Abstract
Aims
To investigate associations between novel human cytochrome P450 (CYP450) combinatory (multigene) and substrate-specific drug metabolism indices, and elements of metabolic syndrome, such as low density lipoprotein cholesterol (LDLc), high density lipoprotein cholesterol (HDLc), triglycerides and BMI, using physiogenomic analysis.
Methods
CYP2C9, CYP2C19 and CYP2D6 genotypes and clinical data were obtained for 150 consecutive, consenting hospital admissions with a diagnosis of major depressive disorder and who were treated with psychotropic medications. Data analysis compared clinical measures of LDLc, HDLc, triglyceride and BMI with novel combinatory and substrate-specific CYP450 drug metabolism indices.
Results
We found that a greater metabolic reserve index score is related to lower LDLc and higher HDLc, and that a greater metabolic alteration index score corresponds with higher LDLc and lower HLDc values. We also discovered that the sertraline drug-specific indices correlated with cholesterol and triglyceride values.
Conclusions
Overall, we demonstrated how a multigene approach to CYP450 genotype analysis yields more accurate and significant results than single-gene analyses. Ranking the individual with respect to the population represents a potential tool for assessing risk of dyslipidemia in major depressive disorder patients who are being treated with psychotropics. In addition, the drug-specific indices appear useful for modeling a variable of potential relevance to an individual’s risk of drug-related dyslipidemia.
Keywords: combinatory genotypes, CYP450, dyslipidemia, physiogenomics, psychopharmacology
Metabolic syndrome, a cluster of conditions associated with increased risk of cardiovascular disease, morbidity and mortality, has long been associated with schizophrenia, and more recently, with second-generation antipsychotics [1–5]. A growing body of evidence is emerging linking metabolic syndrome elements of obesity, hypertension, and glucose and lipid alterations with depression and antidepressant pharmacotherapy [6–8].
Major depressive disorder (MDD) has been associated with increased risk of obesity, hypertension, and diabetes, but not with dyslipidemia [7,9,10]. Patients with MDD are at higher risk of developing Type II diabetes, and patients with diabetes mellitus exhibit a high rate of depression [11,12]. MDD patients more frequently have elevated measures of body fat, including waist circumference [3], visceral fat [13] and BMI [14]. The relationship between hypertension and depression is unclear; some studies demonstrate a strong correlation, while others show little or no association [7,15–17].
Although dyslipidemia has not been associated with MDD, certain antidepressant treatments have been shown to be positively correlated with elevated low density lipoprotein cholesterol (LDLc) and triglycerides and diminished high density lipoprotein cholesterol (HDLc) serum levels [10]. Interestingly, there appears to be inter-individual variation in the appearance and extent of dyslipidemia as a side effect of antidepressant treatment. One study found that depressed patients with elevated cholesterol levels were more likely to be fluoxetine nonresponders [18]. Another study concluded that, compared with responders, patients with treatment-resistant depression demonstrated higher baseline triglycerides [19]. Increases in total serum cholesterol were associated with imipramine and doxepin, while decreases in HDLc and elevated triglycerides were associated with treatment with imipramine, amitriptyline and nortriptyline [10,20]. Further studies have found that selective serotonin reuptake inhibitors were associated with increases in LDLc [21–23], although other studies could not reproduce this association [20,24]. Mirtazapine and duloxetine were also found to affect serum lipid and triglyceride levels [25–27].
The cytochrome P450 (CYP450) isoenzyme system is responsible for 70–80% of all phase-I-dependent metabolism in approximately 40–45% of all marketed drugs [28,29]. Accordingly, the high carrier prevalence of altered (non-reference) CYP450 alleles has significant implications for healthcare management. The majority of the population carries single nucleotide polymorphisms on CYP2C9, CYP2C19 or CYP2D6 genes. A pilot study found that the presence of polymorphisms across multiple genes (combinatory polymorphism) is particularly elevated in psychiatric patients [30]. Multiple psychoactive medications are principally metabolized by combinations of products of the CYP450 gene family [31,32]. The value of DNA typing to assess the risk for, or in some cases to evaluate retrospectively drug side effects and treatment resistance has been documented in various case reports and studies [33–36]. Uninformed prescribing of psychotropics to patients with highly compromised biochemical activity for the CYP450 isoenzymes may expose 50% of patients to preventable, severe side effects [36].
In this research we examined associations between CYP2C9, CYP2C19 and CYP2D6 combinatory genotypes and dyslipidemia in 150 psychiatric inpatients. Physiogenomic methods were used to quantify the genotypes according to the CYP450 combinatory and drug-specific metabolism indices described previously, namely the drug metabolism reserve index (metabolic reserve), drug metabolism alteration index (metabolic alteration), allele alteration index and gene alteration index [37,38]. This multigene physiogenomic analysis revealed significant correlations between all four indices and elevated LDLc, HDLc and LDLc:HDLc ratio. The investigators also present evidence supporting the utility of drug-specific indices when assessing side-effect risk for particular psychotropic medications. Our physiogenomics approach has previously elucidated new pharmacological mechanisms related to statin neuromuscular side effects [39,40], anti-psychotic-induced metabolic derangements [41] and thiazolidinedion-related weight gain [42], as well as gene associations with variability in diet-induced weight loss [43,44].
Methods
Sample collection & cohort description
The sample cohort consisted of 150 consecutive, consenting participants of the ages 18–78 (median 40); 39% male, 61% female with a diagnosis of MDD and treated with psychotropic medications through the inpatient psychiatric services at the Institute of Living at Hartford Hospital (CT, USA), admitted January–March, 2007. Self-reported ethnicities were 65% Caucasian, 28% Hispanic and 7% African–American. Data obtained included demographic, clinical and treatment information. Clinical data were acquired through a questionnaire given to patients at the time of enrolment. Treatment data were retrieved from paper and electronic medical records as well as questionnaire responses. Laboratory data, including lipids, glucose and triglycerides, were determined upon admission. HDLc cholesterol methodology was immunoturbidmetric Roche Cobas; LDLc cholesterol was calculated using the Friedewald formula. All specimens were obtained prior to breakfast being eaten. All 150 patients were treated with psychotropics during hospitalization. A total of 98% received antidepressants (45% received more than one during hospitalization, 17% concurrently, mean = 1.49). A total of 65% were taking antipsychotics (12% multiple antipsychotics). Demographic and psychotropic medication data for the entire cohort (n = 150) and the lipid cohort (n = 96) are summarized in Table 1. An overview of prescriptions by history and hospitalization at Institute of Living is provided in Table 2. The study was approved by the Hartford Hospital IRB and each patient signed a statement of informed consent that included permission to use the sample for CYP450 genetic testing.
Table 1.
Demographic and psychotropic data for the full study cohort (n = 150) as well as the individuals who had data for both LDLc and HDLc (n = 96).
| n | Age | Gender | Ethnicity | Number of antidepressants | Number of antipsychotics |
|---|---|---|---|---|---|
| 150 | Min.: 18 | Male: 39% | Caucasian: 65% | Total: 226 | Total: 116 |
| Max.: 78 | Female: 61% | Hispanic: 28% | Average per patient: 1.49 | Average per patient: 0.77 | |
| Median: 40 | African–American: 7% | Treated with AD: 98% | Treated with AP: 65% | ||
| Concurrent AD: 17% | Concurrent AP: 3% | ||||
|
| |||||
| 96 | Min.: 18 | Male: 32% | Caucasian: 69% | Total: 145 | Total: 72 |
| Max.: 78 | Female: 68% | Hispanic: 27% | Average per patient: 1.51 | Average per patient: 0.75 | |
| Median: 43 | African–American: 4% | Treated with AD: 97% | Treated with AP: 63% | ||
| Concurrent AD: 19% | Concurrent AP: 2% | ||||
The number of ADs and APs indicate the total number of prescriptions given in each cohort during Institute of Living hospitalization.
AD: Antidepressant; AP: Antipsychotic; HDLc: High density lipoprotein cholesterol; LDLc: Low density lipoprotein cholesterol; Max.: Maximum; Min.: Minimum.
Table 2.
Antidepressant, antipsychotic and anticonvulsant therapy for the 96 patients for whom LDLc and HDLc lipid measures were available.
| Drug | CYP2C9 | CYP2C19 | CYP2D6 | History | At IOL |
|---|---|---|---|---|---|
| Antidepressant therapy | |||||
| Amitriptyline | 0.25 | 0.25 | 1 | 3 | NA |
| Bupropion | 0.25 | 0 | 0.25 | 32 | 22 |
| Citalopram | 0 | 1 | 0.25 | 10 | 31 |
| Doxepin | 0 | 0 | 1 | 1 | NA |
| Duloxetine | 0 | 0 | 1 | 13 | NA |
| Escitalopram | 0 | 1 | 0 | 21 | 0 |
| Fluoxetine | 1 | 0.25 | 1 | 21 | 15 |
| Fluvoxamine | 0 | 0 | 1 | 4 | NA |
| Mirtazapine | 0.25 | 0 | 1 | 5 | 6 |
| Nortriptyline | 0 | 0.25 | 1 | 1 | NA |
| Paroxetine | 0 | 0 | 1 | 23 | 2 |
| Phenelzine | 0 | 0 | 0 | 1 | NA |
| Selegiline | 0 | 0.25 | 0.25 | 1 | NA |
| Sertraline | 1 | 1 | 0.25 | 24 | 8 |
| Trazodone | 0 | 0.25 | 1 | 11 | 24 |
| Venlafaxine | 0.25 | 0.25 | 1 | 20 | 16 |
| Other | NA | NA | NA | 0 | 21 |
| Antipsychotic therapy | |||||
| Aripiprazole | 0 | 0 | 1 | 4 | 7 |
| Chlorpromazine | 0 | 0 | 1 | 1 | NA |
| Haloperidol | 0 | 0 | 1 | 3 | NA |
| Olanzapine | 0 | 0 | 0.25 | 6 | 4 |
| Quetiapine | 0 | 0 | 0.25 | 12 | 28 |
| Risperidone | 0 | 0 | 1 | 10 | 26 |
| Ziprasidone | 0 | 0 | 0 | 3 | 3 |
| Other | NA | NA | NA | 0 | 4 |
| Anticonvulsant therapy | |||||
| Divalproex | 0 | 0 | 0 | 4 | NA |
| Gabapentin | 0 | 0 | 0 | 3 | 15 |
| Lamotrigine | 0 | 0 | 0 | 4 | 10 |
| Oxcarbazepine | 0 | 0 | 0 | 2 | 4 |
| Topiramate | 0 | 0 | 0 | 1 | 5 |
| Valproic acid | 0.25 | 0.25 | 0 | 0 | 6 |
The three gene columns for CYP2C9, CYP2C19 and CYP2D6 represent the coefficients for each gene used to calculate the drug-specific metabolism indices. A value of 1 indicates a major metabolic pathway, a value of 0.25 a minor metabolic pathway and a value of 0 indicates that the gene is not involved in the drug’s metabolism. The ‘History’ column shows all drugs administered prior to hospitalization and the ‘At IOL’ column shows drugs administered during hospitalization at the IOL. NA values represent values that are either not applicable (in the case of the gene coefficients for ‘Other’ drugs) or values that are not available in our dataset.
IOL: Institute of Living; NA: Not applicable.
Clinical data corrections
LDLc and HDLc data were available for 96 of the 150 patients and triglyceride (TG) data were available for 98. A total of 147 patients had data for presence or absence of hyperlipidemia and 136 had data for BMI. Hyperlipidemia was determined through patient medical records: a physician diagnosis of lipid metabolism (ICD codes 272.0–272.9) indicates the presence of hyperlipidemia, otherwise it is considered absent. All clinical data were corrected for the covariates of age, gender and ethnicity. BMI was found to vary significantly with HDLc and TG, but not LDLc. Following covariance correction, 91 patients had valid values for HDLc and 93 had corrected values for LDLc. One outlier in the TG data (TG = 642 mg/dl) was excluded, leaving 92 corrected values for triglycerides. One outlier in the LDLc cohort (LDLc = 297 mg/dl) and one outlier in the LDLc:HDLc ratio (LDLc:HDLc = 6.8) were excluded from the analysis, yielding 95 samples for LDLc analysis and 90 corrected values for LDLc:HDLc analysis. Excluding outliers, LDLc ranged from 49 to 214 mg/dl with an average of 114.9 mg/dl. HDLc ranged from 30 to 111 mg/dl with an average value of 53.15 kg/m2. Triglycerides ranged from 39 to 322 mg/dl with a mean of 129.1 mg/dl and BMI ranged from 16.91 to 56.24 kg/m2 with an average of 28.36 kg/m2.
CYP450 combinatory & drug-specific metabolism indices
Analysis in this study was done using the CYP450 combinatory and drug-specific metabolism indices described in our methodologies paper [38]. In summary, there are four combinatory indices: the drug metabolism reserve (metabolic reserve) index, the drug metabolism alteration (metabolic alteration) index, the allele alteration index and the gene alteration index. Each index is created by assigning alleles of the CYP2C9, CYP2C19 and CYP2D6 genes a numerical value corresponding to its functional status relative to the reference allele, which is scored as one in the metabolic reserve index and zero in the alteration indices. The final combinatory index value is a summation of the six allelic scores for the given index across the three genes. As such, the indices are termed ‘combinatory’ as a result of the three CYP450 genes contributing equally to the final index. The drug-specific indices apply coefficients to each of the three contributing gene scores depending on the pharmacodynamic properties of a particular enzyme substrate. In this study, the substrates of interest are psychotropic drugs. An isoenzyme that is a major metabolic pathway for a given drug receives a coefficient of 1.0, while an isoenzyme not involved in the drug metabolism or activation has a coefficient of 0.0. Isoenzymes that constitute a minor metabolic pathway for a drug’s metabolism were given a score of 0.25, consistent with the American Pharmacists Association’s designation of a minor metabolic pathway as responsible for less than 30% of total metabolism [45].
Single nucleotide polymorphism assays
All 150 patients were genotyped to determine CYP2C9, CYP2C19 and CYP2D6 polymorphisms. Blood samples were collected into tubes containing either ethylenediaminetetraacetic acid or citrate and were extracted from lymphocytes using the Qiagen EZ-1 robotic DNA isolation procedure. DNA typing was performed at the Genomas Laboratory of Personalized Health at Hartford Hospital (CT, USA). The Genomas Laboratory of Personalized Health is a high-complexity clinical DNA testing center licensed by the Connecticut Department of Public Health (CL-0644) and certified by the Centers for Medicare and Medicaid Services (ID #07D1036625) under Clinical Laboratory Improvement Amendments.
The Tag-It™ Mutation Detection assays (Luminex Corporation, Austin, TX, USA) were utilized for DNA typing of 5, 7 and 18 alleles in genes CYP2C9, CYP2C19 and CYP2D6, respectively, as previously described. These assays employed PCR to amplify the desired gene selectively without co-amplifying pseudogenes or other closely related sequences. In addition, the kit employs a PCR strategy to amplify fragments that are characteristic of unique genomic rearrangements in order to detect the presence of the deletion and duplication alleles in these genes. The kits use multiplexed allele-specific primer extension to identify small nucleotide variations including single base changes and deletions of one or three bases on the Luminex xMAP™ system [46].
Physiogenomic plot
To demonstrate the association of a CYP450 combinatory index [38] value with any given quantitative phenotype we plotted the index value as a function of phenotype using the locally weighted scatter plot smooth fit. Locally weighted scatter plot smooth is a method to smooth data using a locally weighted linear regression [47,37]. At each point in the plot, a quadratic polynomial is fitted to the data in the vicinity of that point. The data are weighted such that they contribute less if they are further away, according to the tricubic function:
where x is the abscissa of the point to be estimated, xi are the data points in the vicinity, and d(x) is the maximum distance of x to xi
Results
All four combinatory indices were found to be significantly correlated with dyslipidemia markers of total cholesterol, LDLc, HDLc and LDLc:HDLc ratio. No significant correlation was found between the combinatory indices and either BMI and triglycerides. Elevated triglycerides were found to correlate directly only with the sertraline drug-specific metabolic and allele alteration indices.
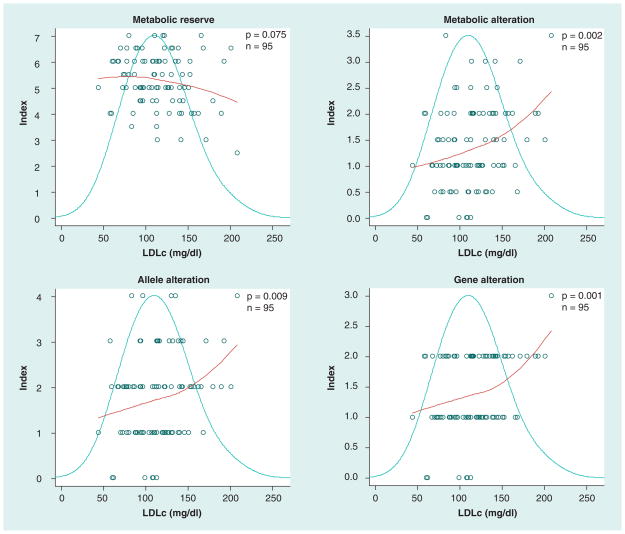
Total cholesterol was found to be higher in patients with low metabolic reserve (p = 0.099) and in those with high metabolic (p = 0.008), allele (p = 0.039) and gene alterations (p = 0.020). Similarly, LDLc was higher in patients with low metabolic reserve and with high alteration (p = 0.075, p = 0.002, p = 0.009 and p = 0.001 for metabolic reserve, metabolic alteration, allele alteration and gene alteration indices, respectively). HDLc demonstrated the opposite pattern; HDLc was lower in patients with low metabolic reserve (p = 0.252) and higher in those with high metabolic (p = 0.073), allele (p = 0.059) and gene alterations (p = 0.011). The LDLc:HDLc ratio was inversely related to metabolic reserve (p = 0.016) and directly related to metabolic alteration (p = 0.005), allele alteration (p = 0.017) and gene alteration (p = 0.001).
Those patients in the top quartile for LDLc (LDLc >139) had an average drug metabolism reserve index value of 4.78 and drug metabolism alteration index value of 1.78 whereas those with LDLc less than or equal to 139 mg/dl had an average drug metabolism reserve index value of 5.40 and drug metabolism alteration value of 1.21 (p = 0.030, and p = 0.003 for reserve and alteration indices, respectively). Those patients in the top quartile for HDLc (HDLc >57) had a mean drug metabolism reserve index score of 5.59 and a mean drug metabolism alteration index of 1.18 compared with those patients with HDLc of less than or equal to 57, who had a mean metabolic reserve index of 5.08 and metabolic alteration index of 1.47 (p = 0.009 and 0.04 for reserve and alteration indices, respectively). In summary, greater drug metabolism reserve yielded higher HDLc and lower LDLc values. A greater number of alterations affecting drug metabolism was associated with higher LDLc and lower HDLc.
The combinatory index ranking curves are also useful in identifying possible extremes in metabolic capacity to correlate with clinical data [38]. For instance, those patients approximately in the bottom 25th percentile for drug metabolism reserve (index ≤4.5) have an average LDLc of 127.2 mg/dl compared with the remainder of the sample whose mean LDLc is 110.10 mg/dl (p = 0.044). Similarly, those individuals approximately in the top 25th percentile for drug metabolism alteration (index >1.5) have an average LDLc of 127.30 mg/dl compared with the bottom 75th percentile who have an average LDLc of 108.90 mg/dl (p = 0.021). Those individuals in the bottom 30% of the metabolic alteration had a 5% chance of having hyperlipidemia compared with those patients in the remaining 70% who had a 23% chance (p = 0.001). Similarly, those individuals in the bottom 50% of the drug metabolism reserve ranking have a 25% chance of exhibiting hyper-lipidemia compared with the 10% chance of those in the top 50% (p = 0.016).
Table 3 provides a summary of the linear correlations between cholesterol and each CYP450 combinatory drug metabolism index. In all models, LDLc and metabolic reserve are inversely related, whereas all alteration indices and LDLc are directly related. Conversely, HDLc and metabolic reserve are directly related, and HDLc and alteration indices are inversely related. These relationships are true for all combinatory as well as single gene indices. Table 3 shows that, in nearly all cases, both the proportion of variability (as indicated by R2) and the significance (as indicated by the p-value) of the single variable linear model correlations are more strongly related to the combinatory indices than to any single gene index. In the case of HDLc, CYP2C9 and CYP2D6 contributed most while CYP2C19 was unrelated. Combining only the CYP2C9 and CYP2D6 indices yielded the greatest significance for HDLc, but not for LDLc. Figures 1 & 2 are physiogenomic plots that demonstrate the relationship between the combinatory drug metabolism indices and cholesterol levels.
Table 3.
Linear models correlating LDLc (left) and HDLc (right) values to the CYP450 combinatory drug metabolism indices as well as CYP2C9, CYP2C19, CYP2D6 and CYP2C9 + CYP2D6 gene-specific indices.
| Linear Models | LDLc | HDLc | ||||||
|---|---|---|---|---|---|---|---|---|
| R2 | Met Res (%) | Met Alt (%) | Allele Alt (%) | Gene Alt (%) | Met Res (%) | Met Alt (%) | Allele Alt (%) | Gene Alt (%) |
| Combinatory | 3.40 | 10.00 | 7.20 | 11.00 | 1.47 | 3.57 | 3.95 | 7.07 |
|
| ||||||||
| CYP2C9 only | 1.60 | 1.60 | 1.10 | 2.00 | 3.83 | 3.83 | 3.57 | 4.70 |
|
| ||||||||
| CYP2C19 only | 0.90 | 0.90 | 0.90 | 1.40 | 0.22 | 0.22 | 0.22 | 0.04 |
|
| ||||||||
| CYP2D6 only | 1.20 | 9.00 | 5.20 | 4.00 | 0.60 | 3.04 | 2.41 | 4.40 |
|
| ||||||||
| p-value | Met Res | Met Alt | Allele Alt | Gene Alt | Met Res | Met Alt | Allele Alt | Gene Alt |
|
| ||||||||
| Combinatory | 0.075 | 0.002 | 0.009 | 0.001 | 0.252 | 0.073 | 0.059 | 0.011 |
|
| ||||||||
| CYP2C9 only | 0.228 | 0.228 | 0.314 | 0.082 | 0.063 | 0.063 | 0.073 | 0.039 |
|
| ||||||||
| CYP2C19 only | 0.355 | 0.355 | 0.355 | 0.257 | 0.662 | 0.662 | 0.662 | 0.856 |
|
| ||||||||
| CYP2D6 only | 0.296 | 0.003 | 0.026 | 0.051 | 0.467 | 0.099 | 0.141 | 0.045 |
The upper section shows the coefficient of determination (R2) for each model as a percentage. The lower section shows the statistical significance of the correlation (p-value). A value in bold indicates the strongest or most significant correlation.
Alt: Alteration; HDLc: High density lipoprotein cholesterol; LDLc: Low density lipoprotein cholesterol; Met: Metabolic; Res: Reserve.
Figure 1. Physiogenomic plots correlating LDLc with the four CYP450 combinatory drug metabolism indices.
Each plot contains three components: the distribution of the phenotype (bell curve), the index score of each individual patient (circles) and the LOESS fit of the index value as a function of phenotype (regression curve). In each of four panels the abscissa represents each patient’s LDLc. The ordinate indicates the index value for the LOESS curve. The p-value is derived from a linear analysis associating the index and clinical variable, and the n indicates the number of samples considered in the analysis. Correction for the comparisons of each index to five phenotypes in this study (LDLc, HDLc, LDLc:HDLc, triglycerides and BMI) requires multiplication of the respective p-value by a factor of five.
LDLc: Low density lipoprotein cholesterol; LOESS: Locally weighted scatter plot smooth.
Figure 2. Physiogenomic plots correlating LDLc:HDLc ratio with the four CYP450 combinatory drug metabolism indices.
Each plot contains three components: the distribution of the phenotype (bell curve), the index score of each individual patient (circles) and the LOESS fit of the index value as a function of phenotype (regression curve). In each of four panels, the abscissa represents each patient’s LDLc:HDLc ratio. The ordinate indicates the index value for the LOESS curve. The p-value is derived from a linear analysis associating the index and clinical variable, and the n indicates the number of samples considered in the analysis. Correction for the comparisons of each index to five phenotypes in this study (LDLc, HDLc, LDLc:HDLc, triglycerides and BMI) requires multiplication of the respective p-value by a factor of five.
LOESS: Locally weighted scatter plot smooth.
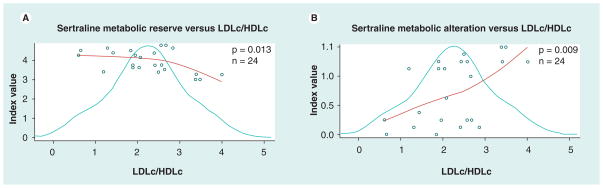
It is also useful to consider the drug-specific indices in order to evaluate the impact of a particular drug. We investigated the correlation between lipid levels and drug-specific indices. Sertraline was selected for substrate-specific analysis because it has been previously associated with dyslipidemia [22,23] and evidences a distributed substrate affinity, including CYP2C9, CYP2C19 and CYP2D6. Furthermore, the sertraline sub-cohort size of 24 patients in this study is relatively large. Sertraline’s metabolism reserve index correlated inversely (p = 0.013, Figure 3a) with the LDLc:HDLc ratio phenotype, as well as to triglycerides (p = 0.042). Sertraline alteration indices correlated positively (p = 0.009, Figure 3b for metabolic alteration) with LDLc:HDLc. Similar results were found for the phenotypes LDLc and HDLc separately (data not shown). By contrast, the combinatory drug metabolism indices demonstrated no significant correlation with lipid values for the 24 patients who had taken sertraline prior to hospitalization, and the CYP2D6 single-gene index (a minor metabolic pathway for sertraline) was particularly unrelated.
Figure 3. Physiogenomic plots correlating LDLc:HDLc ratio with sertraline-specific CYP450 combinatory indices.
Each plot contains three components: the distribution of the phenotype (bell curve), the index score of each individual patient (circles) and the LOESS fit of the index value as a function of phenotype (regression curve). The p-value is derived from a linear analysis associating the index and clinical variable, and the n indicates the number of samples considered in the analysis. (A) Shows the inverse correlation between sertraline metabolic reserve and LDLc:HDLc ratio, and (B) shows the direct relationship between sertraline metabolic alteration and LDLc:HDLc ratio.
HDLc: High density lipoprotein cholesterol; LOESS: Locally weighted scatter plot smooth.
Discussion
This study highlights the clinical relevance and benefits of benchmarking innate drug metabolism reserve using the CYP450 combinatory approach described in our index methodologies paper [38]. We found that lipid measures of LDLc, HDLc and the LDLc:HDLc ratio varied significantly with combined drug metabolism reserve and alteration index values. Notably, these correlations could not be found to the same degree when using a single-gene index. In this MDD cohort, average cholesterol levels were not significantly elevated, which is consistent with earlier studies that found no link between MDD and elevated lipids [9]. However, dyslipidemia has been associated with psychiatric pharmacotherapy [8,10,21,20]. Our findings build upon these observations by demonstrating that drug intolerance, as a result of decreased innate liver enzyme capacity, may result in an increased lipid effect in some patients.
Owing to the timing of the cholesterol measurements upon admission it is difficult to associate dyslipidemia with any particular psychotropic. In such an instance, the combinatory drug metabolism indices are particularly useful. By quantifying innate drug metabolism capacity, they allow for analysis of drug response and effect when numerous enzymes and substrates are relevant to the analysis. Table 2 indicates that a wide variety of psychotropics were administered to this cohort. CYP2D6 serves as a primary metabolic pathway for many of the most prescribed drugs, including fluoxetine, paroxetine, trazodone, venlafaxine and risperidone. However, CYP2C9 and/or CYP2C19 are major metabolizers for citalopram, escitalopram, fluoxetine and sertraline. Table 3 shows that particularly for LDLc, CYP2D6 is often the strongest contributor to the model, consistent with the CYP2D6 isoenzyme’s primary role in psychotropic metabolism. However, CYP2D6 scores alone never supersede in significance the combinatory analysis.
The flexibility of the CYP450 combinatory drug metabolism indices allow for a rigorous and thorough analytical approach. In Table 3 we demonstrate how each combinatory index can be broken down into single-gene indices. In the case of HDLc, we were able to note that CYP2C9 and CYP2D6 gene-specific indices were the primary contributors to the relationship, and therefore create a new combined index omitting the less-relevant CYP2C19 and generating a more accurate model. Moreover, we demonstrate in this study how drug-specific indices, which utilize the metabolism pathway coefficients shown in Table 2, are useful in predicting the effect of a particular drug. While the lack of a treatment timeframe weakens any conclusive determination of causality, it is notable that the sertraline-specific indices correlated more closely with dyslipidemia phenotypes than the combinatory indices for patients treated with this drug prior to hospitalization. Since lipid tests were performed at the time of hospitalization, it remains possible that other medications administered prior to this may have influenced the lipid values in our dataset. A future study could be specifically designed to investigating the drug-specific indices and their relation to cardiometabolic markers.
Beyond the issues surrounding the unspecific timing of drug prescription prior to lipid level evaluation, this study has further limitations that must be addressed. First, diet and patient compliance were not measured as covariates, and could contribute to drug response and dyslipidemia. Ideally, steady-state plasma concentrations and quantitative lifestyle covariates would be considered. In the case of those patients overdosed to psychotropics by virtue of their low functional metabolic reserve, a concomitant assay of all present drugs would have been informative. In addition, the minor metabolic pathway coefficients in the drug-specific indices have been estimated to have a value of 0.25 for the purposes of this study. Precise pharmacologically assayed values between 0.0 and 1.0 could be determined for each drug and metabolic pathway. Covariates such as drug interactions and drug properties such as self-inhibition or active metabolites should also be modeled into the indices as described in our methodology manuscript [38]. Indeed, it is known that certain antipsychotics, such as clozapine and olanzapine, have inherent metabolic side effects that could exacerbate dyslipidemias in patients so treated, which could be accounted for by drug covariates [4]. Finally, our sample size, particularly in the case of the drug-specific analyses, would need to be larger in order to draw conclusions with a higher degree of confidence. Nevertheless, the strength of our statistical associations, the consistency with previously published works, and the congruence with models of pharmacokinetic and biological activity render our discoveries novel and noteworthy.
In conclusion, the results of this study demonstrate the utility and clinical relevance of the CYP450 combinatory drug metabolism indices. Ranking an individual relative to a population, as described by our group [38], represents a potential tool for assessing risk of dyslipidemia in MDD patients being treated with antidepressants and antipsychotics. In addition, the drug-specific indices appear promising as quantitative measures assisting in prescription by modeling a variable of potential relevance to an individual’s risk of drug-related dyslipidemia. The clinical benefits of CYP450 genotyping in psychiatry are growing increasingly clear, and in this case, considering an array of genes together has proven the most effective in evaluating risk of adverse drug reactions. By nature, the CYP450 combinatory indices are easily generalized, allowing for the addition of further genes, such as CYP1A2, CYP3A4, and CYP3A5, as well as new alleles such as CYP2C19*17. Research indicates that the appearance of any one of the many components of metabolic syndrome significantly increases the likelihood of developing metabolic syndrome and eventually dangerous and costly cardiovascular disease [48]. Any information to assist in predicting which psychotropic medications may result in dyslipidemia for a given patient would represent a substantial step forward in controlling these widespread, debilitating and costly side effects. In this study, the CYP450 combinatory indices and substrate specific indices have proven more effective than any single gene score when determining risk of dyslipidemia in depressed patients treated with psychotropics. We anticipate that this combinatory approach will continue to yield significant clinical pharmacogenetic applications.
Executive summary.
Metabolic syndrome, a cluster of cardiovascular risk factors, has been associated with mental illness and psychotropics.
Multiple psychoactive medications are principally metabolized by combinations of isoenzymes coded by the CYP450 gene family.
We examined associations between CYP2C9, CYP2C19 and CYP2D6 combinatory genotypes and dyslipidemia in 150 psychiatric hospitalized patients with a diagnosis of major depressive disorder treated with psychotropic medications (98% received antidepressants, and 65% received antipsychotics).
Physiogenomic methods were used to quantify the genotypes according to CYP450 combinatory and drug-specific metabolism indices (metabolism reserve, metabolism alteration, allele alteration and gene alteration) described in the companion paper.
The multi-gene physiogenomic analysis revealed significant correlations among all four combinatory indices and dyslipidemia measures (LDLc, HDLc, LDLc:HDLc ratio). Patients with a greater drug metabolism reserve evidenced lower LDLc, lower LDLc:HDLc ratio and higher HDLc values. Conversely, patients with a greater number of alterations affecting drug metabolism evidenced higher LDLc, higher LDLc:HDLc ratio and lower HDLc.
Those patients in the top quartile for LDLc (LDLc > 139) had an average drug metabolism reserve index value of 4.78 and drug metabolism alteration index value of 1.78 whereas those with LDLc ≤ 139 mg/dl had an average drug metabolism reserve index value of 5.40 and drug metabolism alteration value of 1.21 (p = 0.030 and p = 0.003 for reserve and alteration indices, respectively).
Those patients in the top quartile for HDLc (HDLc > 57 mg/dl) had a mean drug metabolism reserve index score of 5.59 and a mean drug metabolism alteration index of 1.18 compared to those patients with HDLc less than or equal to 57 who had a mean metabolic reserve index of 5.08 and metabolic alteration index of 1.47 (p = 0.009 and 0.04 for reserve and alteration indices, respectively).
Sertraline has a distributed substrate affinity, including CYP2C9, CYP2C19 and CYP2D6 isoenzymes, and therefore was selected for drug-specific analysis. Sertraline’s metabolism reserve index correlated inversely, and its alteration indeces directly, with the LDLc:HDLc ratio phenotype.
The combinatory index ranking curves are useful in benchmarking innate drug metabolism reserve and identifying patients at the extremes of metabolic capacity to correlate with clinical outcomes.
Our results show that combinatory CYP450 genotyping and corresponding quantitative indices of pharmacogenetic functional status have potential clinical utility in psychiatry for evaluating the risk of iatrogenic cardiometabolic effects of psychotropic treatment.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Financial & competing interests disclosure
G Ruaño, A Windemuth, M Kocherla, K Gorowski and D Villagra are shareholders or were employees of Genomas, Inc. This study was supported by NIH Grant 2 R44 MH073291 to G Ruaño and by Hartford Hospital Research Program funds. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
- 1.Newcomer JW. Second-generation (atypical) antipsychotics and metabolic effects: a comprehensive literature review. CNS Drugs. 2005;19(Suppl 1):1–93. doi: 10.2165/00023210-200519001-00001. [DOI] [PubMed] [Google Scholar]
- 2.Newcomer JW, Haupt DW. The metabolic effects of antipsychotic medications. Can J Psychiatry. 2006;51(8):480–491. doi: 10.1177/070674370605100803. [DOI] [PubMed] [Google Scholar]
- 3.Goethe JW, Szarek BL, Woolley SB, Caley CF. Metabolic abnormalities in psychiatric inpatients. Presented at: American Psychiatric Association meeting; San Diego, CA, USA. 19–24 May 2007. [Google Scholar]
- 4.Goethe JW. Metabolic syndrome in psychiatric inpatients receiving antipsychotics. Presented at: American Psychiatric Association meeting; Atlanta, GA, USA. 21–26 May 2005. [Google Scholar]
- 5.Goethe JW, Szarek BL, Caley CF, et al. Signs and symptoms associated with the metabolic syndrome in psychiatric inpatients receiving antipsychotics: a retrospective chart review. J Clin Psychiatry. 2007;68(1):22–28. doi: 10.4088/jcp.v68n0103. [DOI] [PubMed] [Google Scholar]
- 6.Kinder LS, Carnethon MR, Palaniappan LP, et al. Depression and the metabolic syndrome in young adults: findings from the Third National Health and Nutrition Examination Survey. Psychosom Med. 2004;66(3):316–322. doi: 10.1097/01.psy.0000124755.91880.f4. [DOI] [PubMed] [Google Scholar]
- 7.Heiskanen TH, Niskanen LK, Hintikka JJ, et al. Metabolic syndrome and depression: a cross-sectional analysis. J Clin Psychiatry. 2006;67(9):1422–1427. doi: 10.4088/jcp.v67n0913. [DOI] [PubMed] [Google Scholar]
- 8.Goethe JW, Szarek BL, Caley CF. Metabolic syndrome in psychiatric inpatients treated for depression. In: Thakore J, Leonard B, editors. Metabolic Effects of Psychotropic Drugs. Karger; Basel, Switzerland: 2009. pp. 90–104. [Google Scholar]
- 9.Brown ES, Varghese FP, McEwen BS. Association of depression with medical illness: does cortisol play a role? Biol Psychiatry. 2004;55(1):1–9. doi: 10.1016/s0006-3223(03)00473-6. [DOI] [PubMed] [Google Scholar]
- 10.Goethe JW, Szarek BL, Caley CF. Metabolic syndrome and depression: a review. Depression: Mind and Body. 2008;3(4):138–149. [Google Scholar]
- 11.Katon WJ, Lin EH, Russo J, et al. Cardiac risk factors in patients with diabetes mellitus and major depression. J Gen Intern Med. 2004;19(12):1192–1199. doi: 10.1111/j.1525-1497.2004.30405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musselman DL, Betan E, Larsen H, et al. Relationship of depression to diabetes Types 1 and 2: epidemiology, biology, and treatment. Biol Psychiatry. 2003;54(3):317–329. doi: 10.1016/s0006-3223(03)00569-9. [DOI] [PubMed] [Google Scholar]
- 13.Lee ES, Kim YH, Beck SH, et al. Depressive mood and abdominal fat distribution in overweight premenopausal women. Obes Res. 2005;13(2):320–325. doi: 10.1038/oby.2005.43. [DOI] [PubMed] [Google Scholar]
- 14.Ohayon MM. Epidemiology of depression and its treatment in the general population. J Psychiatr Res. 2007;41(3–4):207–213. doi: 10.1016/j.jpsychires.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. JAMA. 2002;287(3):356–359. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- 16.Almeida OP, Flicker L, Norman P, et al. Association of cardiovascular risk factors and disease with depression in later life. Am J Geriatr Psychiatry. 2007;15(6):506–513. doi: 10.1097/01.JGP.0000246869.49892.77. [DOI] [PubMed] [Google Scholar]
- 17.Scalco AZ, Scalco MZ, Azul JB, et al. Hypertension and depression. Clinics (Sao Paulo) 2005;60(3):241–250. doi: 10.1590/s1807-59322005000300010. [DOI] [PubMed] [Google Scholar]
- 18.Sonawalla SB, Papakostas GI, Petersen TJ, et al. Elevated cholesterol levels associated with nonresponse to fluoxetine treatment in major depressive disorder. Psychosomatics. 2002;43(4):310–316. doi: 10.1176/appi.psy.43.4.310. [DOI] [PubMed] [Google Scholar]
- 19.Papakostas GI, Petersen T, Sonawalla SB, et al. Serum cholesterol in treatment-resistant depression. Neuropsychobiology. 2003;47(3):146–151. doi: 10.1159/000070584. [DOI] [PubMed] [Google Scholar]
- 20.Kopf D, Westphal S, Luley CW, et al. Lipid metabolism and insulin resistance in depressed patients: significance of weight, hypercortisolism, and antidepressant treatment. J Clin Psychopharmacol. 2004;24(5):527–531. doi: 10.1097/01.jcp.0000138762.23482.63. [DOI] [PubMed] [Google Scholar]
- 21.Raeder MB, Bjelland I, Emil VS, et al. Obesity, dyslipidemia, and diabetes with selective serotonin reuptake inhibitors: the Hordaland Health Study. J Clin Psychiatry. 2006;67(12):1974–1982. doi: 10.4088/jcp.v67n1219. [DOI] [PubMed] [Google Scholar]
- 22.Lara N, Baker GB, Archer SL, et al. Increased cholesterol levels during paroxetine administration in healthy men. J Clin Psychiatry. 2003;64(12):1455–1459. doi: 10.4088/jcp.v64n1209. [DOI] [PubMed] [Google Scholar]
- 23.Bailey DL, Le Melledo JM. Effects of selective serotonin reuptake inhibitors on cholesterol levels in patients with panic disorder. J Clin Psychopharmacol. 2003;23(3):317–319. doi: 10.1097/00004714-200306000-00016. [DOI] [PubMed] [Google Scholar]
- 24.Peter H, Tabrizian S, Hand I. Serum cholesterol in patients with obsessive compulsive disorder during treatment with behavior therapy and SSRI or placebo. Int J Psychiatry Med. 2000;30(1):27–39. doi: 10.2190/APWF-N1XU-Y7A0-TCBW. [DOI] [PubMed] [Google Scholar]
- 25.Stimmel GL, Dopheide JA, Stahl SM. Mirtazapine: an antidepressant with noradrenergic and specific serotonergic effects. Pharmacotherapy. 1997;17(1):10–21. [PubMed] [Google Scholar]
- 26.Hardy T, Sachson R, Shen S, et al. Does treatment with duloxetine for neuropathic pain impact glycemic control? Diabetes Care. 2007;30(1):21–26. doi: 10.2337/dc06-0947. [DOI] [PubMed] [Google Scholar]
- 27.Montgomery SA. Safety of mirtazapine: a review. Int Clin Psychopharmacol. 1995;10(Suppl 4):37–45. doi: 10.1097/00004850-199512004-00006. [DOI] [PubMed] [Google Scholar]
- 28.Laika B, Leucht S, Heres S, et al. Intermediate metabolizer: increased side effects in psychoactive drug therapy. The key to cost-effectiveness of pretreatment CYP2D6 screening? Pharmacogenomics J. 2009;9(6):395–403. doi: 10.1038/tpj.2009.23. [DOI] [PubMed] [Google Scholar]
- 29.Ingelman-Sundberg M, Rodriguez-Antona C. Pharmacogenetics of drug-metabolizing enzymes: implications for a safer and more effective drug therapy. Philos Trans R Soc Lond B Biol Sci. 2005;360(1460):1563–1570. doi: 10.1098/rstb.2005.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruaño G, Villagra D, Rahim U-S, et al. Increased carrier prevalence of deficient CYP2C9, CYP2C19 and CYP2D6 alleles in depressed patients referred to a tertiary psychiatric hospital. Personalized Medicine. 2008;5(6):579–587. doi: 10.2217/17410541.5.6.579. [DOI] [PubMed] [Google Scholar]
- 31.Black JL, III, O’Kane DJ, Mrazek DA. The impact of CYP allelic variation on antidepressant metabolism: a review. Expert Opin Drug Metab Toxicol. 2007;3(1):21–31. doi: 10.1517/17425255.3.1.21. [DOI] [PubMed] [Google Scholar]
- 32.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part II. Clin Pharmacokinet. 2009;48(12):761–804. doi: 10.2165/11318070-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 33.Ruaño G, Blair CL, Bower B, et al. Somatic complications of psychotropic medications in a patient with multiple CYP2 drug metabolism deficiencies. Conn Med. 2007;71(4):197–200. [PubMed] [Google Scholar]
- 34.Kirchheiner J, Rodriguez-Antona C. Cytochrome P450 2D6 genotyping: potential role in improving treatment outcomes in psychiatric disorders. CNS Drugs. 2009;23(3):181–191. doi: 10.2165/00023210-200923030-00001. [DOI] [PubMed] [Google Scholar]
- 35.Kirchheiner J, Nickchen K, Bauer M, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry. 2004;9(5):442–473. doi: 10.1038/sj.mp.4001494. [DOI] [PubMed] [Google Scholar]
- 36.Kootstra-Ros JE, Van Weelden MJ, Hinrichs JW, et al. Therapeutic drug monitoring of antidepressants and cytochrome p450 genotyping in general practice. J Clin Pharmacol. 2006;46(11):1320–1327. doi: 10.1177/0091270006293754. [DOI] [PubMed] [Google Scholar]
- 37.Ruaño G, Windemuth A, Holford T. Physiogenomics: integrating systems engineering and nanotechnology for personalized medicine. In: Bronzino JD, editor. The Biomedical Engineering Handbook. 3. CRC Press; FL, USA: 2005. [Google Scholar]
- 38.Villagra D, Goethe J, Schwartz HI, et al. Novel drug metabolism indices for pharmacogenetic functional status based on combinatory genotyping of CYP2C9, CYP2C19 and CYP2D6 genes. Biomark Med. 2011;5(4):427–438. doi: 10.2217/bmm.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruaño G, Thompson PD, Windemuth A, et al. Physiogenomic analysis links serum creatine kinase activities during statin therapy to vascular smooth muscle homeostasis. Pharmacogenomics. 2005;6(8):865–872. doi: 10.2217/14622416.6.8.865. [DOI] [PubMed] [Google Scholar]
- 40.Ruaño G, Thompson PD, Windemuth A, et al. Physiogenomic association of statin-related myalgia to serotonin receptors. Muscle Nerve. 2007;36(3):329–335. doi: 10.1002/mus.20871. [DOI] [PubMed] [Google Scholar]
- 41.Ruaño G, Goethe JW, Caley C, et al. Physiogenomic comparison of weight profiles of olanzapine- and risperidone-treated patients. Mol Psychiatry. 2007;12:474–482. doi: 10.1038/sj.mp.4001944. [DOI] [PubMed] [Google Scholar]
- 42.Ruaño G, Bernene J, Windemuth A, et al. Physiogenomic comparison of edema and BMI in patients receiving rosiglitazone or pioglitazone. Clin Chim Acta. 2009;400(1–2):48–55. doi: 10.1016/j.cca.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Ruaño G, Windemuth A, Kocherla M, et al. Physiogenomic analysis of weight loss induced by dietary carbohydrate restriction. Nutr Metab (Lond) 2006;3:20. doi: 10.1186/1743-7075-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seip RL, Volek JS, Windemuth A, et al. Physiogenomic comparison of human fat loss in response to diets restrictive of carbohydrate or fat. Nutr Metab (Lond) 2008;5:4. doi: 10.1186/1743-7075-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lacy CF, Armstrong LL, Goldman MP, Lance LL, editors. Drug Information Handbook: A Comprehensive Resource for All Clinicians and Healthcare Professionals. Lexi-Comp; OH, USA: 2009. [Google Scholar]
- 46.Gordon J, Merante F, Weiss S, Zastawny R. Pharmacogenetic P-450 screening using the tag-it universal bead-based array platform. In: Wong SH, Linder MW, Valdes R Jr, editors. Pharmacogenomics and Proteomics: Enabling the Practice of Personalized Medicine. AAC Press; Washington DC, USA: 2006. [Google Scholar]
- 47.Cleveland WS, Devlin SJ. Locally weighted regression: an approach to regression analysis by local fitting. J Am Stat Assoc. 1988;83(403):596–610. [Google Scholar]
- 48.Kahn R, Buse J, Ferrannini E, et al. The metabolic syndrome: time for a critical appraisal: joint statement from the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care. 2005;28(9):2289–2304. doi: 10.2337/diacare.28.9.2289. [DOI] [PubMed] [Google Scholar]