Abstract
Objective
We investigated genetic variants predictive of muscular side effects in patients treated with statins. We utilized a physiogenomic approach to prototype a multi-gene panel correlated with statin-induced myalgia.
Background
Statin induced myalgia occurs in ∼10% of lipid clinic outpatients. Its clinical manifestation may depend in part upon gene variation from patient to patient.
Methods
We genotyped 793 patients (377 with myalgia and 416 without) undergoing statin therapy at four U.S. outpatient clinic sites to evaluate 31 candidate genes from the literature for their association with statin-induced common myalgia.
Results
Three previously hypothesized candidate genes were validated: COQ2 (rs4693570) encoding para-hydroxybenzoate-polyprenyltransferase, which participates in the biosynthesis of coenzyme Q10 (p<0.000041); ATP2B1 (rs17381194) which encodes a calcium transporting ATPase involved in calcium homeostasis (p<0.00079); and DMPK (rs672348) which encodes a protein kinase implicated in myotonic dystrophy (p<0.0016).
Conclusions
The candidate genes COQ2, ATP2B1, and DMPK, representing pathways involved in myocellular energy transfer, calcium homeostasis, and myotonic dystonia, respectively, were validated as markers for the common myalgia observed in patients receiving statin therapy. The three genes integrated into a physiogenomic predictive system could be relevant to myalgia diagnosis and prognosis in clinical practice.
Keywords: statins, pharmacogenetics, physiogenomics, myalgia, myopathy, dyslipidemia, personalized medicine
Introduction
Statins are the most prescribed drugs in the United States(1) and the world(2). Atorvastatin, simvastatin, and rosuvastatin comprise 85% of the prescriptions written in the U.S.(1) The success of these drugs in preventing cardiovascular events (3) has fostered increasingly aggressive usage and dosing. The main clinical side effect of statins is statin-induced myopathy. Statin-associated myopathy includes myositis with marked CK elevations and frank rhabdomyolysis, but also include the myalgias, weakness, and muscle cramps all with or without increases in serum creatine kinase activity(4). Statin myalgia is frequent and reported in 3-20% of patients on statins(4;5). It often requires alteration of therapy, and may reduce compliance to treatment with statins.
The mechanism(s) by which statins induce muscle complaints is not known. The etiologies that have been suggested include decreased sarcolemmal or sarcoplasmic reticular cholesterol, decreased production of coenzyme Q10 (CoQ10) or ubiquinone, decreased production of prenylated proteins, changes in fat metabolism, increased sarcolemmal uptake of cholesterol or phytosterols, failure to replace damaged muscle protein via the ubiquitin pathway, disruption of sarcolemmal calcium homeostasis, inflammation, and inhibition of selenoprotein synthesis(4;6). Statin-associated muscle complaints appear to cluster in families(5) and genetic contributors have been suggested (6-9), but the variety of possible mechanisms, the difficulty in objectively measuring myalgia, and the fact that prior studies have used relatively small sample sizes have made identification of genetic factors difficult. Physiological genomics (physiogenomics) is an engineering approach that considers physiological and genetic factors as system inputs and phenotype as output(10). The approach is most valuable when applied to responses to an intervention, and has been successfully applied to statin treatment (7;8).
Consequently, we performed a physiogenomic study to validate candidate gene associations that examined, a priori, the relationship of genes with known or hypothetical roles in myalgia in a group of 793 outpatients undergoing statin therapy. The diagnosis of myalgia was ascertained by clinicians experienced in statin therapy, and the study sample was enriched with patients diagnosed with statin-induced myalgia in order to enhance statistical power.
Subjects and Methods
Study Design and Sample
The Statin Induction and Neuro-Myopathy (SINM) study is a non-interventional, cross-sectional study of neuromuscular side effects in 793 patients treated for hyperlipidemia. Patients were recruited through the Preventive Cardiology Clinic (CT-b, n=214) Hartford Hospital and other affiliated clinics in Hartford, CT (CT-a, N=182), the Cardiovascular Research Institute and a clinic at the San Francisco General Hospital (SF, N=353), and the Lipid Clinic at the Rogosin Institute, Rockefeller University, New York (NY, N=39). Patients provided written informed consent, including permission to use the sample in genomic studies, as approved by the IRBs of all participating institutions.
Definition of Phenotype and Covariates
Patients were assessed for myopathy by a qualified investigator and were classified as suffering myalgia or “muscle pain” or not. A myalgia score of 1 was assigned to patients with definite myalgia when any of the following criteria could be established from the patient medical chart: (1) muscle pain began concurrently with the initiation of statin therapy; (2) muscle pain coincided with an increase in statin dose; (3) muscle pain resolved when the inducing statin was switched to another statin; or (4) muscle pain resolved when statin therapy was discontinued. A myalgia score of 0 was assigned when none of the criteria were met.
For association testing, the dichotomous myalgia score was adjusted for age, gender, heritage, and study site by logistic regression. The probability derived from this regression was then used as a quantitative trait in the association analysis. Heritage was defined as combination of race and ethnicity with Hispanic ethnicity treated as a category in addition to the groups that include European Americans, African American, Asian American, and Native American.
Genotyping
Blood samples for DNA contained either ethylenediamine tetraacetic acid or citrate, and DNA was extracted from leukocytes in 3.5 mL or more of whole blood using a DNA isolation kit (PureGene Gentra®, Qiagen, Valencia, CA). The extracted DNA was genotyped using the Human Hap 1M OmniQuad Genotyping BeadChip of Illumina (San Diego, CA) based on the Infinium Total-Genome Genotyping platform. An iScan scanner was used to read the fluorescence signals from the chip and the raw data processed using the GenomeStudio software. Careful quality control was performed, assuring that all loci were called in at least 99% of individuals. SNPs with allele frequency of less than 3% were excluded. We also determined the genetic gender by the presence or absence of a) heterozygote signals of markers on the X chromosome and b) any signal of markers on the Y chromosome. Comparison with clinically reported gender identified 15 suspect reports, which patients were not considered as part of the overall cohort.
As a further quality control measure, we investigated the genetic population structure using high resolution genome-wide allelic dissimilarity analysis. Allelic dissimilarity was calculated as
between any pair of individuals, involving the genotypes gik for subject i at locus k, for all loci. genotypes gik were 0 for reference homozygotes, 1 for heterozygotes, and 2 for variant homozygotes. Analysis of the resulting distance matrix permitted identification of sample duplications, and unreported family relationships among patients. By comparing the population structure as determined genetically with race and ethnic groups reported clinically, we determined that population structure was adequately accounted for by adjusting the risk factors for reported race and ethnicity. This was confirmed by comparing the score distribution with the null distribution using a q-q plots and genomic inflation factor. The final number of samples used after quality control was 793, the final number of SNPs was 865,483.
Statistical analysis
The myalgia score was adjusted for covariates as described above, and then analyzed quantitatively using linear regression vs. marker allele count (0, 1, or 2, indicating the number of alternative alleles). The regression p-values were converted to log-scores according to the formula s = - log10 p. Effect sizes were transformed using the logistic function f(z) = 1 / (1 +e-z) to yield probablility of myalgia according to logistic regression. In order to guard against false positive associations caused by non-normal phenotype distribution, small numbered genotype groups and other potential reasons, we also performed a non-parametric permutation analysis, which requires more computation but results in a different set of p-values free of bias caused by divergence from distribution assumptions. The two sets of p-values were compared to identify any statistical anomalies.
For confirmation, multiple SNPs were tested for each candidate locus. Since the candidate genes were derived from biological consideration and did not generally have a genetic marker associated with them, we defined a locus center as the genomic location in the middle between the first and last gene-associated SNP on the array. Association scores for each SNP within a 400 kb interval around the center were adjusted by dividing the p-value by the number of closer SNPs tested. In addition, the 31 candidates derived from previous studies(6-9) were evaluated for statistical significance against a Bonferroni corrected alpha of p < 0.05/31, corresponding to a log-score of 2.8 or greater.
Results
Clinical characteristics
Patients ranged in age from 12 to 90 yr (mean ± sd = 56.8 ± 13.7) and there were 632 European Americans, 21 African Americans, 16 Hispanic Latinos, 34 Asian Americans, and 50 patients of unknown ethnogeographic origin. Characteristics of patients according to myalgia status are shown in Table 1. Simvastatin, atorvastatin, rosuvastatin, pravastatin, lovastatin, fluvastatin and over the counter Chinese red rice yeast qualified as statin treatment in 116, 320, 150, 40, 10, 8, and 17 patients, respectively. The remaining 132 patients could not be unambiguously assigned to one statin, because of a history of multiple statin use (71%) or because the statin was not known (29%). Myalgia fraction was heterogeneously distributed across intake site, by drug, and across heritage (Table 2). Extensive medical histories were available in Ct-b and NY samples but not others. Prevalences for myalgia and no myalgia patients, respectively, in this subset of 253 patients were, for coronary artery disease: 27.2% vs. 31.8%; for congestive heart failure: 2.6% vs. 0%; and for peripheral vascular disease: 5.0% vs. 5.4% (all NS). Current smokers comprised 8.2% of the myalgia group and 11.2% of the no myalgia group (NS). Diabetes prevalence was 13.5% in the patients with myalgia versus 44.4% in patients without myalgia (p<1e-6).
Table 1.
Characteristics of patients with myalgia and without myalgia.
| Characteristic | N | Group | ||
|---|---|---|---|---|
| Myalgia n=377 |
No myalgia N=416 |
P value | ||
| Men | 497 | 38.7% | 61.3% | 1e-6 |
| Women | 296 | 56.0% | 44.0% | |
| Age (yrs) | 793 | 58.1 ± 11.3 | 55.6 ± 14.4 | 0.01 |
| BMI (kg/m2) | 642 | 28.1 ± 4.6 | 27.2 ± 5.6 | 0.04 |
| Total chol (mg/dL) | 632 | 243.6 ± 73.9 | 186.2 ± 72.4 | 1e-21 |
| LDL chol (mg/dL) | 600 | 155.6 ± 59.9 | 110.8 ± 63.6 | 1e-17 |
| HDL chol (mg/dL) | 637 | 51.6 ± 15.2 | 49.4 ± 17.8 | NS |
| Triglycerides (mg/dL) | 635 | 223 ± 341 | 173 ± 306 | 0.05 |
| Highest CK (IU/L) | 232* | 258 ± 427 | 178 ± 164 | 0.04 |
excludes one patient whose highest CK was 41,258 IU/L
Table 2. Prevalence of statin induced myalgia in the study population.
| Category | Descriptor | N | Myalgia fraction |
|---|---|---|---|
| Heritage | Caucasian | 672 | 0.45 |
| African American | 21 | 0.60 | |
| Hispanic Latino | 16 | 0.50 | |
| Asian | 34 | 0.42 | |
| other | 50 | 0.37 | |
|
| |||
| Site | CT-a* | 187 | 0.24 |
| CT-b† | 214 | 0.78 | |
| SF‡ | 353 | 0.35 | |
| NY§ | 39 | 0.61 | |
|
| |||
| Drug | simvastatin | 116 | 0.38 |
| atorvastatin | 320 | 0.29 | |
| rosuvastatin | 150 | 0.67 | |
| pravastatin | 40 | 0.34 | |
| lovastatin | 10 | 0.50 | |
| fluvastatin | 8 | 0.86 | |
| Chinese RedYeast Rice | 17 | 0.67 | |
| unknown | 132 | 0.64 | |
Key:
CT-a, patients treated at a primary care outpatient clinic in Connecticut
CT-b, patients treated at a lipid specialty outpatient clinic in Connecticut
SF, patients treated at lipid specialty outpatient clinics in San Francisco
NY, patients treated at a lipid specialty outpatient clinic in New York
The strongest covariate was intake site (R2 = 20%), because some sites were seeing patients particularly because of myalgia complaints (Table 2). Gender and age were also significant covariates, with an increase in myalgia probability by 15% for female gender, and an increase of 0.3% for each year of age. Treatment was significant as well, with patients on rosuvastatin and fluvastatin having a higher incidence of myalgia by 21% and 41% respectively. None of the other statin groups had a significantly different incidence of myalgia. We draw no conclusions from the drug dependence because there is a confounding association between site and treatment, and because treatment was not randomized to eliminate prescription bias. All covariates were adjusted for in the association tests.
Candidate-gene associations and validation
Of the 31 candidate genes, 9 were found to have an adjusted score above 1.3 (p<0.05) and 3 were statistically significant after adjustment for 31 comparisons. The SNPs and genes are rs4693570 which is located 100kb downstream from COQ2 (p < 3·10-5), rs2960336 located in ATP2B1, intron 9 (p = 0.001), and rs672348 located in DMPK intron 1 (p = 0.002) (Table 3) (9;11). All of the markers represent non-coding SNPs.
Table 3.
Candidate genes and validations. The score column lists log-scores of association for each locus, adjusted for testing of multiple SNPs within each locus. Scores greater than 1.3 (n=31, p<0.05) are shown. Scores 2.8 or greater indicate statistically significant associations after Bonferroni-adjustment for testing of multiple genes (highlighted and bold).
| Group | Chr | Symbol | SNP Id | Score | gene |
|---|---|---|---|---|---|
| Serotonin receptors (20) | 11 | HTR3B | rs1176746 | Ns | 5-hydroxytryptamine (serotonin) receptor 3B |
| 10 | HTR7 | rs11596518 | Ns | 5-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled) | |
|
| |||||
| Pharmacokinetic(21) | 12 | SLCO1B1 | rs7959887 | Ns | solute carrier organic anion transporter family, member 1B1 |
|
| |||||
| Vascular genes (7) | 3 | AGTR1 | rs1492099 | Ns | angiotensin II receptor, type 1 |
| 7 | NOS3 | rs1800779 | 1.4 | nitric oxide synthase 3 (endothelial cell) | |
|
| |||||
| Fuel processing and energy transfer (19,24) | 11 | PYGM | rs625172 | 2.0 | phosphorylase, glycogen, muscle |
| 17 | ACE | rs4344 | Ns | angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | |
| 17 | GAA | rs2304836 | Ns | glucosidase, alpha; acid | |
| 11 | CPT1A | rs2924674 | Ns | carnitine palmitoyltransferase 1A (liver) | |
| 22 | CPT1B | rs7238 | Ns | carnitine palmitoyltransferase 1B (muscle) | |
| 1 | CPT2 | rs1288335 | Ns | carnitine palmitoyltransferase II | |
| 1 | AMPD1 | rs2268699 | Ns | adenosine monophosphate deaminase 1 (isoform M) | |
| 4 | COQ2 | rs4693570 | 4.4 | para-hydroxybenzoate-polyprenyltransferase | |
| 9 | APTX | rs10813916 | Ns | aprataxin | |
|
| |||||
| Diseases (muscular dystrophy, rippling muscle disease) (19, 24) | X | DMD | rs5935419 | Ns | dystrophin |
| 18 | AQP4 | rs151251 | Ns | aquaporin 4 | |
| 19 | DMPK | rs672348 | 2.8 | dystrophia myotonica-protein kinase | |
| 8 | FAM82B | rs1542081 | Ns | family with sequence similarity 82, member B | |
| 3 | CAV3 | rs11916022 | Ns | caveolin 3 | |
|
| |||||
| Calcium transport (19) | 12 | ATP2B1 | rs17381194 | 3.1 | ATPase, Ca++ transporting, plasma membrane 1 |
| 16 | ATP2A1 | rs205356 | Ns | ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 | |
| 17 | ATP2A3 | rs758641 | Ns | ATPase, Ca++ transporting, ubiquitous | |
| 19 | RYR1 | rs2960336 | Ns | ryanodine receptor 1 (skeletal) | |
| 1 | RYR2 | rs10925416 | Ns | ryanodine receptor 2 (cardiac) | |
| 3 | ATP2B2 | rs6442177 | 1.5 | ATPase, Ca++ transporting, plasma membrane 2 | |
| 3 | ATP2C1 | rs218492 | 1.5 | ATPase, Ca++ transporting, type 2C, member 1 | |
| 7 | RYR3 | rs6460575 | 1.7 | ryanodine receptor 3 | |
| X | ATP2B3 | rs2269414 | Ns | ATPase, Ca++ transporting, plasma membrane 3 | |
| 16 | ATP2C2 | rs247808 | Ns | ATPase, Ca++ transporting, type 2C, member 2 | |
| 12 | ATP2A2 | rs17187412 | Ns | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | |
| 1 | ATP2B4 | rs3753036 | 1.8 | ATPase, Ca++ transporting, plasma membrane 4 | |
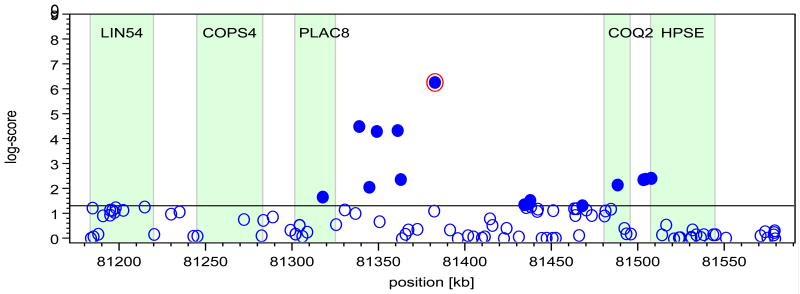
To exemplify the approach for each candidate gene, Figure 1 shows the genomic environment and the SNP effect on myalgia for the COQ2 gene. The locus shows linkage disequilibrium, indicated by the fact that there are other associated SNPs in the direct vicinity of the index SNP. The panel on the right shows the SNP effect magnitude, as provided by the regression coefficient with 95% confidence intervals, in subgroups taking the three major statins.
Figure 1.
Genomic environment and effect on myalgia for COQ2. The plot (left) shows log-scores of association for all SNPs versus chromosomal location within 200 kb of the index SNP location, and bar graph (right) shows the effect of the variant allele on the probability of myalgia in subgroups of patients taking one of the three major statins. Gene names are LIN54, C. elegans, homolog of; COPS4; PLAC8, placenta specific gene 8; COQ2, para-hydroxybenzoate-polyprenyltransferase; HPSE, heparanase. For bar graph, abbreviations for statin names are SIM, simvastatin; ATO, atorvastatin, and ROS, rosuvastatin.
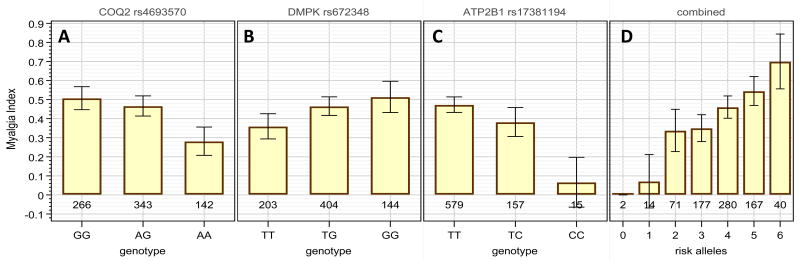
The genotypic effect of the markers on the probability of myalgia is detailed in Figure 2. There is a clear tendency for the risk allele (COQ2-G, DMPK-G, and ATP2B1-T) to be dominant (panels A-C), and there is a substantial effect of each allele on the probability of developing myalgia. The combined effect of the three markers is shown in panel D, showing that of the 2 subjects with no risk alleles, none have myalgia, and of the 40 subjects with the full complement of 6 risk alleles, 70% have myalgia.
Figure 2.
Effect of the validated SNP associations on the probability of myalgia. Panels A-C: the fraction of myalgia patients among the three different genotypes for each marker. Gene names are COQ2, para-hydroxybenzoate-polyprenyltransferase; DMPK, myotonin, protein kinase; ATP2B1, Plasma membrane calcium-transporting ATPase 1). Panel D: the fraction of myalgia patients among groups having 0, 1, 2, 3, 4, 5, or 6 of the risk alleles, regardless from which of the three genes. 95% confidence intervals are given for the true fraction based on the patient count in each group.
Discussion
With the cost of genotyping arrays decreasing rapidly, it is becoming economical to perform total-genome genotyping even when only a small number of candidate genes are to be studied. Samples can be genotyped with the total genome array prior to candidate selection (i.e. pre-genotyped), and then multiple candidate studies can be performed using that data, without further DNA analysis. This approach is demonstrated by the present study, where we utilize a total genome array to successfully interrogate candidate genes for associations.
The present study used a physiogenomic approach to examine genetic associations with statin myalgia in 377 patients diagnosed with statin myalgia and 416 asymptomatic statin treated patients. To our knowledge this is the largest study to date examining genetic factors associated with statin myalgia across a range of statins.
In tests of a priori hypotheses, we confirmed associations between myalgia and 3 candidate genes, including COQ2, which encodes an enzyme in CoQ10 production pathway; ATP2B1, which encodes for plasma membrane calcium-transporting ATPase 1, an enzyme that participates in calcium removal from the sarcolemma; and DMPK, a gene encoding for myotonin-protein kinase that has been associated with myotonic dystrophy. Brief descriptions of the genes and hypothetical roles in myalgia follow.
COQ2
The COQ2 gene encodes mitochondrial para-hydroxybenzoate-polyprenyltransferase, an enzyme that catalyzes a reaction in the biosynthesis of CoQ10, specifically, the prenylation of parahydroxybenzoate with an all-trans polyprenyl group (12). CoQ10 serves as a redox carrier in the mitochondrial respiratory chain and is a lipid-soluble antioxidant. CoQ10 blood concentrations are reduced by statins(13), and CoQ10 deficiency has been postulated as a cause of statin myopathy(13;14). CoQ10, known also as ubiquinone, is a decaprenyl benzoquinone with redox properties critical to its function as a cofactor in mitochondrial electron transport and therefore cellular energy transfer. Statin-associated myonecrosis is known to affect preferentially the mitochondrial poor Type II muscle fibers(15). Serum CoQ10 levels also decrease with statin therapy, a phenomonen attributable to its lipid solubility and transport in low density lipoproteins(13). COQ2 genetic variants have previously been found to be more frequent in statin-myalgic patients(14), but the present study offers to our knowledge the first independent confirmation of this association. CoQ10 supplementation has been studied in two small clinical trials (16;17) without definitive results. Nevertheless, the present finding suggests that factors affecting CoQ10 production may contribute to statin myopathy.
ATP2B1
The ATP2B1 gene encodes plasma membrane calcium-transporting ATPase 1, which belongs to the family of P-type primary ion transport ATPases characterized by the formation of an aspartyl phosphate intermediate during the reaction cycle. These enzymes remove calcium ions from the cell interior and play a critical role in intracellular calcium homeostasis. Statins decrease Ca2+ ATPase (18) activity contributing to increased sarcoplasmic Ca2+. In animal studies this transient rise in cystosolic Ca2+ caused by statins has been associated with membranolysis (19). The identification of an association between a gene contributing to intracellular calcium regulation, ATP2B1, and myalgia is consistent with prior studies suggesting a role for the ryanodine receptor, which also participates in intracellular calcium regulation, and statin-induced muscle injury(20).
DMPK
DMPK encodes myotonin-protein kinase (MT-PK), a serine-threonine kinase that has been implicated in myotonic dystrophy type I(21), a inherited multisystemic disease characterized by wasting of the muscles (muscular dystrophy), cataracts, heart conduction defects, endocrine changes, and myotonia(22). Statins have previously exposed mild forms of muscular dystrophy caused by various genes(6).
The associations determined in our study confirm connections between common myalgia and genes involved in biochemical pathways that have been previously implicated in statin mylagia. Other candidate genes tested here were not validated. One of these, the SLCO1B1 gene, demonstrates variants that are strongly associated to elevated CK during simvastatin therapy(9;23). Our study observed a different phenotype and tested only a limited number of patients receiving simvastatin. The present study detected no association of SLCO1B1 to myalgia probably as a consequence of its broader set of statins and phenotype.
Chemical and pharmacokinetic properties differ among statins, and variants of genes encoding transporters ABCG2 and ABCB1 and metabolic enzymes CYP2C8 and UGT1A3 involved in the disposition of statins have been posed as risk factors for statin-induced myopathy. However all of these are statin-specific genes, while our approach focused on investigation of class-wide genes identified by comprehensive reviews. With limited numbers of patients receiving any given statin therapy, the present study was not powered to test drug-specific associations.
The present study confirmed 3 previously identified markers. Among the 377 patients diagnosed clinically as having statin myalgia, all have at least one risk allele from the 3 validated genes. The SINM study sought to identify genetic markers for statin myopathy that can be used to predict which patients are most likely to develop myopathic complaints during statin treatment and to aid in objectifying the diagnosis of statin myopathy. Combining the COQ2, ATP2B1, and DMPK markers into a physiogenomic panel to represent candidate pathways(24) and deriving a score, establishes a prototype system to predict the onset of myalgia and to aid in diagnosis. These results are encouraging. Despite subjectivity in diagnosis of statin myalgia, the study identified previously suspected candidate genes with logical relationships to muscle metabolism as contributing to statin myalgia.
We plan future studies including placebo controlled, blinded treatments to diagnose statin myalgia quantitatively and to ascertain other genetic determinants. Markers predictive of statin induced low density lipoprotein cholesterol lowering have been identified by many groups (25). Validation of markers for statin safety and efficacy is essential for development of the tools to bring genetically personalized medicine into the mainstream of cardiovascular management(26).
Study Limitations
There are limitations to the study. The retrospective and multisite nature of the study did not permit us to standardize collection of clinical data beyond history of statin use and subsequent myalgia. We were unable to determine if muscle symptoms were different among patients with COQ2, ATB2B1 or DMPK mutations as breaking the cohort into myalgia subcategories would reduce the power to detect genetic associations. This is however a significant potential correlation that could be identified in future studies. Another limitation of our study was the inability to assign unambiguously some patients to one specific statin. This was either due to the fact that that there was a history of multiple statin use in a majority of these patients or retrospective review of limited medical record was unable to identify the particular statin prescribed. However, we did not aim to derive drug specific markers and therefore this limitation may not compromise our findings. The study also included patients taking Chinese red rice yeast (CRRY). Different preparations of CRRY may contain variable amounts of Manacolin K. Other compounds in Chinese red rice yeast may possibly have pharmacological effects.
The diagnosis of statin myalgia by experienced physicians in this study was based on criteria previously defined by us(8) which represent a subset of all statin myopathies(4;27). We focused on the largest subset, aptly termed common myalgia. We identified several genes logically related to muscle metabolism suggesting that the true relationship may be even stronger for myalgia sub-phenotypes. Our sample size is relatively small yet to our knowledge it is the largest genetic study on common myalgia as experienced by outpatients receiving statin therapy. The study included multiple ethnicities, and used heritage as a covariate to account for the population structure thus introduced. The present paper is therefore confirmatory for selected candidate genes.
Conclusion
In summary, we employed total genome arrays which allow association queries targeted to candidate genes drawn from mechanistic studies and biological models. Using a priori hypothesis testing in a subset of genes, we validated COQ2, ATP2B1, and DMPK as candidate genes for statin induced myalgia. Each gene has a possible physiological relationship to muscle metabolism and thereby to statin related muscular complaints. SNPs in these three genes may be integrated into a predictive and diagnostic physiogenomic system for the assessment of statin myalgia in clinical practice.
Supplementary Material
Acknowledgments
Funding Support: Supported by the NIH Small Business Innovation Research Grant 1 R44 HL091697-01 “DNA Diagnostic System for Statin Safety and Efficacy,” and in part by 1 R01 HL081893-01 “The Effect of Statins on Skeletal Muscle Function.” The ClinicalTrials.gov identifier for the SINM study is NCT00767130. Dr. Ruaño is Principal Investigator for the R44 NIH grant and Dr. Thompson for the R01 NIH grant.
Footnotes
Statement of Originality: We confirm that the paper is not under consideration elsewhere, and that none of the paper's contents have been previously published. All authors have read and approved the manuscript.
Conflict of Interest: Neither Dr. Thompson, Dr. Kane, Dr. Wu, or Dr. Gordon have consulting or other financial arrangements with Genomas. Drs. Ruaño and Windemuth are employees and shareholders of Genomas, Inc. Mr. Kocherla and Ms. Bogaard are full time employees of Genomas, Inc. Drs. Holford and Seip are consultants to Genomas, Inc. Dr. Thompson has received speaking honoraria or consulting fees from GlaxoSmithKline, Merck, Roche, Pfizer, Astra Zeneca B. Braun, Schering-Plough, Takeda, and Abbott.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Findlay S. The statin drugs. Prescription and price trends October 2005 to December 2006 and potential cost savings to Medicare from increased use of lower cost statins. Consumer Reports. 2007 [Google Scholar]
- 2.Statins: The World Market, 2009-2024. London, UK: Visiongain, Ltd.; 3-3-2009. [Google Scholar]
- 3.Waters DD. What the statin trials have taught us. Am J Cardiol. 2006;98:129–34. doi: 10.1016/j.amjcard.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 4.Thompson PD, Clarkson P, Karas RH. Statin-associated myopathy. JAMA. 2003;289:1681–90. doi: 10.1001/jama.289.13.1681. [DOI] [PubMed] [Google Scholar]
- 5.Bruckert E, Hayem G, Dejager S, Yau C, Begaud B. Mild to moderate muscular symptoms with high-dosage statin therapy in hyperlipidemic patients--the PRIMO study. Cardiovasc Drugs Ther. 2005;19:403–14. doi: 10.1007/s10557-005-5686-z. [DOI] [PubMed] [Google Scholar]
- 6.Ghatak A, Faheem O, Thompson PD. The genetics of statin-induced myopathy. Atherosclerosis. 2010;210:337–43. doi: 10.1016/j.atherosclerosis.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 7.Ruaño G, Thompson PD, Windemuth A, et al. Physiogenomic analysis links serum creatine kinase activities during statin therapy to vascular smooth muscle homeostasis. Pharmacogenomics. 2005;6:865–72. doi: 10.2217/14622416.6.8.865. [DOI] [PubMed] [Google Scholar]
- 8.Ruaño G, Thompson PD, Windemuth A, et al. Physiogenomic association of statin-related myalgia to serotonin receptors. Muscle Nerve. 2007;36:329–35. doi: 10.1002/mus.20871. [DOI] [PubMed] [Google Scholar]
- 9.Link E, Parish S, Armitage J, et al. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789–99. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 10.Ruaño G, Windemuth A, Holford T. Physiogenomics: Integrating Systems Engineering and Nanotechnology for Personalized Medicine. In: Bronzino JD, editor. The Biomedical Engineering Handbook. 3rd. CRC Press; 2005. pp. 28-1–28-9. [Google Scholar]
- 11.Vladutiu GD. Genetic predisposition to statin myopathy. Curr Opin Rheumatol. 2008;20:648–55. doi: 10.1097/BOR.0b013e328314b7b4. [DOI] [PubMed] [Google Scholar]
- 12.Forsgren M, Attersand A, Lake S, et al. Isolation and functional expression of human COQ2, a gene encoding a polyprenyl transferase involved in the synthesis of CoQ. Biochem J. 2004;382:519–26. doi: 10.1042/BJ20040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcoff L, Thompson PD. The role of coenzyme Q10 in statin-associated myopathy: a systematic review. J Am Coll Cardiol. 2007;49:2231–7. doi: 10.1016/j.jacc.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 14.Oh J, Ban MR, Miskie BA, Pollex RL, Hegele RA. Genetic determinants of statin intolerance. Lipids Health Dis. 2007;6:7. doi: 10.1186/1476-511X-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westwood FR, Bigley A, Randall K, Marsden AM, Scott RC. Statin-induced muscle necrosis in the rat: distribution, development, and fibre selectivity. Toxicol Pathol. 2005;33:246–57. doi: 10.1080/01926230590908213. [DOI] [PubMed] [Google Scholar]
- 16.Caso G, Kelly P, McNurlan MA, Lawson WE. Effect of coenzyme q10 on myopathic symptoms in patients treated with statins. Am J Cardiol. 2007;99:1409–12. doi: 10.1016/j.amjcard.2006.12.063. [DOI] [PubMed] [Google Scholar]
- 17.Young JM, Florkowski CM, Molyneux SL, et al. Effect of coenzyme Q(10) supplementation on simvastatin-induced myalgia. Am J Cardiol. 2007;100:1400–3. doi: 10.1016/j.amjcard.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 18.Baker SK, Tarnopolsky MA. Statin myopathies: pathophysiologic and clinical perspectives. Clin Invest Med. 2001;24:258–72. [PubMed] [Google Scholar]
- 19.Mosieniak G, Figiel I, Kaminska B. Cyclosporin A, an immunosuppressive drug, induces programmed cell death in rat C6 glioma cells by a mechanism that involves the AP-1 transcription factor. J Neurochem. 1997;68:1142–9. doi: 10.1046/j.1471-4159.1997.68031142.x. [DOI] [PubMed] [Google Scholar]
- 20.Draeger A, Monastyrskaya K, Mohaupt M, et al. Statin therapy induces ultrastructural damage in skeletal muscle in patients without myalgia. J Pathol. 2006;210:94–102. doi: 10.1002/path.2018. [DOI] [PubMed] [Google Scholar]
- 21.Musova Z, Mazanec R, Krepelova A, et al. Highly unstable sequence interruptions of the CTG repeat in the myotonic dystrophy gene. Am J Med Genet A. 2009;149A:1365–74. doi: 10.1002/ajmg.a.32987. [DOI] [PubMed] [Google Scholar]
- 22.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voora D, Shah SH, Spasojevic I, et al. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol. 2009;54:1609–16. doi: 10.1016/j.jacc.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilke RA, Mareedu RK, Moore JH. The Pathway Less Traveled: Moving from Candidate Genes to Candidate Pathways in the Analysis of Genome-Wide Data from Large Scale Pharmacogenetic Association Studies. Curr Pharmacogenomics Person Med. 2008;6:150–9. doi: 10.2174/1875692110806030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seip RL, Duconge J, Ruaño G. Genotype-Guided Statin Therapy. In: Wu AHB, Yeo J, editors. Pharmacogenomic Testing in Current Clinical Practice: Implementation in the Clinical Laboratory. New York: Humana Press, Springer Science+Business Media; 2010. pp. 155–74. [Google Scholar]
- 26.Winkelmann BR, Herrington D. Pharmacogenomics--10 years of progress: a cardiovascular perspective. Pharmacogenomics. 2010;11:613–6. doi: 10.2217/pgs.10.68. [DOI] [PubMed] [Google Scholar]
- 27.Pasternak RC, Smith SC, Jr, Bairey-Merz CN, Grundy SM, Cleeman JI, Lenfant C. ACC/AHA/NHLBI Clinical Advisory on the Use and Safety of Statins. Stroke. 2002;33:2337–41. doi: 10.1161/01.str.0000034125.94759.41. [DOI] [PubMed] [Google Scholar]
- 28.Lanktree MB, Anand SS, Yusuf S, Hegele RA. Replication of genetic associations with plasma lipoprotein traits in a multiethnic sample. J Lipid Res. 2009;50:1487–96. doi: 10.1194/jlr.P900008-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.