Abstract
The autotaxin (ATX) enzyme exhibits lysophospholipase D activity responsible for the conversion of lysophosphatidyl choline to lysophosphatidic acid (LPA). ATX and LPA have been linked to the initiation of atherosclerosis, cancer invasiveness, and neuropathic pain. ATX inhibition therefore offers currently unexploited therapeutic potential, and substantial interest in the development of ATX inhibitors is evident in the recent literature. Here we report the performance-based comparison of ligand-based pharmacophores developed on the basis of different combinations of ATX inhibitors in the training sets against an extensive database of compounds tested for ATX inhibitory activity, as well as with docking results of the actives against a recently reported ATX crystal structure. In general, pharmacophore models show better ability to select active ATX inhibitors binding in a common location when the ligand-based superposition shows a good match to the superposition of actives based on docking results. Two pharmacophore models developed on the basis of competitive inhibitors in combination with the single inhibitor crystallized to date in the active site of ATX were able to identify actives at rates over 40%, a substantial improvement over the <10% representation of active site-directed actives in the test set database.
Keywords: Autotaxin, pharmacophore, docking
1. Introduction
Autotaxin (ATX) is a 125kDa extracellular enzyme that facilitates numerous biological processes.[1–3] ATX was first identified in 1992 as a potent autocrine motility-stimulating factor isolated from the human A2058 melanoma cell line.[4] ATX is a member of the nucleotide pyrophosphatase phosphodiesterase (NPP) family based on the comparison of its sequence similarities and enzymatic properties.[5, 6]
ATX is found in several biological fluids and tissues, including the blood, kidney, and brain, where it contributes to normal development.[7–9] ATX exerts its function through its ability to hydrolyze lysophosphatidylcholine (LPC), as a lysophospholipase D (lysoPLD) enzyme, to produce the bioactive lipid lysophosphatidic acid (LPA) and is responsible for the majority of LPA production in blood.[3, 10–12] A variety of biological processes are mediated by LPA including angiogenesis, chemotaxis, smooth muscle contraction, brain development, and cell proliferation, migration, and survival with its primary effects being growth-related.[2, 13–15] Other important effects elicited by LPA include cellular differentiation, proliferation, stimulation of inflammation and suppression of apoptosis.[16–22] Many of these diverse signaling processes are stimulated through the activation of G-coupled protein receptors (GCPRs) specific to LPA.[19, 20, 23, 24]
Recent literature links ATX expression and LPA production with the promotion and proliferation of various cancers including melanomas, renal cell carcinomas, metastatic breast and ovarian cancers, thyroid carcinomas, Hodgkin lymphomas, neuroblastomas, and invasive glioblastoma multiforme. [25–34] ATX, through its production of LPA, is also thought to play a critical role in a variety of other human diseases, including obesity, diabetes, rheumatoid arthritis, neuropathic pain, multiple sclerosis, and Alzheimer’s disease.[35–43] Given the role of ATX in human disease, it has become an attractive drug target for pharmacological therapeutic development.
Until recently, an obstacle to developing potent inhibitors for ATX has been the lack of a three-dimensional protein structure. Therefore, ligand-based modeling has been of value for this system. Recently, a number of nonlipid small molecule inhibitors of ATX have been published using indirect structural data and the enzyme mechanism as guides.[1, 12, 35, 44–48] Preceding these small molecules, the only known ATX inhibitors were metal chelators and various lipid analogs that lacked structural diversity and characteristics typical of orally bioavailable compounds.[49–54] Lipid-based analogues also possess high numbers of rotatable bonds, limiting their value for ligand-based computational modeling techniques.[55] Crystallographic structures of ATX were reported in January 2011, and now provide a context in which to re-interpret results obtained using ligand-based methods.[56, 57]
In this paper, we examine the correspondence between ligand-based pharmacophore models selected on the basis of performance against a test set of compounds with known ATX inhibitory activity and the superpositions obtained upon docking the same ligands into a crystallographic structure of ATX. North et al. illustrated the use of pharmacophores, based on moderately potent ATX inhibitors, to be a dynamic tool in identification of several novel ATX inhibitors.[55] This was accomplished in two steps. First, specific points in space occupied by shared functional groups of known inhibitors were identified. Such points represent features necessary for biological interactions between ATX and its inhibitors. Second, database searching using these pharmacophores produced several novel inhibitors with potencies in the hundred nanomolar range. Using the inhibitors discovered by these prior pharmacophore models, along with additional published and in-house data on lipid and small molecule inhibitors of ATX, a database was compiled using the Molecular Operating Environment (MOE) software and updated pharmacophore models were developed using four combinations of input compounds (training sets). The pharmacophore models with the highest overlap and accuracy scores for each training set were then evaluated against the larger complete database (test set) to choose the pharmacophore model with the highest hit rate for comparison against docked positions of actives from the training set. The current work differs from that described by North et al.[55] in that inactive compounds and subsequently identified inhibitors with greater potency were included. Additionally, the pharmacophore models selected based on performance showed remarkable correspondence with docked conformations of the training compounds, suggesting that performance-based pharmacophore selection assists in the identification of the bioactive conformation.
2. Methods
2.1. Database generation
To develop and validate a pharmacophore model(s) to use for the identification of prospective ATX inhibitors, a database of known inhibitors was assembled. Compounds reported solely in the patent literature[58–60] were not included as activities in many of these patents are described based on exceeding threshold values and thus cannot always be unambiguously compared with our criterion for activity. All structures were input and ionized as expected at pH 7.4, based on the typical assay conditions used for ATX inhibition. Due to the relatively open and solvent-accessible nature of the binding pocket in the crystallographic structures of ATX, this is expected to be a reasonable estimate of the likely ionization state of bound inhibitors. The database contained fields for the compound structure, a molecule ID, % ATX inhibition, IC50, Ki (representing affinity for the enzyme), and Ki’ (representing affinity for the enzyme:substrate complex) with the identified substrate and concentrations from the literature noted. Identity of substrate was required as a variety of substrates have been used to characterize ATX inhibition, including the natural substrate LPC (usually assayed using an Amplex Red based system that couples choline production to a fluorescent readout[3]), FS-3 (an internally quenched substrate that gives rise to a fluorescent signal upon enzymatic hydrolysis[61]), and para-nitrophenyl thymidine monophosphate (which gives a UV-absorbing product upon enzymatic hydrolysis[3]). In total, this database contained 457 compounds drawn from the literature[1, 12, 35, 44–48, 55, 62–65] and unpublished in-house assays, of which 111 compounds were deemed active according to their percent inhibition (>50% ATX inhibition at 10 µM regardless of substrate) or IC50 values (≤ 10 µM). This cutoff for active inhibitors was chosen to be consistent with our previously published pharmacophores.[55] A conformational search was performed for each of the 457 compounds using the MMFF94x force field,[66] within the Molecular Operating Environment software package (MOE, Chemical Computing Group, Montreal, Canada),[67] to generate a multi-conformer database (test set) representing a range of conformations accessible to each molecule. This was important as the bioactive conformation of a molecule is not necessarily the lowest energy conformation. The search was performed using default settings with the following exceptions, 1) the method was changed to stochastic, 2) the rejection limit was changed to 10, and 3) the conformation limit was changed to 10. Up to 10 of the lowest energy conformations for each molecule were collected into the test set database.
2.2. Pharmacophore generation
Different ATX inhibitors have been characterized as showing varying inhibition mechanisms based on Michaelis-Menten kinetic studies,[1, 44, 55] although the majority of reported inhibitors have not been characterized to this extent. Pharmacophore model development based on compounds sharing a common biological activity assumes that the compounds interact with their biological target through a common set of interactions at a single site. Therefore, three different training sets containing small numbers of active and inactive compounds were compiled (Figure 1, sets A–C) and used to generate pharmacophore models prior to the availability of the crystallographic structure of ATX. Our hope was that one or more of the sets chosen would include compounds sharing a common interaction site. One additional set (Figure 1, set D) was compiled after the crystallographic structure became available and included actives showing competitive inhibition by Michaelis-Menten kinetics or binding at the active site by crystallography. The active compounds were chosen for their structural diversity and high percent inhibition of ATX. Corresponding inactive compounds were chosen for their structural similarity to the actives and low percent inhibition. Selection of the inactive molecules similar in shape to the active molecules assured steric clashes were not the sole reason for their inability to bind. The compounds chosen for all sets lacked known Ki’ values, which would indicate a mechanism other than competitive inhibition and a site of interaction distinct from the enzyme active site. However, not all chosen actives had been characterized for their mechanism of inhibition. The active molecules from each training set were flexibly aligned to one another and shared functional groups identified using the elucidation option in MOE. The elucidation is an automatic tool that develops pharmacophore queries based on a set of molecules with shared functional features. The default settings were used with the exception that the conformational search method was changed to stochastic, and the activity field was changed to % inhibition with >50 designating actives. This step was different than our previous pharmacophore modeling study due to the elucidation feature being new in MOE 2009.10,[67] whereas North et al.[55] used the MOE 2008.09 version of the software.
Figure 1.
Training set compounds used in pharmacophore elucidations.
2.3. Pharmacophore performance assessment
The top ten models as ranked by the overlap of active compounds in the training set were further validated using the in-house ATX inhibitor conformational database as a test set. Searches were performed in MOE[67] using the pharmacophore search option with default settings except for hit entries being changed to select, and hits changed to best per molecule. The models were tested to determine their accuracy in determining active and inactive ATX inhibitors. The metrics used to compare performance based on the training set and test set, summarized in Table 1, were overlap and accuracy for the molecules in the training set, as well as matches, true positives, true negatives, false positives, false negatives, hit rate, recovery percent, miss percent, and negative accuracy for the molecules in the test set. The overlap and accuracy scores were generated in the elucidation and are characterized as the score of the alignment of training set actives and the accuracy of the query in separating training set actives and inactives, respectively. Matches were defined as compounds the model chose as active from the test set conformational database. True positives were defined as matches that have been experimentally confirmed as active. True negatives were defined as experimentally confirmed inactives that were not selected as matches by the pharmacophore model. False positives were defined as matches experimentally demonstrated to be inactive. False negatives were defined as compounds failing to match the pharmacophore that experiments indicate are active. The hit rate is a percent of matches that are active (true positives divided by the sum of true positives and false positives, multiplied by one hundred). The recovery percent is the percent of actives in the test set selected by the pharmacophore model (true positives divided by the sum of true positives and false negatives, multiplied by one hundred). The miss percent is the percent of actives in the test set not selected as actives by the pharmacophore model (false negatives divided by the sum of true positives and false negatives, multiplied by one hundred). The recovery percent and miss percent should sum to one hundred. The negative accuracy is the percentage of inactives in the test set not selected as active by the pharmacophore model (true negatives divided by the sum of true negatives and false positives, multiplied by one hundred). Using these metrics, the pharmacophore with the maximized molecular grouping of overlap, accuracy, and hit rate from each of the four training sets was evaluated against the other three and the leading pharmacophore of the four chosen for further refinements.
Table 1.
Pharmacophore model performance metrics.
| Metrics calculated based on the training set | |
|---|---|
| Overlap | Score reflecting volume and functional group overlap of actives, maximum equal to number of actives |
| Accuracy | Fraction of compounds correctly matched (actives) and not matched (inactives) |
| Metrics calculated based on the test set | |
| Matches | Compounds selected by the pharmacophore model |
| True positives | Matches confirmed by experimental assay to be active |
| True negatives | Non-matches confirmed by experimental assay to be inactive |
| False positives | Matches confirmed by experimental assay to be inactive |
| False negatives | Non-matches confirmed by experimental assay to be active |
| Hit rate | Percent of matches that are true positives |
| Recovery percent | Percent of total actives (true positives + false negatives) in the matches list |
| Miss percent | Percent of total actives not in the matches list |
| Negative accuracy | Fraction of inactives not matching the pharmacophore model |
2.4. Docking
Protein DataBank (PDB)[68] entry 3NKM[56] was used as the source of the ATX structure for docking. The system was simplified by removing all non-protein atoms other than the catalytic site zinc ions. Hydrogen atoms were added and chelating histidine residues were tautomerized to the form with an available lone pair directed toward the appropriate zinc ion using the MOE[67] software. Active molecules shown in Figure 1 were constructed using the MOE software in the ionization states expected at pH 7.4. All files were prepared for docking using Raccoon coupled with MGL Tools version 1.5.4 and flexible docking was performed using Autodock Vina.[69] The docking box was centered at x, y, z coordinates 21.383, 36.532, 7.403 near the sidechain of N212 and covered a grid of 40, 30, and 30 Å in the x, y, and z directions.
3. Results
For the first time we have compiled data on all currently published ATX inhibitors. The resultant in-house database (test set) contains 3710 conformations of 457 compounds. A threshold of 50% inhibition at 10 µM or IC50 < 10 µM divides these compounds into 111 actives and 346 inactives. Using 4–8 inhibitors selected from this database, four training sets were developed from different combinations of input compounds. Numerous ligand-based pharmacophore models were developed for each training set, and the top 10 plus selected other models chosen based on metrics calculated using the training set molecules were further evaluated against the complete test set to compute the test set performance metrics. The pharmacophore models were then compared with the superposition of active training set compounds generated by docking to evaluate how well the test set performance metrics recapitulated the bioactive conformations of the active compounds.
3.1. Training Set A
Training set A includes two active compounds (KM04131 and PF8380) with activities in the sub-micromolar range, as well as two inactive compounds (ChemBridge 6035829 and 703705) as shown in Figure 1. The elucidation procedure generated 13896 pharmacophore models with 4–5 pharmacophore points each. Table 2 summarizes the test set search results of the top ten pharmacophores (sorted by overlap score) as well as the first pharmacophore sorted by overlap score with the highest accuracy resulting from training set A .
Table 2.
Performance of the 10 initially best-ranked and subsequent most accurate pharmacophore models from training set A.
| Training Set Metrics | Test Set Metrics | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| # | Overlap | Accuracy | p | Hits | TPa | FP | Hit rateb | Recovery % |
Negative Accuracy |
| 1 | 1.5269 | 0.50 | 0.0000 | 186 | 47 (16) | 139 | 20.2 (8.6) | 18.5 | 59.8 |
| 2 | 1.5098 | 0.75 | −0.1419 | 49 | 11 (7) | 38 | 18.3 (14.3) | 3.4 | 89.0 |
| 3 | 1.5062 | 0.75 | −0.1419 | 68 | 16 (9) | 52 | 19.0 (13.2) | 5.2 | 85.0 |
| 4 | 1.5052 | 0.75 | −0.1419 | 218 | 69 (28) | 149 | 24.0 (12.8) | 25.9 | 56.9 |
| 5 | 1.5038 | 0.50 | 0.0000 | 159 | 42 (15) | 117 | 20.9 (9.4) | 15.5 | 66.2 |
| 6 | 1.5005 | 0.75 | −0.1419 | 174 | 51 (16) | 123 | 22.7 (9.2) | 18.6 | 64.5 |
| 7 | 1.4953 | 0.50 | 0.0000 | 159 | 34 (14) | 125 | 17.6 (8.8) | 13.3 | 63.9 |
| 8 | 1.4902 | 0.50 | 0.0000 | 243 | 74 (27) | 169 | 23.3 (11.1) | 29.5 | 51.2 |
| 9 | 1.4869 | 0.75 | −0.1419 | 253 | 82 (40) | 171 | 24.5 (15.8) | 31.9 | 50.6 |
| 10 | 1.4850 | 0.75 | −0.1419 | 242 | 72 (37) | 170 | 22.9 (15.2) | 29.0 | 50.9 |
| 14 | 1.4771 | 1.00 | −0.5826 | 67 | 17 (11) | 50 | 20.2 (16.4) | 5.4 | 85.5 |
The parenthesized numbers denote true positives that share a common docking site.
The parenthesized numbers indicate hit rate based on 50 actives that share a common docking site with the training set actives.
The 10 pharmacophore models with the highest overlap scores showed training set accuracies ranging from 0.5 to 0.75. None of these models were able to identify active compounds from the test set at a hit rate substantially better than the 24% representation of actives within the 457 test compounds. It is notable, however, that the only models showing negative accuracy values above 80% were model 14 with a training set accuracy of 1.00 and models 2 and 3 with training set accuracies of 0.75.
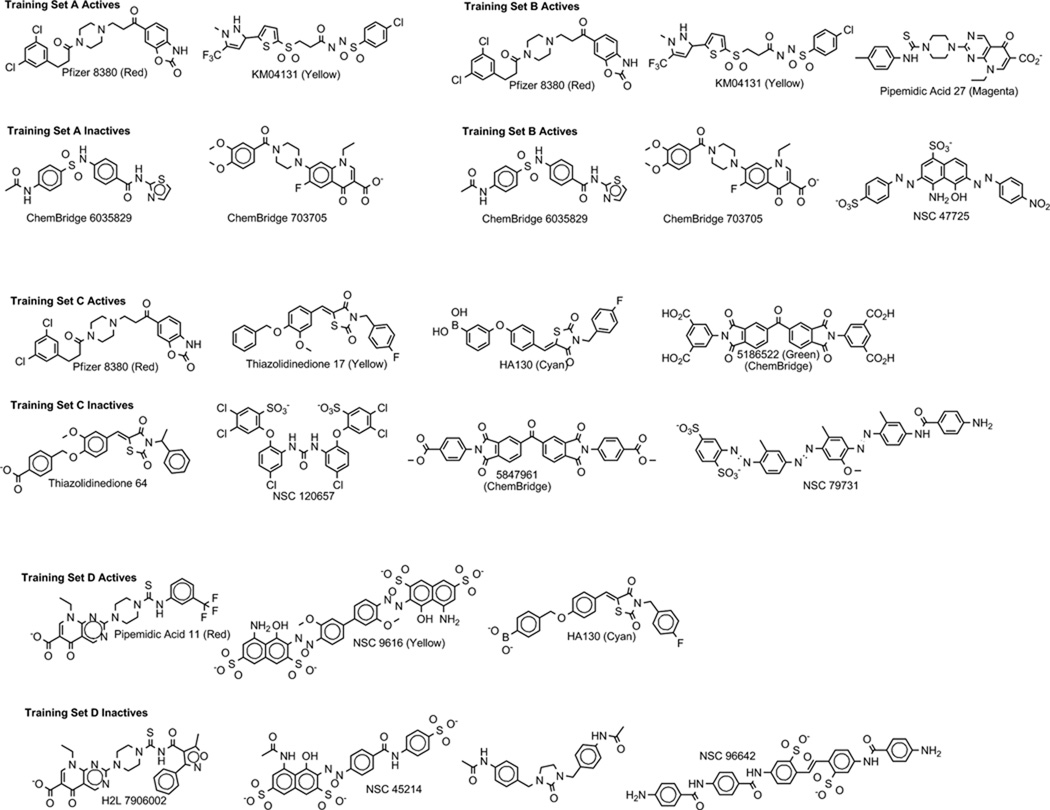
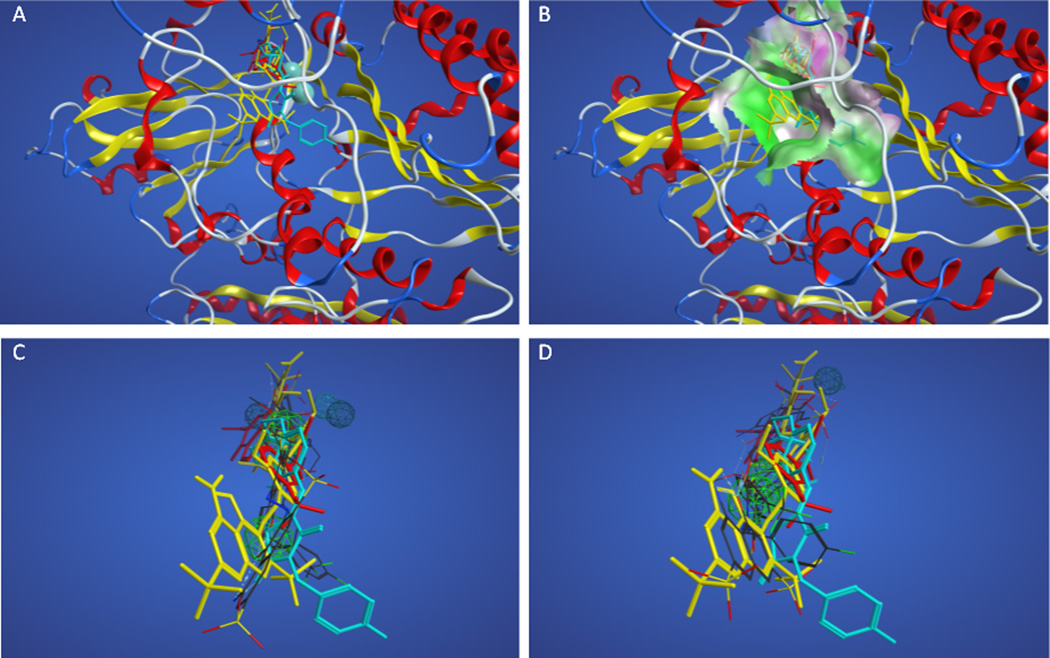
Due to the relatively poor performance of the pharmacophores resulting from training set A, the 111 actives (including the training set actives) were docked into the autotaxin crystal structure once it became available. Figures 2A and 2B show the superposition of the docked training set actives in the enzyme, supporting the idea that these actives share a binding site and are good candidates for the development of a pharmacophore model. The dichloro-substituted aromatic ring of PF8380 and the heterocyclic ring of KM04131 are superposed in a hydrophobic pocket formed by F273, A217, A304, and L216. The aromatic groups at the opposite ends of each structure are superposed in a more polar pocket, although lacking specific hydrogen bonding interactions. The binding site for these actives is distant from the divalent metal cations at the active site, with the closest atom in either ligand greater than 8 Å from either metal ion. Figures 2C–2D show how the superpositions of actives with the greatest negative accuracies (#2 and #14 in Table 2) compare with the docked superposition. The pharmacophore features and distances between the features for these two models are shown in Tables 3 and 4. The overall volume overlap for #2 is quite good, and pharmacophore points overlap with (or project from) appropriate chemical functionality both in the ligand-based superposition and in the docked superposition. The overall volume overlap for #14 is poor due to a self-interacting folded conformation identified in the ligand-based superposition that does not superpose well on the more extended conformation of the docked structures.
Figure 2.
Superpositions of training set A actives. Panel A. Superposition of actives based on docking with PF8380 shown in red sticks and KM04131 in yellow sticks. Panel B. Superposition of actives colored as in panel A based on docking showing nearby enzyme surface colored green for lipophilic and magenta for hydrophilic regions. Panels C–D. Docked superposition of actives colored as in panel A over ligand-based superposition of actives from Table 2 model #2 (Panel C) and model #14 (Panel D).
Table 3.
Pharmacophore features and inter-feature distances (Å) for model 2 from training set A.
| 2: Hydrophobic | 3: Hydrophobic | 4: Acceptor* | 5: Acceptor* | |
|---|---|---|---|---|
| 1: Aromatic/Pi | 6.85 | 9.95 | 5.22 | 6.64 |
| 2: Hydrophobic | 7.89 | 6.83 | 4.05 | |
| 3: Hydrophobic | 11.92 | 10.15 | ||
| 4: Acceptor* | 8.28 |
Denotes site points with which the noted chemical functionality could interact.
Table 4.
Pharmacophore features and inter-feature distances (Å) for model 14 from training set A.
| 2: Hydrophobic | 3: Hydrophobic | 4: Hydrophobic | 5: Acceptor* | |
|---|---|---|---|---|
| 1: Aromatic/Pi | 7.74 | 6.08 | 6.45 | 6.58 |
| 2: Hydrophobic | 3.42 | 5.54 | 9.45 | |
| 3: Hydrophobic | 7.15 | 6.05 | ||
| 4: Hydrophobic | 11.60 |
Denotes site points with which the noted chemical functionality could interact.
The entire set of 111 active compounds included a total of 50 compounds that docked in a common volume with the training set A actives. This common volume is defined to be within 4.5 Å of any atom of the two training set actives. The pharmacophore models derived from these training molecules should only be expected to identify the 50 actives that share a common binding site (10.9% of the test set database). Table 2 shows true positives and hit rates recalculated on the basis of defining actives as only the 50 actives sharing a binding site. This reanalysis indicates that all except one of the models that showed significant discrimination within the training set (models 2, 3, 4, 6, 9, 10 and 14) have hit rates that exceed the 10.9% representation of commonly-docked actives in the test set.
3.2. Training Set B
Training set B added one active, pipemidic acid 27, and inactive, NSC 47725, compound to those included in training set A (Figure 1) in the hope that improved performance would be obtained. The added active compound was chosen due to its lack of a flexible linker between two aromatic end groups in the hopes that self-interacting conformations such as that shown in Figure 2D would be prevented. The elucidation procedure with this training set generated 1683 pharmacophore models with 4–5 pharmacophore points. Table 5 summarizes the test set search results of the top ten pharmacophores (sorted by overlap score) as well as the first pharmacophore sorted by overlap score with the highest accuracy resulting from training set B.
Table 5.
Performance of the 10 initially best-ranked and subsequent most accurate pharmacophore models from training set B.
| Training Set Metrics | Test Set Metrics | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| # | Overlap | Accuracy | p | Hits | TP | FP | Hit rate | Recovery % |
Negative Accuracy |
| 1 | 2.0221 | 0.6667 | −0.1232 | 246 | 71 | 175 | 22.4 | 29.3 | 49.4 |
| 2 | 1.9688 | 0.6667 | −0.1232 | 231 | 63 | 168 | 21.4 | 26.1 | 51.4 |
| 3 | 1.9603 | 0.6667 | −0.1232 | 340 | 95 | 245 | 21.8 | 48.5 | 29.2 |
| 4 | 1.9518 | 0.5 | 0 | 273 | 70 | 203 | 20.4 | 32.4 | 41.3 |
| 5 | 1.9502 | 0.6667 | −0.1232 | 250 | 61 | 189 | 19.6 | 28.0 | 45.4 |
| 6 | 1.9481 | 0.6667 | −0.1232 | 264 | 65 | 199 | 19.8 | 30.7 | 42.5 |
| 7 | 1.9424 | 0.5 | 0 | 256 | 67 | 189 | 20.7 | 29.9 | 45.4 |
| 8 | 1.9321 | 0.6667 | −0.1232 | 288 | 79 | 209 | 21.5 | 36.6 | 39.6 |
| 9 | 1.9294 | 0.6667 | −0.1232 | 300 | 88 | 212 | 22.7 | 39.6 | 38.7 |
| 10 | 1.9288 | 0.5 | 0 | 263 | 72 | 191 | 21.5 | 31.7 | 44.8 |
| 31 | 1.8731 | 1 | −0.9523 | 98 | 29 | 69 | 22.8 | 9.5 | 80.1 |
The 10 pharmacophore models with the highest overlap scores showed training set accuracies ranging from 0.5 to 0.67. The 31st model as ranked by overlap score showed a training set accuracy of 1. All models failed to identify actives in the larger test set at a hit rate higher than the 24% representation of actives in the set.
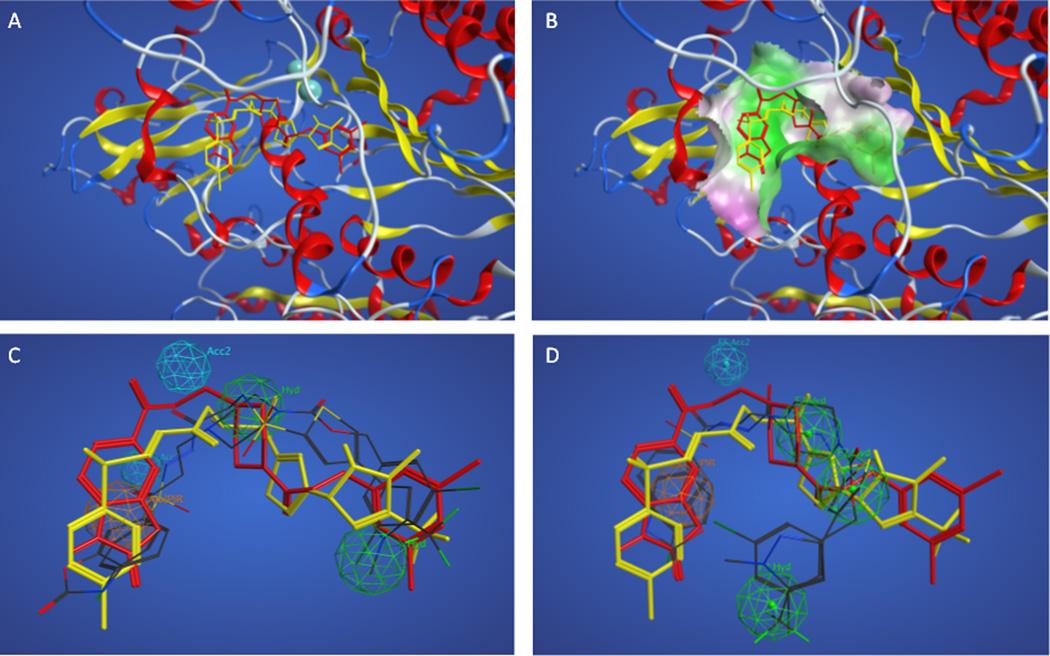
Due to the relatively poor performance of the pharmacophores resulting from training set B, the docking results for these actives were compared. Figures 3A and 3B show the superposition of the docked actives in the enzyme, indicating that the added active compound shares only a portion of the binding site occupied by the two actives originally selected for training set A, making the combined set of three compounds poor candidates for the development of a pharmacophore model. Figure 3C shows how the superposition of actives with the highest overlap score (#1 in Table 5) compares with the docked superposition. Table 6 shows the pharmacophore features and distances between features of this model. The overall volume overlap is biased toward the two compounds that dock into a common site, and three of four pharmacophore points overlap with (or project from) appropriate chemical functionality in the ligand-based superposition but only relate well to two of three compounds in the docked superposition. Figure 3D shows how the superposition of actives with the greatest accuracy (#31 in Table 5) compares with the docked superposition, and is similar to the superposition shown in Figure 3C. Table 7 shows the pharmacophore features and distances between features of this model. The selection of the more rigid pipemidic acid 27 as an active compound successfully selected against self-interacting conformations, but led to poor results due to the divergence of the binding site for this active compared to the other two actives in the training set.
Figure 3.
Superpositions of training set B actives. Panel A. Superposition of actives based on docking with PF8380, KM04131, and pipemidic acid 27 shown in red, yellow and magenta sticks, respectively. Panel B. Superposition of actives colored as in panel A based on docking showing nearby enzyme surface colored green for lipophilic and magenta for hydrophilic regions. Panels C–D. Docked superposition of actives colored as in panel A over ligand-based superposition of actives from Table 3 model #1 (Panel C) and model #31 (Panel D).
Table 6.
Pharmacophore features and inter-feature distances (Å) for model 1 from training set B.
| 2: Hydrophobic | 3: Hydrophobic | 4: Acceptor* | |
|---|---|---|---|
| 1: Aromatic/Pi | 11.69 | 6.94 | 4.71 |
| 2: Hydrophobic | 7.39 | 13.42 | |
| 3: Hydrophobic | 6.50 |
Denotes site points with which the noted chemical functionality could interact.
Table 7.
Pharmacophore features and inter-feature distances (Å) for model 31 from training set B.
| 2: Hydrophobic | 3: Hydrophobic | 4: Acceptor* | 5: Acceptor* | |
|---|---|---|---|---|
| 1: Hydrophobic | 3.13 | 10.48 | 6.92 | 11.31 |
| 2: Hydrophobic | 11.68 | 8.96 | 12.37 | |
| 3: Hydrophobic | 6.87 | 5.61 | ||
| 4: Acceptor* | 5.45 |
Denotes site points with which the noted chemical functionality could interact.
3.3. Training Set C
Training set C included one compound from training sets A and B, Pfizer 8380, and added three other active, thiazolidinedione 17, HA130 and ChemBridge5186522, and four inactive, thiazolidinedione 64, NSC 120657, ChemBridge5847961 and NSC 79731, compounds (Figure 1). The elucidation procedure with this training set generated 64 pharmacophore models with 4–5 pharmacophore points. Table 8 summarizes the test set search results of the top ten pharmacophores (sorted by overlap score) as well as the first pharmacophore sorted by overlap score with the highest accuracy resulting from training set C.
Table 8.
Performance of the 10 initially best-ranked and subsequent most accurate pharmacophore models from training set C.
| Training Set Metrics | Test Set Metrics | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| # | Overlap | Accuracy | p | Hits | TP | FP | Hit rate | Recovery % |
Negative Accuracy |
| 1 | 2.6127 | 0.5 | 0 | 289 | 78 | 211 | 21.3 | 36.6 | 39.0 |
| 2 | 2.6025 | 0.5 | 0 | 321 | 82 | 239 | 20.3 | 43.4 | 30.9 |
| 3 | 2.5972 | 0.625 | −0.1154 | 173 | 60 | 113 | 25.8 | 20.5 | 67.3 |
| 4 | 2.5426 | 0.5 | 0 | 285 | 80 | 205 | 21.9 | 36.2 | 40.8 |
| 5 | 2.5225 | 0.625 | −0.1154 | 207 | 68 | 139 | 24.7 | 24.7 | 59.8 |
| 6 | 2.5216 | 0.625 | −0.1154 | 271 | 75 | 196 | 21.7 | 33.3 | 43.4 |
| 7 | 2.5173 | 0.625 | −0.1154 | 220 | 65 | 155 | 22.8 | 25.4 | 55.2 |
| 8 | 2.5037 | 0.5 | 0 | 237 | 61 | 176 | 22.4 | 26.4 | 49.1 |
| 9 | 2.5031 | 0.5 | 0 | 367 | 106 | 261 | 22.4 | 24.7 | 24.6 |
| 10 | 2.4078 | 0.625 | −0.1154 | 219 | 62 | 157 | 22.0 | 27.0 | 54.6 |
| 42 | 2.2121 | 0.75 | −0.3507 | 116 | 32 | 84 | 21.6 | 27.0 | 90.8 |
The 10 pharmacophore models with the highest overlap scores showed training set accuracies ranging from 0.5 to 0.625. The 42nd model as ranked by overlap score showed a training set accuracy of 0.75. All models failed to identify actives in the larger test set at a hit rate notably higher than the 24% representation of actives in the set.
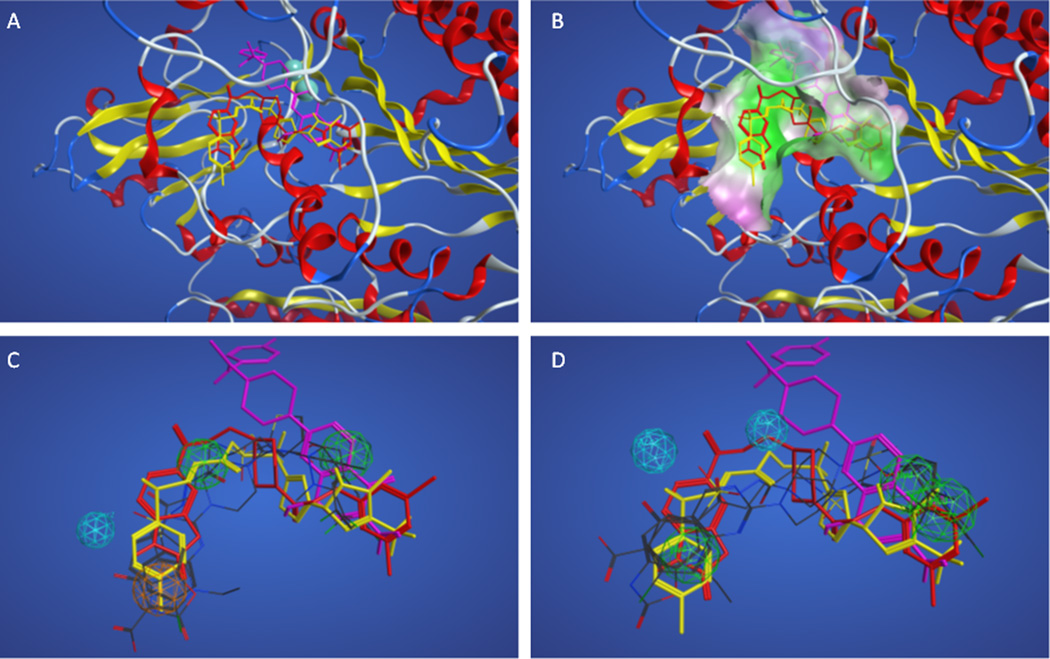
Due to the relatively poor performance of the pharmacophores resulting from training set C, the docking results for these actives were compared. Figures 4A and 4B show the superposition of the docked actives in the enzyme, indicating that the active compounds share only a portion of the binding site (left front side as shown), and diverge into four different subpockets, making these four active compounds poor candidates for the development of a ligand-based pharmacophore model. Figure 4C shows how the superposition of actives with the highest overlap score (#1 in Table 8) compares with the docked superposition. The overall volume overlap is poor due to the poor overlap of docked actives. Figure 3D shows how the superposition of actives with the greatest accuracy (#42 in Table 8) compares with the docked superposition, and shows equally poor correlation between the ligand-based superposition and the docked superposition. Tables 9 and 10 show the pharmacophore features and distances between features for these two models from training set C.
Figure 4.
Superpositions of training set C actives. Panel A. Superposition of actives based on docking with PF8380, thiazolidinedione 17, HA130, and 5186522 shown in red, yellow, cyan, and green sticks, respectively. Panel B. Superposition of actives colored as in panel A based on docking showing nearby enzyme surface colored green for lipophilic and magenta for hydrophilic regions. Panels C–D. Docked superposition of actives colored as in panel A over ligand-based superposition of actives from Table 3 model #1 (Panel C) and model #42 (Panel D).
Table 9.
Pharmacophore features and inter-feature distances (Å) for model 1 from training set C.
| 2: Hydrophobic | 3: Hydrophobic | 4: Acceptor* | |
|---|---|---|---|
| 1: Aromatic/Pi | 7.20 | 12.00 | 4.80 |
| 2: Hydrophobic | 6.00 | 7.20 | |
| 3: Hydrophobic | 12.00 |
Denotes site points with which the noted chemical functionality could interact.
Table 10.
Pharmacophore features and inter-feature distances (Å) for model 31 from training set C.
| 2: Hydrophobic | 3: Hydrophobic | 4: Acceptor* | 5: Acceptor* | |
|---|---|---|---|---|
| 1: Hydrophobic | 6.69 | 12.35 | 8.72 | 11.92 |
| 2: Hydrophobic | 5.66 | 5.50 | 5.92 | |
| 3: Hydrophobic | 7.73 | 3.63 | ||
| 4: Acceptor* | 4.95 |
Denotes site points with which the noted chemical functionality could interact.
3.4. Training Set D
The active compounds for training set D were selected on the basis of either Michaelis-Menten kinetic studies confirming a competitive mechanism of action (pipemidic acid 11 and NSC 9616) or on the basis of crystallographic evidence of binding at the active site (HA 130) (Figure 1). The elucidation procedure with this training set generated 8025 pharmacophore models with 4–5 pharmacophore points. Table 11 summarizes the test set search results of the top ten pharmacophores (sorted by overlap score) as well as the first pharmacophore sorted by overlap score with the highest accuracy resulting from training set D.
Table 11.
Performance of the 10 initially best-ranked and subsequent more accurate pharmacophore models from training set D.
| Training Set Metrics | Test Set Metrics | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| # | Overlap | Accuracy | p | Hits | TPa | FP | Hit rateb | Recovery % |
Negative Accuracy |
| 1 | 1.7396 | 0.5714 | −0.0802 | 141 | 42 (7) | 99 | 29.8 (5.0) | 37.8 | 71.4 |
| 2 | 1.7357 | 0.7143 | −0.2581 | 205 | 71 (23) | 134 | 34.6 (11.2) | 63.9 | 61.3 |
| 3 | 1.7357 | 0.7143 | −0.2581 | 205 | 71 (23) | 134 | 34.6 (11.2) | 63.9 | 61.3 |
| 4 | 1.7319 | 0.5714 | −0.0802 | 27 | 12 (11) | 15 | 44.4 (40.7) | 10.8 | 95.7 |
| 5 | 1.7302 | 0.7143 | −0.2581 | 14 | 6 (6) | 8 | 42.9 (42.9) | 5.4 | 97.7 |
| 6 | 1.7175 | 0.7143 | −0.2581 | 67 | 17 (8) | 50 | 25.4 (11.9) | 15.3 | 85.5 |
| 7 | 1.7171 | 0.7143 | −0.2581 | 16 | 4 (4) | 12 | 25.0 (25.0) | 3.6 | 96.5 |
| 8 | 1.717 | 0.5714 | −0.0802 | 30 | 5 (4) | 25 | 16.7 (13.3) | 4.5 | 92.8 |
| 9 | 1.7162 | 0.7143 | −0.2581 | 24 | 6 (5) | 18 | 25.0 (20.8) | 5.4 | 94.8 |
| 10 | 1.7094 | 0.5714 | −0.0802 | 248 | 67 (17) | 181 | 27.0 (6.9) | 60.4 | 47.7 |
| 135 | 1.6193 | 0.8571 | −0.5714 | 208 | 75 (27) | 133 | 36.1 (13.0) | 67.6 | 61.6 |
| 1350 | 1.4033 | 1.0000 | −1.1433 | 101 | 35 (25) | 66 | 34.7 (24.8) | 31.5 | 80.9 |
The parenthesized numbers denote true positives that dock at the active site.
The parenthesized numbers indicate hit rate based on 36 actives that dock at the active site.
The 10 pharmacophore models with the highest overlap scores showed training set accuracies ranging from 0.57 to 0.71 The 135th model as ranked by overlap score showed a training set accuracy of 0.86. A model with accuracy of 1.0 was ranked as the 1350th model based on overlap score. This set of pharmacophore models included two exhibiting hit rates above 40% (entries 4 and 5 in Table 11), a substantial enrichment over the 24% representation of active compounds in the test set.
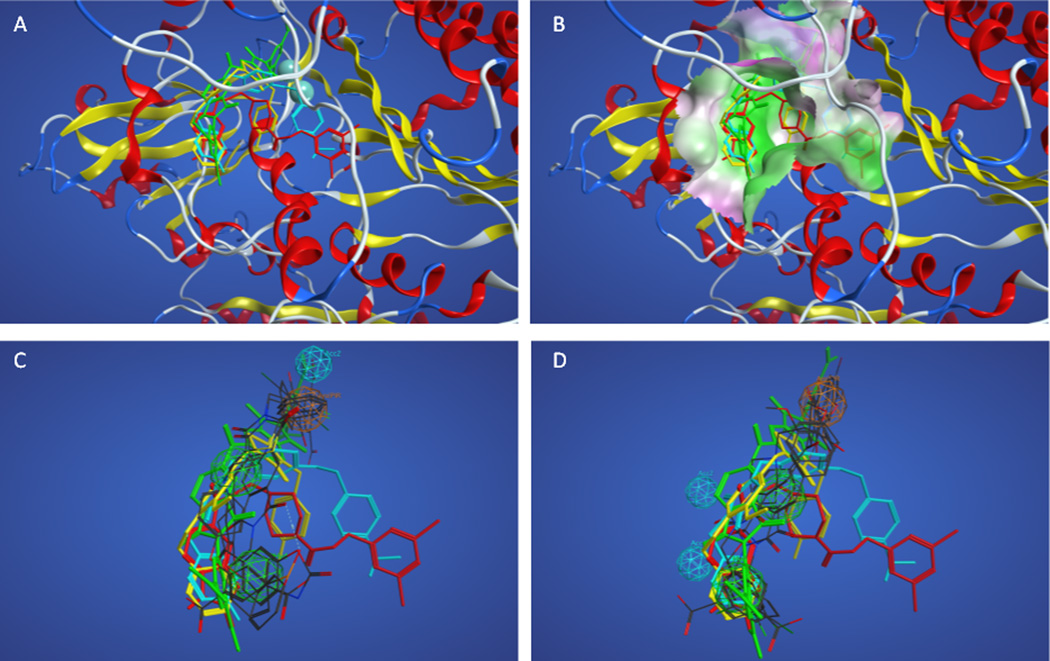
Although good performance was observed for some pharmacophores resulting from training set D, the docking results for actives in this training set were compared to evaluate whether the selection of compounds on the basis of similar behavior in Michaelis-Menten kinetic assays and active site crystallization yielded better overlap of actives. Figures 5A and 5B show the superposition of the docked actives in the enzyme, indicating that the active compounds share a common volume adjacent to the divalent metal cations (back left as shown) with the varying sizes of the active compounds producing varied occupancy of subsites distant from the divalent metal cations. Figure 5C and 5D show how the superposition of actives with the highest overlap score (#1 in Table 11) and highest hit rate (#4 in Table 11) compare with the docked superposition. The overall volume of overlap is quite good in both cases. Tables 12 and 13 show the pharmacophore features and distances between features for these two models.
Figure 5.
Superpositions of training set D actives. Panel A. Superposition of actives based on docking with pipemidic acid 11 and NSC 9616 as well as the crystallographic position of HA130 shown in red, yellow, and cyan sticks, respectively. Panel B. Superposition of actives colored as in panel A showing nearby enzyme surface colored green for lipophilic and magenta for hydrophilic regions. Panels C–D. Docked superposition of actives colored as in panel A over ligand-based superposition of actives from Table 3 model #1 (Panel C) and model #4 (Panel D).
Table 12.
Pharmacophore features and inter-feature distances (Å) for model 1 from training set D.
| 2: Hydrophobic | 3: Acceptor* | 4: Acceptor* | 5: Metal Ligator* | |
|---|---|---|---|---|
| 1: Hydrophobic | 6.65 | 15.18 | 15.16 | 16.21 |
| 2: Hydrophobic | 10.32 | 10.57 | 11.24 | |
| 3: Acceptor* | 5.75 | 1.65 | ||
| 4: Acceptor* | 4.78 |
Denotes site points with which the noted chemical functionality could interact.
Table 13.
Pharmacophore features and inter-feature distances (Å) for model 4 from training set D.
| 2: Hydrophobic | 3: Hydrophobic | 4: Acceptor* | 5: Metal Ligator* | |
|---|---|---|---|---|
| 1: Acceptor* | 2.83 | 7.40 | 8.97 | 8.97 |
| 2: Hydrophobic | 4.64 | 15.86 | 15.86 | |
| 3: Hydrophobic | 11.64 | 11.64 | ||
| 4: Acceptor* | 0.00 |
Denotes site points with which the noted chemical functionality could interact.
The entire set of 111 active compounds included a total of 36 compounds that docked in the active site, defined as compounds having atoms within 4.5 Å of one of the divalent metal cations. The pharmacophore models derived from training set D molecules should only be expected to identify the 36 actives that share a common binding site (7.9% of the test set database). Table 5 shows true positives and hit rates recalculated on the basis of defining actives as only the 36 actives sharing a binding site. This reanalysis indicates that the two models exhibiting hit rates above 40% based on the original classification of actives were, in fact, identifying actives that target the active site and retained hit rates over 40% when re-analyzed based on active site targeting. Entries 4 and 5 perform at a level far in excess of the 7.9% representation of active site-directed actives in the test set.
4. Discussion
The pharmacophores reported here represent a second-generation set of pharmacophore models for ATX inhibition. The first-generation pharmacophore models,[55] were constructed using the first reported non-lipid ATX inhibitors, [48] which showed potencies in the low to mid micromolar range. These previous models proved able to identify ATX inhibitors from the NCI database at rates ranging from 19 – 40%. A total of 106 compounds were selected for screening and 33 were experimentally confirmed to inhibit at least 50% of ATX activity at a 10 µM concentration. The most potent inhibitors exhibited competitive inhibition. These compounds are included in the multi-conformer test database. The pharmacophores reported here were developed using both active and inactive training molecules, including actives showing 10–100 fold improved potencies over those used to develop the first generation pharmacophores. Second generation pharmacophores trained using actives that co-localize to a common region when docked to ATX have also shown impressive hit rates. It is therefore instructive to compare performance of the current active site directed pharmacophores (derived from training set D) and our previous, first generation pharmacophore models on the same multi-conformer test set. Table 14 summarizes the pharmacophore features and distances between features for the first generation pharmacophore models.
Table 14.
Pharmacophore features and inter-feature distances (Å) for first generation pharmacophores.
| 2: Acceptor | 3: Aromatic | 4: Acceptor | 5: Aromatic | 6: Donor | |
|---|---|---|---|---|---|
| 1: Anion | 11.96 | 15.04 | 14.82 | 11.82 | 8.86 |
| 2: Acceptor | 3.77 | 4.58 | 3.66 | 5.68 | |
| 3: Aromatic | 3.97 | 5.97 | 9.25 | ||
| 4: Acceptor | 3.66 | 7.93 | |||
| 5: Aromatic | 4.32 |
The features of the first generation pharmacophores have several notable differences when compared to the features of model 4 obtained from training set D (Table 13). In particular, aromatic features appear, rather than the more general hydrophobic features. Additionally, anionic and hydrogen bond donor features are unique to the first generation pharmacophores.
Table 15 summarizes the performance of these first-generation models against the test database. These results show that the five and six point first-generation pharmacophores match very few members of the test set, and only one hit identified by these pharmacophores is a true positive. The four-point pharmacophores match between 9 and 50 structures from the test database, with hit rates ranging from 11 to 28%. The majority of matches are a subset of the 33 active structures screened based on searches in the NCI database with these pharmacophores. The matches fail to include the entire set of 33 experimentally confirmed active compounds selected from these searches for several reasons. First, the NCI database searches were limited to six distance ranges (a limitation of the web interface). Six distances ranges are sufficient to unambiguously define the position of four points in space, but not five or six points. The searches with the five and six point pharmacophores were therefore incompletely constrained and may have identified compounds that did not actually match the completely constrained pharmacophore. In contrast, the searches performed in the test set were performed in MOE, using the entire set of features and distances for each pharmacophore. Thus some compounds identified in the NCI searches with the incompletely constrained pharmacophores may be excluded in these searches. Second, the set of conformations stored in the NCI database and the set of conformations stored in our multi-conformer test set are unlikely to be exact matches. The pharmacophores may have failed to select compounds that could match due to under-representation of the conformational space of some compounds in the multi-conformer test set. The failure of these pharmacophores to match additional actives in this test set is not surprising given the specific requirement for one or two aromatic features. LPA analogs that show ATX inhibition are included in the test set, but most lack aromatic features and would not be selected by the first generation pharmacophores. Additional actives (such as PF8380) lack an anionic functional group and would not be selected as matches by the first-generation pharmacophore models.
Table 15.
Pharmacophore features and inter-feature distances (Å) for first generation pharmacophores. The 6 point pharmacophore includes all features from Table 14. The 5 point pharmacophores 5a, 5b, 5c, and 5d are lacking the single point 2, 3, 4, and 5, respectively. Pharmacophore 4a lacks features 4 and 5. Pharmacophore 4b lacks features 3 and 4. Pharmacophore 4c lacks features 2 and 5. Pharmacophore 4d lacks features 2 and 3.
| Test Set Metrics | ||||||
|---|---|---|---|---|---|---|
| # | Hits | TPa | FP | Hit rateb | Recovery % |
Negative Accuracy |
| 6 point | 0 | 0 | 0 | 0 | 0 | 100 |
| 5a | 2 | 0 | 2 | 0 | 0 | 99.4 |
| 5b | 5 | 0 | 5 | 0 | 0 | 98.6 |
| 5c | 1 | 0 | 1 | 0 | 0 | 99.7 |
| 5d | 2 | 1 | 1 | 50 | 0.9 | 99.7 |
| 4a | 9 | 1 | 8 | 11 | 0.9 | 97.7 |
| 4b | 23 | 5 | 18 | 22 | 4.5 | 94.8 |
| 4c | 9 | 2 | 7 | 22 | 1.8 | 98.0 |
| 4d | 50 | 14 | 36 | 28 | 12.6 | 89.6 |
5. Conclusions
We have compiled an extensive database of compounds tested for their inhibition of ATX phospholipase D activity. This database includes 111 actives defined as those showing greater than 50% inhibition at a 10 µM concentration or an IC50 less than 10 µM. Thirty-six of these actives are placed by Autodock Vina near the divalent metal cations at the active site, and 50 are placed at an alternate common binding site distant from those divalent metal cations. Prior to the availability of the ATX crystallographic structures, the only experimental evidence linking different actives to a common binding site was inhibition mechanism determined by Michaelis-Menten kinetics. Unfortunately, mechanism of inhibition is reported for a relatively small proportion of the 111 actives represented in our comprehensive database. We have demonstrated that ligand-based pharmacophore models developed using actives that dock in common locations show excellent correspondence with the docked positions of those actives (Figure 2C and Figure 5C,D). Pharmacophore model 2 from training set A showed slight enrichment of actives that dock distantly from the metal ions (14.3% of hitlist versus 10.9% representation of such actives in the test set, Table 2). Pharmacophore models 4 and 5 from training set D showed substantial enrichment of actives that dock at the active site (40.7 and 42.9% of hitlist versus 7.9% representation of such actives in the test set, Table 5). Pharmacophore model 4 from training set D showed superior performance at selecting a diverse set of actives from the test set relative to previously published first-generation pharmacophore models (compare Table 5 and Table 15). These results highlight the importance of careful training set selection to support the development of computational tools to guide lead discovery efforts for ATX, due to its very large binding pocket and extended hydrophobic tunnel leading into that binding pocket,[56, 57] as noted based on crystallographic results.
Highlights.
- A comprehensive database of over 450 ATX inhibitor structures and corresponding assay data has been compiled.
- Ligand based pharmacophore models based on four different training sets have been developed and validated against the comprehensive test database.
- The 111 active compounds from the comprehensive database have been docked against the recently reported ATX crystal structure and have been compared with the ligand-based pharmacophore models.
- Pharmacophore models based on active site-directed inhibitors show the greatest hit rate enhancement.
Acknowledgements
This material is based upon work supported by the National Science Foundation under CHE-0851880 and by the National Institutes of Health under HL 084007. Special thanks to the Chemical Computing Group for the MOE program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hoeglund AB, Howard AL, Wanjala IW, Pham TC, Parrill AL, Baker DL. Characterization of non-lipid autotaxin inhibitors. Bioorg Med Chem. 2010;18:769–776. doi: 10.1016/j.bmc.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 2.Okudaira S, Yukiura H, Aoki J. Biological roles of lysophosphatidic acid signaling through its production by autotaxin. Biochimie. 2010;92:698–706. doi: 10.1016/j.biochi.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Tokumura A, Majima E, Kariya Y, Tominaga K, Kogure K, Yasuda K, et al. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J Biol Chem. 2002;277:39436–39442. doi: 10.1074/jbc.M205623200. [DOI] [PubMed] [Google Scholar]
- 4.Stracke ML, Krutzsch HC, Unsworth EJ, Arestad A, Cioce V, Schiffmann E, et al. Identification, purification, and partial sequence analysis of autotaxin, a novel motility-stimulating protein. J Biol Chem. 1992;267:2524–2529. [PubMed] [Google Scholar]
- 5.Bollen M, Gijsbers R, Ceulemans H, Stalmans W, Stefan C. Nucleotide pyrophosphatases/phosphodiesterases on the move. Crit Rev Biochem Mol Biol. 2000;35:393–432. doi: 10.1080/10409230091169249. [DOI] [PubMed] [Google Scholar]
- 6.Murata J, Lee HY, Clair T, Krutzsch HC, Arestad AA, Sobel ME, et al. cDNA cloning of the human tumor motility-stimulating protein, autotaxin, reveals a homology with phosphodiesterases. J Biol Chem. 1994;269:30479–30484. [PubMed] [Google Scholar]
- 7.Tanaka M, Okudaira S, Kishi Y, Ohkawa R, Iseki S, Ota M, et al. Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. J Biol Chem. 2006;281:25822–25830. doi: 10.1074/jbc.M605142200. [DOI] [PubMed] [Google Scholar]
- 8.van Meeteren LA, Ruurs P, Stortelers C, Bouwman P, van Rooijen MA, Pradere JP, et al. Autotaxin, a secreted lysophospholipase D, is essential for blood vessel formation during development. Mol Cell Biol. 2006;26:5015–5022. doi: 10.1128/MCB.02419-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferry G, Giganti A, Coge F, Bertaux F, Thiam K, Boutin JA. Functional invalidation of the autotaxin gene by a single amino acid mutation in mouse is lethal. FEBS Lett. 2007;581:3572–3578. doi: 10.1016/j.febslet.2007.06.064. [DOI] [PubMed] [Google Scholar]
- 10.Umezu-Goto M, Kishi Y, Taira A, Hama K, Dohmae N, Takio K, et al. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J Cell Biol. 2002;158:227–233. doi: 10.1083/jcb.200204026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Y, Meier KE. Lysophospholipase D and its role in LPA production. Cell Signal. 2004;16:975–981. doi: 10.1016/j.cellsig.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Albers HM, Dong A, van Meeteren LA, Egan DA, Sunkara M, van Tilburg EW, et al. Boronic acid-based inhibitor of autotaxin reveals rapid turnover of LPA in the circulation. Proc Natl Acad Sci U S A. 2010;107:7257–7262. doi: 10.1073/pnas.1001529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Im DS. New intercellular lipid mediators and their GPCRs: an update. Prostaglandins Other Lipid Mediat. 2009;89:53–56. doi: 10.1016/j.prostaglandins.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Tigyi G, Parrill AL. Molecular mechanisms of lysophosphatidic acid action. Prog Lipid Res. 2003;42:498–526. doi: 10.1016/s0163-7827(03)00035-3. [DOI] [PubMed] [Google Scholar]
- 15.Osborne N, Stainier DY. Lipid receptors in cardiovascular development. Annu Rev Physiol. 2003;65:23–43. doi: 10.1146/annurev.physiol.65.092101.142235. [DOI] [PubMed] [Google Scholar]
- 16.Moolenaar WH. LPA: a novel lipid mediator with diverse biological actions. Trends Cell Biol. 1994;4:213–219. doi: 10.1016/0962-8924(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 17.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 18.Kingsbury MA, Rehen SK, Ye X, Chun J. Genetics and cell biology of lysophosphatidic acid receptor-mediated signaling during cortical neurogenesis. J Cell Biochem. 2004;92:1004–1012. doi: 10.1002/jcb.20061. [DOI] [PubMed] [Google Scholar]
- 19.Radeff-Huang J, Seasholtz TM, Matteo RG, Brown JH. G protein mediated signaling pathways in lysophospholipid induced cell proliferation and survival. J Cell Biochem. 2004;92:949–966. doi: 10.1002/jcb.20094. [DOI] [PubMed] [Google Scholar]
- 20.Meyer zu Heringdorf D, Jakobs KH. Lysophospholipid receptors: signalling, pharmacology and regulation by lysophospholipid metabolism. Biochim Biophys Acta. 2007;1768:923–940. doi: 10.1016/j.bbamem.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 21.Pua TL, Wang FQ, Fishman DA. Roles of LPA in ovarian cancer development and progression. Future Oncol. 2009;5:1659–1673. doi: 10.2217/fon.09.120. [DOI] [PubMed] [Google Scholar]
- 22.Bourgoin SG, Zhao C. Autotaxin and lysophospholipids in rheumatoid arthritis. Curr Opin Investig Drugs. 2010;11:515–526. [PubMed] [Google Scholar]
- 23.Moolenaar WH. Bioactive lysophospholipids and their G protein-coupled receptors. Exp Cell Res. 1999;253:230–238. doi: 10.1006/excr.1999.4702. [DOI] [PubMed] [Google Scholar]
- 24.Parrill AL. Lysophospholipid interactions with protein targets. Biochim Biophys Acta. 2008;1781:540–546. doi: 10.1016/j.bbalip.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.David M, Wannecq E, Descotes F, Jansen S, Deux B, Ribeiro J, et al. Cancer cell expression of autotaxin controls bone metastasis formation in mouse through lysophosphatidic acid-dependent activation of osteoclasts. PLoS One. 2010;5:e9741. doi: 10.1371/journal.pone.0009741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng Y, Kakehi Y, Nouh MA, Tsunemori H, Sugimoto M, Wu XX. Gene expression profiles of lysophosphatidic acid-related molecules in the prostate: relevance to prostate cancer and benign hyperplasia. Prostate. 2009;69:283–292. doi: 10.1002/pros.20879. [DOI] [PubMed] [Google Scholar]
- 27.Kishi Y, Okudaira S, Tanaka M, Hama K, Shida D, Kitayama J, et al. Autotaxin is overexpressed in glioblastoma multiforme and contributes to cell motility of glioblastoma by converting lysophosphatidylcholine to lysophosphatidic acid. J Biol Chem. 2006;281:17492–17500. doi: 10.1074/jbc.M601803200. [DOI] [PubMed] [Google Scholar]
- 28.Hoelzinger DB, Mariani L, Weis J, Woyke T, Berens TJ, McDonough WS, et al. Gene expression profile of glioblastoma multiforme invasive phenotype points to new therapeutic targets. Neoplasia. 2005;7:7–16. doi: 10.1593/neo.04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kehlen A, Englert N, Seifert A, Klonisch T, Dralle H, Langner J, et al. Expression, regulation and function of autotaxin in thyroid carcinomas. Int J Cancer. 2004;109:833–838. doi: 10.1002/ijc.20022. [DOI] [PubMed] [Google Scholar]
- 30.Black EJ, Clair T, Delrow J, Neiman P, Gillespie DA. Microarray analysis identifies Autotaxin, a tumour cell motility and angiogenic factor with lysophospholipase D activity, as a specific target of cell transformation by v-Jun. Oncogene. 2004;23:2357–2366. doi: 10.1038/sj.onc.1207377. [DOI] [PubMed] [Google Scholar]
- 31.Yang SY, Lee J, Park CG, Kim S, Hong S, Chung HC, et al. Expression of autotaxin (NPP-2) is closely linked to invasiveness of breast cancer cells. Clin Exp Metastasis. 2002;19:603–608. doi: 10.1023/a:1020950420196. [DOI] [PubMed] [Google Scholar]
- 32.Nam SW, Clair T, Campo CK, Lee HY, Liotta LA, Stracke ML. Autotaxin (ATX), a potent tumor motogen, augments invasive and metastatic potential of ras-transformed cells. Oncogene. 2000;19:241–247. doi: 10.1038/sj.onc.1203263. [DOI] [PubMed] [Google Scholar]
- 33.Zhang G, Zhao Z, Xu S, Ni L, Wang X. Expression of autotaxin mRNA in human hepatocellular carcinoma. Chin Med J (Engl) 1999;112:330–332. [PubMed] [Google Scholar]
- 34.Kawagoe H, Stracke ML, Nakamura H, Sano K. Expression and transcriptional regulation of the PD Ialpha/autotaxin gene in neuroblastoma. Cancer Res. 1997;57:2516–2521. [PubMed] [Google Scholar]
- 35.Parrill AL, Baker DL. Autotaxin inhibitors: a perspective on initial medicinal chemistry efforts. Expert Opin Ther Pat. 2010;20:1619–1625. doi: 10.1517/13543776.2010.533658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pradere JP, Tarnus E, Gres S, Valet P, Saulnier-Blache JS. Secretion and lysophospholipase D activity of autotaxin by adipocytes are controlled by N-glycosylation and signal peptidase. Biochim Biophys Acta. 2007;1771:93–102. doi: 10.1016/j.bbalip.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Ferry G, Tellier E, Try A, Gres S, Naime I, Simon MF, et al. Autotaxin is released from adipocytes catalyzes lysophosphatidic acid synthesis activates preadipocyte proliferation. Up-regulated expression with adipocyte differentiation and obesity. J Biol Chem. 2003;278:18162–18169. doi: 10.1074/jbc.M301158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inoue M, Xie W, Matsushita Y, Chun J, Aoki J, Ueda H. Lysophosphatidylcholine induces neuropathic pain through an action of autotaxin to generate lysophosphatidic acid. Neuroscience. 2008;152:296–298. doi: 10.1016/j.neuroscience.2007.12.041. [DOI] [PubMed] [Google Scholar]
- 39.Inoue M, Ma L, Aoki J, Ueda H. Simultaneous stimulation of spinal NK1 and NMDA receptors produces LPC which undergoes ATX-mediated conversion to LPA, an initiator of neuropathic pain. J Neurochem. 2008;107:1556–1565. doi: 10.1111/j.1471-4159.2008.05725.x. [DOI] [PubMed] [Google Scholar]
- 40.Inoue M, Ma L, Aoki J, Chun J, Ueda HAutotaxin. a synthetic enzyme of lysophosphatidic acid (LPA), mediates the induction of nerve-injured neuropathic pain. Mol Pain. 2008;4:6. doi: 10.1186/1744-8069-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagai J, Uchida H, Matsushita Y, Yano R, Ueda M, Niwa M, et al. Autotaxin and lysophosphatidic acid1 receptor-mediated demyelination of dorsal root fibers by sciatic nerve injury and intrathecal lysophosphatidylcholine. Mol Pain. 6:78. doi: 10.1186/1744-8069-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao C, Fernandes MJ, Prestwich GD, Turgeon M, Di Battista J, Clair T, et al. Regulation of lysophosphatidic acid receptor expression and function in human synoviocytes: implications for rheumatoid arthritis? Mol Pharmacol. 2008;73:587–600. doi: 10.1124/mol.107.038216. [DOI] [PubMed] [Google Scholar]
- 43.Kehlen A, Lauterbach R, Santos AN, Thiele K, Kabisch U, Weber E, et al. IL-1 beta- and IL-4-induced down-regulation of autotaxin mRNA and PC-1 in fibroblast-like synoviocytes of patients with rheumatoid arthritis (RA) Clin Exp Immunol. 2001;123:147–154. doi: 10.1046/j.1365-2249.2001.01432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoeglund AB, Bostic HE, Howard AL, Wanjala IW, Best MD, Baker DL, et al. Optimization of a Pipemidic Acid Autotaxin Inhibitor. J Med Chem. 2010;53:1056–1066. doi: 10.1021/jm9012328. [DOI] [PubMed] [Google Scholar]
- 45.Albers HM, van Meeteren LA, Egan DA, van Tilburg EW, Moolenaar WH, Ovaa H. Discovery and optimization of boronic acid based inhibitors of autotaxin. J Med Chem. 2010;53:4958–4967. doi: 10.1021/jm1005012. [DOI] [PubMed] [Google Scholar]
- 46.Braddock D. Small molecule inhibitors of autotaxin, and methods of use for the treatment of cancer. USA: Yale University; 2009. 82 pp. Application: WO WO. [Google Scholar]
- 47.Saunders LP, Ouellette A, Bandle R, Chang WC, Zhou H, Misra RN, et al. Identification of small-molecule inhibitors of autotaxin that inhibit melanoma cell migration and invasion. Mol Cancer Ther. 2008;7:3352–3362. doi: 10.1158/1535-7163.MCT-08-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parrill AL, Echols U, Nguyen T, Pham TC, Hoeglund A, Baker DL. Virtual screening approaches for the identification of non-lipid autotaxin inhibitors. Bioorg Med Chem. 2008;16:1784–1795. doi: 10.1016/j.bmc.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 49.Jiang G, Xu Y, Fujiwara Y, Tsukahara T, Tsukahara R, Gajewiak J, et al. Alpha-substituted phosphonate analogues of lysophosphatidic acid (LPA) selectively inhibit production and action of LPA. ChemMedChem. 2007;2:679–690. doi: 10.1002/cmdc.200600280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui P, Tomsig JL, McCalmont WF, Lee S, Becker CJ, Lynch KR, et al. Synthesis and biological evaluation of phosphonate derivatives as autotaxin (ATX) inhibitors. Bioorg Med Chem Lett. 2007;17:1634–1640. doi: 10.1016/j.bmcl.2006.12.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gududuru V, Zeng K, Tsukahara R, Makarova N, Fujiwara Y, Pigg KR, et al. Identification of Darmstoff analogs as selective agonists and antagonists of lysophosphatidic acid receptors. Bioorg Med Chem Lett. 2006;16:451–456. doi: 10.1016/j.bmcl.2005.08.096. [DOI] [PubMed] [Google Scholar]
- 52.Baker DL, Fujiwara Y, Pigg KR, Tsukahara R, Kobayashi S, Murofushi H, et al. Carba analogs of cyclic phosphatidic acid are selective inhibitors of autotaxin and cancer cell invasion and metastasis. J Biol Chem. 2006;281:22786–22793. doi: 10.1074/jbc.M512486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Meeteren LA, Ruurs P, Christodoulou E, Goding JW, Takakusa H, Kikuchi K, et al. Inhibition of autotaxin by lysophosphatidic acid and sphingosine 1-phosphate. J Biol Chem. 2005;280:21155–21161. doi: 10.1074/jbc.M413183200. [DOI] [PubMed] [Google Scholar]
- 54.Clair T, Koh E, Ptaszynska M, Bandle RW, Liotta LA, Schiffmann E, et al. L-histidine inhibits production of lysophosphatidic acid by the tumor-associated cytokine, autotaxin. Lipids Health Dis. 2005;4:5. doi: 10.1186/1476-511X-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.North EJ, Howard AL, Wanjala IW, Pham TC, Baker DL, Parrill AL. Pharmacophore development and application toward the identification of novel, small-molecule autotaxin inhibitors. J Med Chem. 2010;53:3095–3105. doi: 10.1021/jm901718z. [DOI] [PubMed] [Google Scholar]
- 56.Nishimasu H, Okudaira S, Hama K, Mihara E, Dohmae N, Inoue A, et al. Crystal structure of autotaxin and insight into GPCR activation by lipid mediators. Nat Struct Mol Biol. 2011;18:205–212. doi: 10.1038/nsmb.1998. [DOI] [PubMed] [Google Scholar]
- 57.Hausmann J, Kamtekar S, Christodoulou E, Day JE, Wu T, Fulkerson Z, et al. Structural basis of substrate discrimination and integrin binding by autotaxin. Nat Struct Mol Biol. 2011;18:198–204. doi: 10.1038/nsmb.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schiemann K, Schultz M, Blaukat A, Kober I, Staehle W. Preparation of carbamoylthiazoles as anticancer agents. Germany: Merck Patent GmbH; 2009. 102 pp. Application: WO WO. [Google Scholar]
- 59.Schiemann K, Schultz M, Staehle W, Kober I, Wienke D, Krier M. Preparation of 2,5-diamino-substituted pyrido[4,3-d]pyrimidines as autotaxin inhibitors useful in treating cancer. Germany: Merck Patent GmbH; 2010. 177 pp. Application: WO WO. [Google Scholar]
- 60.Staehle W, Kober I, Schiemann K, Schultz M, Wienke D. Preparation of benzo[b][1,6]naphthyridines as inhibitors of autotaxin for the treatment of tumors. Germany: Merck Patent GmbH; 2010. 142 pp. Application: WO WO. [Google Scholar]
- 61.Ferguson CG, Bigman CS, Richardson RD, van Meeteren LA, Moolenaar WH, Prestwich GD. Fluorogenic phospholipid substrate to detect lysophospholipase D/autotaxin activity. Org Lett. 2006;8:2023–2036. doi: 10.1021/ol060414i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gierse JK, Thorarensen A, Beltey K, Bradshaw-Pierce E, Cortes-Burgos L, Hall T, et al. A Novel Autotaxin Inhibitor Reduces Lysophosphatidic Acid Levels in Plasma and the Site of Inflammation. J Pharmacol Exp Ther. 2010;334:310–317. doi: 10.1124/jpet.110.165845. [DOI] [PubMed] [Google Scholar]
- 63.Prestwich GD, Gajewiak J, Zhang H, Xu X, Yang G, Serban M. Phosphatase-resistant analogues of lysophosphatidic acid: agonists promote healing, antagonists and autotaxin inhibitors treat cancer. Biochim Biophys Acta. 2008;1781:588–594. doi: 10.1016/j.bbalip.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parrill AL, Baker DL. Autotaxin inhibition: challenges and progress toward novel anti-cancer agents. Anticancer Agents Med Chem. 2008;8:917–923. doi: 10.2174/187152008786847765. [DOI] [PubMed] [Google Scholar]
- 65.Moulharat N, Fould B, Giganti A, Boutin JA, Ferry G. Molecular pharmacology of adipocyte-secreted autotaxin. Chem Biol Interact. 2008;172:115–124. doi: 10.1016/j.cbi.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 66.Halgren TA, Merck Molecular Force Field I. Basis, Form, Scope, Parameterization, and Performance of MMFF94*. J Comput Chem. 1996;17:490–519. [Google Scholar]
- 67.MOE. Montreal: Chemical Computing Group; 2010. [Google Scholar]
- 68.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]