Abstract
DNA sequencing of 268 individuals drawn from four US populations carrying two unresolved DRB1*14 alleles differing only outside the antigen recognition site identified DRB1*1454 in the majority. A database of 4222 human leukocyte antigen (HLA)-matched hematopoietic stem cell transplantation donor–recipient pairs was queried to determine the number likely mismatched for DRB1*140101/DRB1*1454 but matched for class I loci. A power calculation suggests that more than 88,000 transplants among European Americans will be needed to identify sufficient 7/8 allele-matched pairs to evaluate the impact of the DRB1*140101/DRB1*1454 mismatch on transplant outcome. Molecular modeling of the HLA-DR interaction with the T-cell receptor and with CD4 suggests that the amino acid substitution distinguishing the two alleles will have minimal impact on allorecognition.
Keywords: allele frequencies, DNA sequencing, hematopoietic stem cell transplantation, human leukocyte antigen-DRB1*14
DRB1*140101 and DRB1*1454 are examples of alleles that share the amino acid sequence of their antigen recognition site (ARS). The two alleles differ for a nonsynonymous nucleotide polymorphism occurring in exon 3, altering codon 112 in the second extracellular domain [TAC (tyrosine) to CAC (histidine)]. Horn et al. suggested that DRB1*1454 is likely to be relatively frequent among cells typed before its discovery as DRB1*1401 because DRB1*1454 shares the H112 with most other DRB1 alleles (1). The first goal of this study was to investigate the frequency of the two DRB1*14 ARS-identical alleles in four US population groups. A second goal was to determine the size of a donor–recipient population needed to evaluate the clinical impact of mismatching these alleles in unrelated donor hematopoietic stem cell transplantation (HSCT). A final goal was to predict the impact of the amino acid substitution distinguishing these allelic products using molecular modeling. Most human leukocyte antigen (HLA) typing focuses on resolving alleles differing in the ARS; the importance of matching for differences outside this functional site is not yet known.
A total of 268 unrelated individuals from the United States previously typed as DRB1*1401 positive prior to the discovery of DRB1*1454 were randomly selected to include 69 Asian Americans, 65 European Americans, 68 African Americans, and 66 Hispanic Americans. A sample size of at least 59 individuals gave us a 95% probability of detecting an allele present at 5% in the population. The ethnic/racial designations were self-described. To distinguish DRB1*1454 from DRB1*140101, it was necessary to amplify a 2100-bp DRB1*14-specific fragment with primers annealing in the first hypervariable region of exon 2 (a sequence specific to DRB1*03, *11, *13, and *14 alleles) and at DRB1-conserved codon 177 in exon 3 (Table 1). If the sample carried DRB1*03, *11, or *13 in addition to DRB1*14, the primer pair would amplify both DRB1 alleles; this occurred in 93 (35%) of the samples. For the 93 samples, a homozygous sequence was observed at codon 112 of exon 3 unless the cell carried DRB1*140101. Table 1 shows the allele frequencies of DRB1*1454 as found within the DRB1*1401-positive populations. Most individuals in all four population groups carried DRB1*1454 and not DRB1*140101. The latter was more frequent in European Americans (9.2%) and Hispanic Americans (15.2%). We sequenced the DRB3 allele in all the samples typed as DRB1*140101 or DRB1*1454 that did not carry a second allele associated with DRB3 (Table 1). Only two haplotypes DRB1*140101-DRB3*0201 and DRB1*1454-DRB3*0202 were observed.
Table 1. DRB1*1454 and DRB1*140101 allelea frequency distributions in four ethnic groups in the United States.
| Asian Americans, Nb (%) | European Americans, N (%) | African Americans, N (%) | Hispanic Americans, N (%) | |
|---|---|---|---|---|
| DRB1 | ||||
| Total | 69 (100) | 65 (100) | 68 (100) | 66 (100) |
| DRB1*140101 | 0 (0) | 6 (9.2) | 1 (1.5) | 10 (15.2) |
| DRB1*1454 | 69 (100) | 59 (90.8) | 67 (98.5) | 56 (84.8) |
| DRB3 | ||||
| Total | 58 (100) | 43 (100) | 31 (100) | 45 (100) |
| DRB1*140101-DRB3*0201 | 0 (0) | 5 (11.6) | 1 (3.2) | 4 (8.9) |
| DRB1*1454-DRB3*0202 | 58 (100) | 38 (88.4) | 30 (96.8) | 41 (91.1) |
PCR, polymerase chain reaction; HLA, human leukocyte antigen.
PCR primers [sense 5′GGAGTACTCTACGTCTGAG (nucleotides 111-129 exon 2); antisense 5′GCTCCACCTGGCAGGTGTAA (nucleotides 597-616 exon 3)] were used to link the sequence of exon 2 to exon 3. The PCR mixture in a final volume of 25 μl consisted of 500 ng genomic DNA, 1× High-Fidelity PCR buffer (Invitrogen, Carlsbad, CA), 50 mM MgCl2, 1.25 mM deoxynucleotide triphosphate (Invitrogen), 10% dimethyl sulfoxide (Sigma, St. Louis, MO), 10 pmol of each PCR primer (Invitrogen), and 0.25 μl Platinum® Taq DNA polymerase High Fidelity (Invitrogen). The PCR cycling program was as follows: an initial denaturation of 2 min at 96°C, followed by 5 cycles of 95°C for 30 s, 62°C for 30 s, and 68°C for 4 min 30 s; followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 4.5 min; and finally at 68°C for 10 min. Cycling was carried out using an Applied Biosystems 2720 thermal cycler (Applied Biosystems, Foster City, CA). Cycle sequencing was performed using the version 3.0 Big Dye™ Terminator Cycle Sequencing Kit (Applied Biosystems) according to the manufacturer's protocols. Sequencing primers included GGAGTACTCTACGTCTGAG (nucleotides 111-129 exon 2), CGCCCCGCGCCGCGCCTCAC (nucleotides 1-19 intron 2), TCCATCCTAAGGTGACTGTA (nucleotides 371-390 exon 3), and CCAGTCCGAGGAACTGTT (nucleotides 572-590 exon 3). Reaction products were identified with an Applied Biosystems Model 3730xl DNA analyzer, and sequence interpretation was performed with Assign software (Conexio Genomics, Applecross, Western Australia). Reference cells included CAP-DL11-2006 (DRB1*140101) and TER 400 (DRB1*1454). DRB3 was identified by sequencing using the HLA-DRB High Resolution Typing System (Applied Biosystems) following manufacturer's protocols.
N, number of individuals tested.
To evaluate the feasibility of studying the impact of mismatching for DRB1*140101/DRB1*1454 in unrelated donor HSCT, we queried the National Marrow Donor Program (NMDP)/Center for International Blood and Marrow Transplant Research (CIBMTR) database for transplant pairs matched for alleles of HLA-A, -B, -C, and -DRB1 carrying the unresolved allele alternative DRB1*140101 or DRB1*1454. The HLA typing of these pairs has already been described (2). We chose to evaluate these single DRB1 allele mismatches in the presence of allele matching at HLA-A, -B, and -C (i.e. 7/8 matches) because outcome studies have shown the importance of matching alleles at these key loci (3) and because methods to adjust for HLA mismatches at multiple loci in evaluating outcome are not yet optimal. Only pairs in which donor and recipient were both European American were evaluated because of the greater probability of finding mismatches for the two DRB1*14 alleles in this group. [The number of transplants of Hispanic patients is limited. In the NMDP/CIBMTR database, the majority of the pairs (83%) are European American/European American, 4.3% have one member of the pair listed as Hispanic American and the second as European American, and 1.7% are Hispanic American/Hispanic American. Pairs in which one member is European American and the second is Asian American (0.3%) or African American (1.1%) are also limited.] From the pool of four thousand two hundred and twenty-two 8/8 matched European American donor–recipient transplant pairs in the NMDP database, we identified only 102 pairs that carried the unresolved DRB1*140101/DRB1*1454 with matching at class I loci and the second DRB1 allele. Because all pairs were typed for DRB3 alleles, we used the strong DRB3 linkage to identify 12 pairs likely to be mismatched for DRB1*140101/DRB1*1454. Based on a 9% expected difference in survival between a 7/8 vs an 8/8 match (3), the analysis would require a database of eighty-eight thousand two hundred and sixty-seven 8/8 matched pairs with 80% power at a significance level of 0.05 (nQuery Advisor 5.0; Statistical Solutions Ltd., Boston, MA) to have sufficient DRB1*140101/*1454 mismatches to determine if this alteration in exon 3 does have an effect on outcome.
Based on the crystal structure of a DR–T-cell receptor complex [protein database (PDB): 1J8H] (4), the amino acid substitution at codon 112 defining the two DRB1*14 ARS-identical alleles lies outside the peptide-binding groove in a loop of the beta2 domain located near the cell surface (Figure 1). The polymorphism does not appear to be in a position to impact the conformation of the antigen-binding site or the interaction with the T-cell receptor. Based on a structure of a class II–CD4 complex (PDB: 1JL4) (5), the residue is near but not in the site for CD4 binding. Substitution of Y112 as found in DRB1*140101 may alter an intra-molecular salt bridge between H112 and E162. Although not appearing to affect CD4 binding, further studies would be required to determine if the affinity of the DR–CD4 interaction is altered. In summary, the impact of a substitution at residue 112 in affecting allorecognition is not obvious from the crystal structures, although it appears unlikely.
Figure 1.
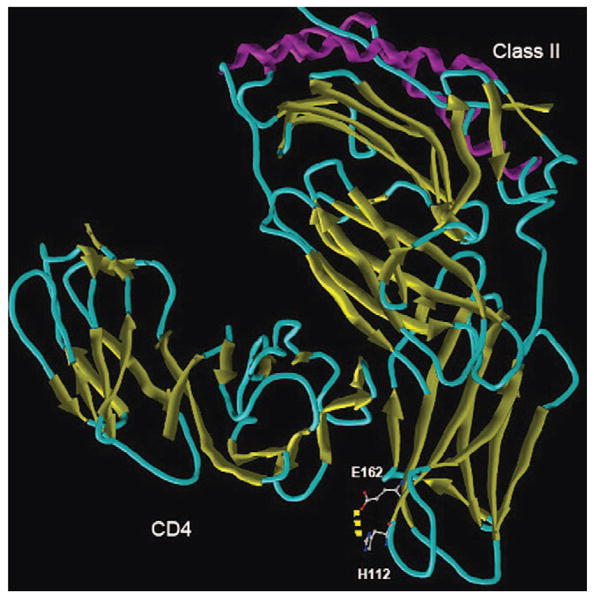
The structure of a class II–CD4 complex showing the position of polymorphism distinguishing DRB1*140101 and DRB1*1454. The alpha helices of the antigen recognition site are colored purple. The X-ray crystal structures of murine I-A/human CD4 [protein database (PDB): 1JL4] and DR/TCR (PDB: 1J8H) were used to model the impact of the H112Y substitution. Missing residues in the DR–TCR complex were reconstructed by homology modeling using modeller (9). Both variant structures were energy minimized and underwent 10 ps molecular dynamics (MD) simulations with a distant-dependent dielectric constant using the sander module of amber 8.0 (10). All the parameters were set to default for the minimization and MD simulations.
It should be noted that mismatching DRB1*140101/DRB1*1454 will also mismatch the DRB3 locus because of the strong linkage observed. Whether this dual mismatch has a greater impact than the DRB1 mismatch alone is not known. The two DRB3 allelic products differ at residues 86 and 164; residue 86 is a difference in the ARS but DRB3 is expressed at lower levels than DRB1 (6). The influence of the secondary DRB loci on transplant outcome has not yet been evaluated.
HLA-DR typing of volunteer hematopoietic stem cell donors at recruitment routinely uses reagents detecting only exon 2 polymorphisms, so DRB1*140101 is not distinguished from DRB1*1454. One issue to be resolved is whether it is necessary to distinguish between these alleles when matching hematopoietic stem cell donor and recipient because adding the typing of exon 3 will increase the time and complexity of the routine testing. We sought to evaluate the frequency at which a mismatch for DRB1*140101/DRB1*1454 will occur in transplants to estimate the number of retrospective transplant pairs needed to evaluate the impact on outcome. The large number of pairs required suggests that we may need to pool multiple HLA mismatches that differ only outside the ARS (e.g. A*02010101/A*0209, B*70201/B*0744) to evaluate impact, but this will require alternative alleles with sufficiently high frequencies to find mismatches in the transplant pairs. For example, another set of DR alleles identical in their ARS but differing elsewhere in the mature protein includes DRB1*120101 and DRB1*1206, but DRB1*1206 is not present at any detectable frequency in US populations (7).
The best alternative to a retrospective study is to determine the functional significance using in vitro assays of alloreactivity to predict the importance of this type of mismatch. An abstract by Oudshoorn et al. showed that in several donor–recipient pairs, a difference between DRB1*140101 and *1454 along with the DRB3 mismatch did not result in a primary mixed lymphocyte culture response (8). At present, data suggest that the clinical relevance is likely to be minimal but warrants additional laboratory investigation of in vitro alloreactivity for these ARS-identical allele combinations.
Acknowledgments
This research is supported by funding from the Office of Naval Research N00014-06-1-0726 (CKH and JN) and N00014-06-1-0704 (NMDP). The CIBMTR is supported by Public Health Service grant U24-CA76518 from the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, and the National Heart, Lung and Blood Institute; Health Resources and Services Administration (Department of Health and Human Services); and others. The views expressed in this article are those of the authors and do not reflect the official policy or position of the National Institutes of Health, Department of the Navy, the Department of Defense, NMDP, or any other agency of the US government.
References
- 1.Horn PA, Albis-Camps M, Verboom M, et al. The nature of diversity of HLA-DRB1 exon 3. TissueAntigens. 2007;70:335–7. doi: 10.1111/j.1399-0039.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- 2.Hurley CK, Fernandez-Vina M, Hildebrand WH, et al. A high degree of HLA disparity arises from limited allelic diversity: analysis of 1775 unrelated bone marrow transplant donor-recipient pairs. Hum Immunol. 2007;68:30–40. doi: 10.1016/j.humimm.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–83. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 4.Hennecke J, Wiley DC. Structure of a complex of the human alpha/beta T cell receptor (TCR) HA1.7, influenza hemagglutinin peptide, and major histocompatibility complex class II molecule, HLA-DR4 (DRA*0101 and DRB1*0401): insight into TCR cross-restriction and alloreactivity. J Exp Med. 2002;195:571–81. doi: 10.1084/jem.20011194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang JH, Meijers R, Xiong Y, et al. Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule. Proc Natl Acad Sci U S A. 2001;98:10799–804. doi: 10.1073/pnas.191124098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emery P, Mach B, Reith W. The different level of expression of HLA-DRB1 and -DRB3 genes is controlled by conserved isotypic differences in promoter sequence. Hum Immunol. 1993;38:137–47. doi: 10.1016/0198-8859(93)90531-5. [DOI] [PubMed] [Google Scholar]
- 7.Lazaro AM, Steiner NK, Cao K, et al. Searching for HLA-DRB1*1206 in volunteer marrow donors in four US population groups. Tissue Antigens. 2006;68:439–41. doi: 10.1111/j.1399-0039.2006.00689.x. [DOI] [PubMed] [Google Scholar]
- 8.Oudshoorn M, Lie JL, Roelen DL, Verduyn W, Claas FHJ. Is an HLA-DRB1*1454/HLA-DRB1*1401 mismatch an acceptable mismatch in hematopoietic stem cell transplantation? Tissue Antigens. 2008;71:315–6. [Google Scholar]
- 9.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 10.Case DA, Cheatham TE, III, Darden T, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]


