |
|---|---|---|
|
Primer for RT–PCR/RT-qPCR | ||
| p1f |
FRMD7-FL-f |
5′-CAAAGCAGGTAAAAAATCCTAAGG-3′ |
| p1r |
FRMD7-FL-r |
5′-ATGTGAGATACCATCAACGCTGT-3′ |
| p2f |
FRMD7-S-f |
5′-TAACAAAGCAGGTAAAAATGGAC-3′ |
| p2r |
FRMD7-S-r |
5′-AAGATGTGAGATACCATCAACG-3′ |
| p3f |
GAPDH-f |
5′- CCCACTCCTCCACCTTTGAC-3′ |
| p3r |
GAPDH-r |
5′- CATACCAGGAAATGAGCTTGACAA-3′ |
|
Primer for cloning | ||
| pAf |
pcDNA3.1-FRMD7-FL-HA-f |
5′-CGCTAGCGCCACCATGCTACATTTAAAAGTGCAGT-3′ |
| pAr |
pcDNA3.1-FRMD7-FL-HA-r |
5′-GCTCGAGTTAAGCGTAATCTGGAACATCGTATGGGTAAGCTAAAAAGTAATTACATG-3′ |
| pBf |
pcDNA3.1-FRMD7-S-Myc-f |
5′-CGCTAGCGCCACCATGCTACATTTAAAAGTGCAGT-3′ |
| pBr |
pcDNA3.1-FRMD7-S-Myc-r |
5′-GCTCGAGTTACAGATCCTCTTCAGAGATGAGTTTCTGCTCAGCTAAAAAGTAATTACATG-3′ |
| pCf |
pEGFP-FRMD7-FL-f |
5′-CGCTAGCATGATTCCCAGAAGAT-3′ |
| pCr |
pEGFP-FRMD7-FL-r |
5′-GCTCGAGAGCTAAAAAGTAATTACA-3′ |
| pDf |
pDsRed-FRMD7-S-f |
5′-CGCTAGCATGATTCCCAGAAGAT-3′ |
| pDr | pDsRed-FRMD7-S-r | 5′-GCTCGAGAGCTAAAAAGTAATTACA-3′ |
Underlined nucleotides mark the HA-tag and Myc-tag.
cDNA clone and plasmid construction
Human FRMD7-FL and FRMD7-S genes were isolated from NT2 cell cDNA (synthesized from the total RNA of natural NT2 cells) by PCR amplification using the following primers: 5′-ATG CTA CAT TTA AAA GTG CAG TTT-3′ and 5′-TTA AGC TAA AAA GTA ATT ACA TGG T-3′. The mouse FRMD7-FL and FRMD7-S genes were cloned from 18-day fetal mouse brain cDNA using the following primers: 5′-GTA TGG ATC CAT GCT CCA TTT AAA AGT G-3′ and 5′-AGC TAA GAA ATA ATT GCA TGG CTT TAG C-3′. PCR was performed in 25 μl of a reaction mixture containing DNA polymerase from Thermococcus kodakaraensis (KOD) DNA polymerase buffer and 1.25 U KOD DNA polymerase (Toyobo, Osaka, Japan). Subsequent cloning of PCR products into the pEASY-Blunt vector (TransGen Biotech, Beijing, China) was performed using the pEASY-Blunt Cloning Kit for Sequencing (TransGen Biotech). Sequence analyses were performed by the Invitrogen Company (Shanghai, China). To generate eukaryotic expression vectors for hFRMD7 splice variants, cloned RT–PCR fragments in pEASY-Blunt were used as the template. The hFRMD7-FL cDNA was sub-cloned into pcDNA3.1(+) in frame with an NH2-terminal HA tag (hFRMD7-FL-HA) and pEGFP-N1 (pEGFP-hFRMD7-FL) with the pAf/pAr and pCf/pCr primer sets, respectively (Table 1). The hFRMD7-S gene was subcloned into pcDNA3.1(+) with an NH2-terminal Myc tag (hFRMD7-S-Myc) and plasmid pDsRed-N1 (pDsRed-hFRMD7-S) using the pBf/pBr and pDf/pDr primer sets, respectively (Table 1). NheI and XhoI were chosen as the cloning sites for all three vectors, and the sequences of all of the expression cassettes were verified.
Cell transfection and immunofluorescence microscopy
For transient transfection experiments, the NT2 cells were placed on glass coverslips or cell culture flasks until they reached approximately 60% confluency and were transfected with FuGENE HD transfection reagent (Roche) at a ratio of 1 μg of DNA to 3 μl of FuGENE HD per coverslip. The cells were processed for immunofluorescence at 48 h post-transfection. For stable overexpression experiments, G418 (1.2 μg/ ml medium; Invitrogen) antibiotic selection was performed for 4 weeks. For the immunofluorescence experiments, the induced or transfected NT2 cells were washed with phosphate-buffered saline (PBS) and fixed for 15 min at room temperature in 4% paraformaldehyde (PFA) in PBS. The coverslips were washed twice with ice-cold PBS, and the cells were permeabilized with PBS containing 0.2% Triton X-100 for 10 min at 4 °C. After washing the cells in PBS three times for 5 min (each wash), a blocking solution containing PBS and 10% normal goat serum was applied for 1 h at room temperature. Afterwards, primary antibodies (monoclonal anti-Myc or anti-HA or antibodies; Abmart Inc., Shanghai, China) diluted in a blocking solution were applied overnight at 4 °C or for 1 h at room temperature. In the double-labeling experiments, the cultures were incubated simultaneously with two primary antibodies. After three washes in PBS, the secondary antibodies (Dylight 488, Dylight 594; Jackson ImmunoResearch Laboratories, Inc. West Grove, PA) were diluted 1:250 in a blocking solution and added for 1 h at room temperature in the dark. The secondary antibody solution was decanted and washed three times with PBS for 5 min each in the dark. Finally, the cells were incubated for 5 min with 0.1 μg/ml DAPI as a nuclear counterstain. After three washes, the coverslips were mounted with a drop of Antifade Mounting Medium (Beyotime Institute of Biotechnology, Suzhou, China) and stored in the dark at −20 °C or 4 °C. The cells were viewed using a Nikon eclipse inverted fluorescence microscope equipped with a Nikon digital camera (Nikon Corporation, Tokyo, Japan). Image overlays and contrast enhancement were performed using Adobe Photoshop CS2 (Adobe Systems Inc. San Jose, CA) software.
Co-immunoprecipitation and immunoblot analyses
For co-immunoprecipitation (co-IP) experiments, NT2 cells were transiently co-transfected with 2 μg of each of the hFRMD7-FL-HA and hFRMD7-S-Myc vectors. At 48 h post-transfection, the total protein been extracted from the cells by using M-PER® Mammalian Protein Extraction Reagent (Thermo Fisher Scientific Inc., Rockford, IL) following the manufacturer's instructions. Then the ProFound™ Co-Immunoprecipitation Kit (Thermo Fisher Scientific Inc.) was used for co-IP assays according to the manufacturer's instructions. Control immunoprecipitations were performed in the presence of non-specific immunoglobulins.
Statistical analyses
All experiments were done in triplicate, and the data are represented as mean±SEM. Statistical significance between groups was determined using the Kolmogorov–Smirnov test and Student’s t-test, as implemented by SPSS13.0 (SPSS Inc., Chicago, IL). Results were considered to be statistically significant when p<0.05.
Results
Identification of a novel FRMD7 splice variant and tissue distribution of two FRMD7 transcripts
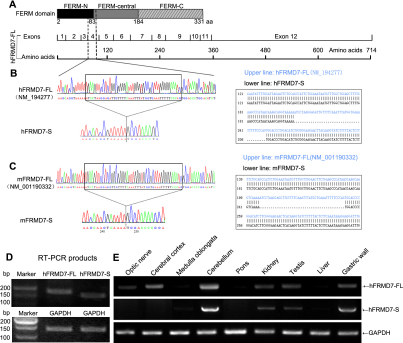
In RT–PCR experiments to clone the hFRMD7-FL gene from NT2 cells, two PCR products were produced. Cloning and sequence analysis of the PCR products indicated that they corresponded to two distinct FRMD7 mRNAs: the expected original hFRMD7-FL mRNA and another unexpected mRNA not described previously, which is referred to herein as hFRMD7-S. This novel transcript, which lacks a 45 bp segment in the 5′ region of exon 4, results in the fusion of the 3′ end of exon 3 to the 5′ end of truncated exon 4; therefore, hFRMD7-S encodes a putative protein of 699 amino acids (Figure 1A,B). We then performed RT–PCR to confirm the presence of mFRMD7-S in 18-day-old fetal mice (Figure 1C). The expression of hFRMD7-FL and hFRMD7-S transcripts was examined in tissues from 16-wpc human fetal tissue using sequence-specific primers (Figure 1D). We detected high expression of hFRMD7-FL in the cerebellum and moderate expression in the cerebral cortex, optic nerve, kidney, testis, and gastric wall. We detected high expression of hFRMD7-S in the cerebellum and moderate expression in the kidney, testis, and gastric wall. However, hFRMD7-S was not expressed in the optic nerve or cerebral cortex. Neither hFRMD7-FL nor hFRMD7-S were expressed in the medulla oblongata, pons, or liver. Taken together, these results revealed that hFRMD7-S exhibited a similar but limited tissue distribution compared to hFRMD7-FL (Figure 1E).
Figure 1.
Cloning of a novel FRMD7 isoform and tissue distribution of the two splice variants. A: Components of the FERM domain and gene structure of hFRMD7-FL. B and C: Sequence comparison between hFRMD7-FL/mFRMD7-FL (B) and hFRMD7-S/mFRMD7-S (C) showing the deletion of 45 bp in the 5′ end of exon 4 in the hFRMD7-S/mFRMD7-S. D: Agarose gel electrophoresis of the RT–PCR products to confirm their size and the identification of a single PCR product. E: Expression levels of hFRMD7-FL and hFRMD7-S transcripts in selected human fetal tissues. The primer sets used in D and E: p1f/p1r, p2f/p2r and p3f/p3r.
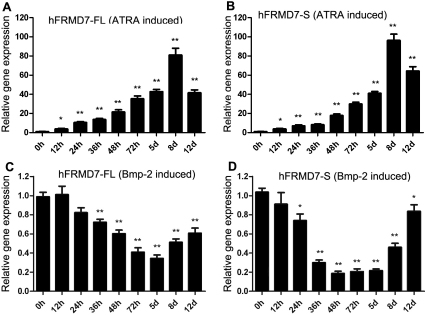
Levels of both hFRMD7 transcripts significantly increased in ATRA-induced differentiating NT2 cells
As shown in Figure 2A,B, the levels of hFRMD7-FL and hFRMD7-S mRNA expression in NT2 cells started to increase at 12 h after ATRA treatment and reached a maximal increase at 8 days (81 fold for hFRMD7-FL and 96 fold for hFRMD7-S relative to the 0 h time point; Figure 2A,B). The control group did not exhibit any change relative to the 0 h time point. Compared to ATRA, BMP-2 had a repressive effect on hFRMD7-FL and hFRMD7-S expression. After 24 h of BMP-2 treatment, the levels of hFRMD7-FL and hFRMD7-S transcripts started to decline relative to the 0 h time point, and continued to decrease after 36 h; after 8 days, the expression levels of hFRMD7-FL and hFRMD7-S transcripts began to increase (Figure 2C,D).
Figure 2.
Effects of ATRA/BMP-2 induction on the expression of FRMD7 splice variants in NT2 cells. A and B: NT2 cells were treated with ATRA (10 µM) or vehicle (0.1% DMSO) for the indicated periods. C and D: NT2 cells were treated with BMP-2 (50 ng/ml) or vehicle (normal saline) for the indicated time periods. Expression levels of hFRMD7-FL and hFRMD7-S transcripts were determined by RT-qPCR. The vehicle-treated cells did not show any significant change (data not shown). The primer sets were used for RT-qPCR: p1f/p1r, p2f/p2r and p3f/p3r. Both the transcript levels are normalized to GAPDH mRNA levels. The data are presented as fold changes relative to 0 h. The experiments were done in triplicate, and the graphs represent the average of three independent experiments (Columns, mean; bars, S.E.M.; *p<0.05, **p<0.01 versus 0 h).
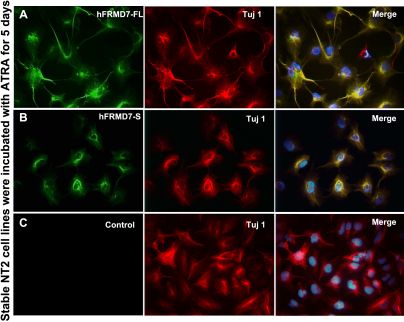
hFRMD7-FL and hFRMD7-S co-localized with class III β-tubulin and exhibited differential effects on neurite development
Stable NT2 cell lines overexpressing hFRMD7-FL-HA or hFRMD7-S-Myc were established for use in experiments to study their role during differentiation. We examined the co-localization of class III β-tubulin, which is not expressed at detectable levels in neural progenitor cells, with the two FRMD7 isoforms by indirect immunofluorescence. Both isoforms exhibited strong co-localization with neuronal class III β-tubulin in differentiating NT2 cells induced with ATRA for 5 days (Figure 3A,B). Having established that the hFRMD7-FL or hFRMD7-S overexpressed cell lines, we next sought to find how this change would affect neurite development in differentiated NT2 cells. Notably, following ATRA treatment for 5 days, a significant increase in the length and number of neurites was observed in NT2 cells stably overexpressing hFRMD7-FL-HA NT2 (Figure 3A,C and Table 2; p<0.05). However, the degree of neurite branching did not differ between the control and hFRMD7-S overexpressing NT2 cells (Figure 3B,C and Table 2; p>0.05).
Figure 3.
Effects of hFRMD7-FL or hFRMD7-S overexpression on neurite outgrowth. NT2 cells stably transfected with hFRMD7-FL-HA (A), hFRMD7-S-Myc (B), or the empty pcDNA3.1(+) vector (C) were treated with ATRA for 5 days. The cells were fixed and immunostained with monoclonal anti-Myc or anti-HA antibodies and a DyLight-488 conjugated secondary antibody. Class III β-tubulin was immunostained using a monoclonal TuJ1 antibody and DyLight-594 secondary antibody. Magnification: 200×.
Table 2. Neurite outgrowth of NT2 cells treated with 10 μM retinoic acid.
| Norm | Control | hFRMD7-S | hFRMD7-FL |
|---|---|---|---|
| Percentage of cells with neurites* |
46.2±2.1 |
47.5±1.3 (ns) |
77.8±1.6 |
| Average neurite length** (μm) | 42.7±2.6 | 43.6±2.1 (ns) | 72.3±1.1 |
Cells were treated with 10 μm retinoic acid, 10% FCS/DMEM-F12 for 5 days, and neurite parameters were quantified. Data are mean±SEM of three independent experiments. At least 250 cells per transfection were counted. *Cells that possess at least one outgrowth of more than one-half the cell body diameter in length. **Neurite length from cell body to distal tip. (ns) stands for Not Significant.
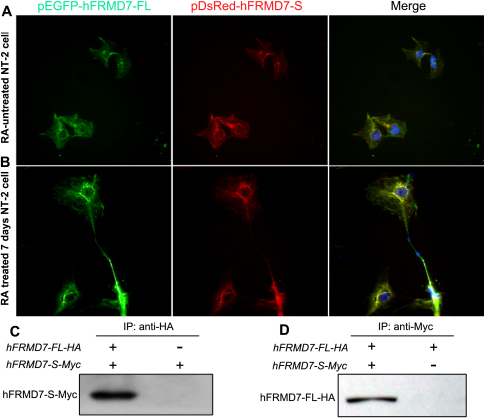
hFRMD7-FL co-localizes and interacts with hFRMD7-S in NT2 cells
To investigate the subcellular co-localization of FRMD7 isoforms and to differentiate between the localization of endogenous FRMD7 and exogenously expressed FRMD7 proteins, we used expression vectors to express hFRMD7-FL or hFRMD7-S differentially tagged with green (EGFP) or red (DsRed) fluorescent protein, respectively. Following transient co-transfection with the fusion constructs encoding pEGFP-hFRMD7-FL and pDsRed-hFRMD7-S, untreated NT2 cells (without ATRA) and NT2 cells treated with ATRA for 7 days were subjected to fluorescence microscopy. In these experiments, EGFP-hFRMD7-FL and DsRed-hFRMD7-S exhibited a high degree of co-localization (Figure 4); both isoforms were primarily localized in cytoplasm and perinuclear regions in undifferentiated NT2 cells (Figure 4A). Similarly, in NT2 cells induced with ATRA for 7 days, EGFP-hFRMD7-FL and DsRed-hFRMD7-S co-localized extensively in the cytoplasm, perinuclear regions, and neurite processes. Notably, the distribution of EGFP-hFRMD7-FL was more abundant in the distal terminals of the neurites relative to that of DsRed-hFRMD7-S (Figure 4B).
Figure 4.
Sub-cellular co-localization and co-immunoprecipitation of hFRMD7-FL with hFRMD7-S. Co-transfection of pEGFP-hFRMD7-FL (green) and pDsRed-hFRMD7-S (red) into RA-untreated (A) or differentiating NT2 cells (B). After 48 h, the cells were fixed and DAPI was used as a nuclear counterstain (blue). Magnification 400×. C and D: Reciprocal co-immunoprecipitation experiments show that hFRMD7-FL-HA co-immunoprecipitates with hFRMD7-S-Myc, and vice versa, in NT2 cells. Cells transfected with only hFRMD7-S-Myc or hFRMD7-FL-HA served as the negative control.
To test for protein–protein interactions, we performed co-IP experiments. We co-expressed HA and Myc NH2-terminally tagged versions of hFRMD7-FL (hFRMD7-FL-HA) and hFRMD7-S (hFRMD7-S-Myc), respectively, in NT2 cells. At 48 h post-transfection, total cell lysates were extracted, and hFRMD7-FL-HA was immunoprecipitated using anti-HA antibodies, followed by immunoblot analysis with anti-Myc antibodies. The results showed that hFRMD7-FL-HA co-immunoprecipitated with hFRMD7-S-Myc (Figure 4C). This finding was confirmed by reciprocal immunoprecipitation experiments, which showed that hFRMD7-S-Myc co-immunoprecipitated with hFRMD7-FL-HA (Figure 4D). The results of these experiments may suggest that hFRMD7-FL and hFRMD7-S either interact directly or are associated with the same protein complex in NT2 cells.
hFRMD7-FL protein stimulates transcription of hFRMD7-S
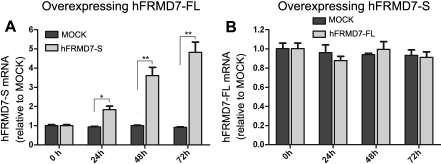
Alternative splicing often impacts the transcription and subsequent translation of the major protein. Therefore, we performed experiments to determine whether overexpression of one FRMD7 splice variant was associated with altered transcription of the other splice variant. Overexpression of hFRMD7-FL or hFRMD7-S was confirmed by RT–PCR and immunoblot analyses (data not shown). We found that the transcription levels of hFRMD7-S exhibited a time-dependent increase in cells overexpressing hFRMD7-FL relative to control cells, with an average of 4.8 fold higher hFRMD7-S expression levels at 72 h (Figure 5A); in contrast, hFRMD7-FL transcription levels did not change significantly in NT2 cells overexpressing hFRMD7-S (Figure 5B).
Figure 5.
Overexpression of hFRMD7-FL stimulates hFRMD7-S transcription. Expression levels of hFRMD7-FL or hFRMD7-S transcripts were monitored by RT-qPCR in transfected NT2 cells at the indicated time points. Cells transfected with the empty vector served as the negative control (MOCK group). A: Expression levels of hFRMD7-S in NT2 cells overexpressing hFRMD7-FL. B: Expression levels of hFRMD7-FL in NT2 cells overexpressing hFRMD7-S. The primer sets were used for RT-qPCR: p1f/p1r, p2f/p2r and p3f/p3r. Both the transcript levels are normalized to GAPDH mRNA levels. The data are presented as the fold changes relative to MOCK group. All of the experiments were done in triplicate, and the graph represents the average (columns, mean; bars, S.E.M.; *p<0.05, **p<0.01 versus MOCK group).
Discussion
The human FRMD7 gene spans a region of 2,145 bp and comprises 12 exons. Until now, no FRMD7 mRNA variants have been described. Herein, we report the discovery of a novel transcript, hFRMD7-S, from the human FRMD7 gene generated by alternative splicing. An increased understanding of the hFRMD7-S variant relative to the original hFRMD7-FL transcript may help clarify the role of FRMD7 splice variants in neuronal development and the pathogenesis of ICN.
We found that the hFRMD7-FL and hFRMD7-S transcripts exhibit a similar tissue distribution, with highest levels in the cerebellum; neither hFRMD7-FL nor hFRMD7-S were expressed in the medulla oblongata or pons. These results corroborate previous studies in mouse models [15]. Further, our previous studies demonstrated that hFRMD7-FL expression is ubiquitous during fetal brain development, but is restricted to regions of eye movement in adults [25]. We may suppose that the FRMD7 transcripts may carry out cerebellum-associated functions (e.g., vestibulo-cerebellar system regulation of normal eye movement). Nevertheless, in the previous work, using immunohistochemistry testing, FRMD7-FL was observed to be highly expressed in the pons and medulla oblongata. Since the size differences in the corresponding mRNAs and protein isoforms are very small (i.e., 2,145 bp for hFRMD7-FL versus 2,100 bp for hFRMD7-S), neither northern blot nor immunoblot analyses were performed to determine endogenous hFRMD7-FL and hFRMD7-S protein expression. The reason for the differences of expression distribution between RNA level and protein level of FRMD7-FL are worth exploring in future studies.
Recent studies suggest that ATRA-induced differentiation is followed by the upregulation of hFRMD7-FL mRNA and protein expression levels in NEURO2A cells [14]. In the current study, we found that ATRA-induced neuronal differentiation of NT2 cells resulted in elevated expression of both hFRMD7-FL and hFRMD7-S transcripts, whereas BMP-2 treatment of NT2 cells had a repressive effect on hFRMD7-FL and hFRMD7-S expression. Further, a recent study demonstrated that downregulation of hFRMD7-FL in NEURO2A cells resulted in altered neurite development [14]. In the current study, we found that hFRMD7-FL overexpression leads to a significant increase in neurite length in differentiating NT2 cells; however, hFRMD7-S overexpression had no effect on this process.
NT2 cells undergo programmed terminal differentiation along a neuronal pathway, resulting in the accumulation of neuronal cells with neurite outgrowths and extended processes within a period of 8 days. Thus, we speculate that both hFRMD7-FL and hFRMD7-S are involved in the process of neurite outgrowth extension. Since hFRMD7-FL and hFRMD7-S share highly similar sequences, their different effect on ATRA-induced NT2 differentiation is of particular interest. Although it is unclear what underlies these differences, our findings suggest that the FRMD7 isoforms are involved in the formation and growth of neurites during neuronal differentiation, and also provide evidence that the hFRMD7-S isoform may play a minor role during this process.
A previous study showed that FARP1 and FARP2, which share homology with FRMD7, are involved in signal transduction between the plasma membrane and cytoskeleton, and that the hFRMD7-FL protein co-localizes with actin in NEURO2A cells [12-14,26,27]. In the current study, we found that hFRMD7-FL and hFRMD7-S splice variants also exhibit strong co-localization with class III β-tubulin, which is considered to be one of the earliest neuron-associated cytoskeletal marker proteins during mammalian development [28,29]. These results give evidence that the FRMD7 isoforms carry out functions associated with microtubules and cytoskeleton-associated proteins or at least play a role in a related molecular pathway.
We found that the hFRMD7-FL isoform co-localized to a high extent with the hFRMD7-S isoform in undifferentiated and differentiating NT2 cells, although the hFRMD7-FL isoform exhibited a more abundant distribution in the distal terminals of axon branches relative to the hFRMD7-S isoform. These differences may be due to different levels of protein translation efficiency. Further, the results of our reciprocal co-IP experiments also suggest that hFRMD7-FL and hFRMD7-S interact in NT2 cells. Taken together, these results uncovered a putative protein complex formed by the two FRMD7 isoforms; however, further studies will be required to confirm this finding.
Notably, we also found that hFRMD7-FL overexpression led to the stimulation of hFRMD7-S gene expression in NT2 cells, whereas overexpression of hFRMD7-S showed no significant effect on hFRMD7-FL expression. These results indicate that hFRMD7-FL might be involved in hFRMD7-S regulation. Considering their different effects on ATRA-induced NT2 differentiation, we speculate that hFRMD7-S plays a less important role of biologic functions. Although the mechanism(s) by which FRMD7-FL regulates hFRMD7-S expression remains elusive, we speculate that the altered FERM domain in hFRMD7-S may explain its inability to regulate hFRMD7-FL expression.
In conclusion, we identified a novel FRMD7 variant generated by alternative mRNA splicing that results in the deletion of 45 base pairs in exon 4. This is the first splice variant of FRMD7 to be reported. The two isoforms share a similar tissue distribution, and they co-localize and interact with each other in NT2 cells. However, the hFRMD7-S isoform showed a slightly limited tissue distribution and a limited function in altering the length and number of neurites in differentiating NT2 cells relative to the hFRMD7-FL isoform. Taken together, although the significance of the 45 bp deletion remains unknown, our observations suggest that the FRMD7 isoforms may play a significant role during neuronal differentiation and development. Therefore, although additional studies will be required to further confirm the association between the two transcripts, the current findings may enhance our understanding of their precise functional roles in the pathogenesis of ICN.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China [81070903 to B.Z.]. We thank the parents who donated the abortions. We also thank all medical staff at the Women's Hospital School Of Medicine Zhejiang University for their dedication in this study.
References
- 1.Thomas S, Proudlock FA, Sarvananthan N, Roberts EO, Awan M, McLean R, Surendran M, Kumar AS, Farooq SJ, Degg C, Gale RP, Reinecke RD, Woodruff G, Langmann A, Lindner S, Jain S, Tarpey P, Raymond FL, Gottlob I. Phenotypical characteristics of idiopathic infantile nystagmus with and without mutations in FRMD7. Brain. 2008;131:1259–67. doi: 10.1093/brain/awn046. [DOI] [PubMed] [Google Scholar]
- 2.Sarvananthan N, Surendran M, Roberts E, Jain S, Thomas S, Shah N, Proudlock FA, Thompson JR, McLean RJ, Degg C, Woodruff G, Gottlob I. The prevalence of nystagmus: the Leicestershire nystagmus survey. Invest Ophthalmol Vis Sci. 2009;50:5201–6. doi: 10.1167/iovs.09-3486. [DOI] [PubMed] [Google Scholar]
- 3.Lee J. Surgical management of nystagmus. J R Soc Med. 2002;95:238–41. doi: 10.1258/jrsm.95.5.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobs JB, Dell’Osso LF. Congenital nystagmus, hypotheses for its genesis and complex waveforms within a behavioral ocular motor system model. J Vis. 2004;4:604–25. doi: 10.1167/4.7.7. [DOI] [PubMed] [Google Scholar]
- 5.Tarpey P, Thomas S, Sarvananthan N, Mallya U, Lisgo S, Talbot CJ, Roberts EO, Awan M, Surendran M, McLean RJ, Reinecke RD, Langmann A, Lindner S, Koch M, Jain S, Woodruff G, Gale RP, Degg C, Droutsas K, Asproudis I, Zubcov AA, Pieh C, Veal CD, Machado RD, Backhouse OC, Baumber L, Constantinescu CS, Brodsky MC, Hunter DG, Hertle RW, Read RJ, Edkins S, O'Meara S, Parker A, Stevens C, Teague J, Wooster R, Futreal PA, Trembath RC, Stratton MR, Raymond FL, Gottlob I. Mutations in FRMD7, a newly identified member of the FERM family, cause X-linked idiopathic congenital nystagmus. Nat Genet. 2006;38:1242–4. doi: 10.1038/ng1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Self J, Lotery A. A review of the molecular genetics of congenital idiopathic nystagmus (CIN). Ophthalmic Genet. 2007;28:187–91. doi: 10.1080/13816810701651233. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, Liu Z, Zhao G, Xie X, Yin X, Hu Z, Xu S, Li Q, Song F, Tian J, Luo W, Ding M, Yin J, Xia K, Xia J. Novel mutations of the FRMD7 gene in X-linked congenital motor nystagmus. Mol Vis. 2007;13:1674–9. [PubMed] [Google Scholar]
- 8.Leto TL, Marchesi VT. A structural model of human erythrocyte protein 4.1. J Biol Chem. 1984;259:4603–8. [PubMed] [Google Scholar]
- 9.Bretscher A, Reczek D, Berryman M. Ezrin: a protein requiring conformational activation to link microfilaments to the plasma membrane in the assembly of cell surface structures. J Cell Sci. 1997;110:3011–8. doi: 10.1242/jcs.110.24.3011. [DOI] [PubMed] [Google Scholar]
- 10.Tepass U. FERM proteins in animal morphogenesis. Curr Opin genet Dev. 2009;19:357–67. doi: 10.1016/j.gde.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Kubo T, Yamashita T, Yamaguchi A, Sumimoto H, Hosokawa K, Tohyama M. A novel FERM domain including guanine nucleotide exchange factor is involved in Rac signaling and regulates neurite remodeling. J Neurosci. 2002;22:8504–13. doi: 10.1523/JNEUROSCI.22-19-08504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang B, Su YS, Sockanathan S. FARP1 promotes the dendritic growth of spinal motor neuron subtypes through transmembrane Semaphorin6A and PlexinA4 signaling. Neuron. 2009;61:359–72. doi: 10.1016/j.neuron.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toyofuku T, Yoshida J, Sugimoto T, Zhang H, Kumanogoh A, Hori M, Kikutani H. FARP2 triggers signals for Sema3A-mediated axonal repulsion. Nat Neurosci. 2005;8:1712–9. doi: 10.1038/nn1596. [DOI] [PubMed] [Google Scholar]
- 14.Betts-Henderson J, Bartesaghi S, Crosier M, Lindsay S, Chen HL, Salomoni P, Gottlob I, Nicotera P. The nystagmus-associated FRMD7 gene regulates neuronal outgrowth and development. Hum Mol Genet. 2010;19:342–51. doi: 10.1093/hmg/ddp500. [DOI] [PubMed] [Google Scholar]
- 15.Self J, Haitchi HM, Griffiths H, Holgate ST, Davies DE, Lotery A. Frmd7 expression in developing mouse brain. Eye (Lond) 2010;24:165–9. doi: 10.1038/eye.2009.44. [DOI] [PubMed] [Google Scholar]
- 16.Andrews PW. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev Biol. 1984;103:285–93. doi: 10.1016/0012-1606(84)90316-6. [DOI] [PubMed] [Google Scholar]
- 17.Pleasure SJ, Page C, Lee VM-Y. Pure, postmitotic, polarized human neurons derived from NTera 2 cells provide a system for expressing exogenous proteins in terminally differentiated neurons. J Neurosci. 1992;12:1802–15. doi: 10.1523/JNEUROSCI.12-05-01802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chadalavada RS, Houldsworth J, Olshen AB, Bosl GJ, Studer L, Chaganti RS. Transcriptional program of bone morphogenetic protein-2-induced epithelial and smooth muscle differentiation of pluripotent human embryonal carcinoma cells. Funct Integr Genomics. 2005;5:59–69. doi: 10.1007/s10142-005-0132-7. [DOI] [PubMed] [Google Scholar]
- 19.Caricasole A. Ward-van, Oostwaard, D., Zeinstra, L., van, den, Eijnden-van, Raaij, A., Mummery, C. Bone morphogenetic proteins (BMPs) induce epithelial differentiation of NT2D1 human embryonal carcinoma cells. Int J Dev Biol. 2000;44:443–50. [PubMed] [Google Scholar]
- 20.Lopez AJ. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 21.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 22.Dredge BK, Polydorides AD, Darnell RB. The splice of life: alternative splicing and neurological disease. Nat Rev Neurosci. 2001;2:43–50. doi: 10.1038/35049061. [DOI] [PubMed] [Google Scholar]
- 23.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-△△CT Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Pu J, Li YZ, Liu ZR, Yan YP, Tian J, Chen S, Zhang BR. Expression and localization of FRMD7 in human fetal brain, and a role for F-actin. Mol Vis. 2011;17:591–7. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuang B, Su YS, Sockanathan S. FARP1 promotes the dendritic growth of spinal motor neuron subtypes through transmembrane Semaphorin6A and PlexinA4 signaling. Neuron. 2009;61:359–72. doi: 10.1016/j.neuron.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolic M. The role of Rho GTPases and associated kinases in regulating neurite outgrowth. Int J Biochem Cell Biol. 2002;34:731–45. doi: 10.1016/s1357-2725(01)00167-4. [DOI] [PubMed] [Google Scholar]
- 28.Katsetos CD, Herman MM, Mörk SJ. Class III beta-tubulin in human development and cancer. Cell Motil Cytoskeleton. 2003;55:77–96. doi: 10.1002/cm.10116. [DOI] [PubMed] [Google Scholar]
- 29.Katsetos CD, Frankfurter A, Christakos S, Mancall EL, Vlachos IN, Urich H. Differential localization of class III, beta-tubulin isotype and calbindin-D28k defines distinct neuronal types in the developing human cerebellar cortex. J Neuropathol Exp Neurol. 1993;52:655–66. doi: 10.1097/00005072-199311000-00013. [DOI] [PubMed] [Google Scholar]