Abstract
Tyrosine kinase 2 (Tyk2) is an integral part of the Janus kinase-signal transducer and activator of transcription (JAK-STAT) pathway which relays intracellular signals of various cytokines. Tyk2 crucially contributes to host defense mechanisms against microbial pathogens and to tumor surveillance but also facilitates immune pathologies. Here we investigated the impact of Tyk2 on the macrophage proteome using the synthetic double-stranded RNA analog polyinosinic acid-polycytidylic acid (poly(I:C)) as a mimicry of viral infections. By means of 2D-DIGE in connection with PMF obtained by MALDI-MS and sequence tag determination by MS/MS we unambiguously identified eighteen protein spots corresponding to sixteen distinct proteins that are regulated by poly(I:C) and differentially expressed between wildtype (WT) and Tyk2-deficient macrophages. The majority of these proteins are functionally assigned to cellular immune responses and to metabolism. We show for selected metabolic enzymes, i.e. triosephosphate isomerase (TIM), ATP-citrate synthase (ACLY) and long-chain-fatty-acid-CoA ligase 4 (ACSL4), that Tyk2 affects protein expression transcriptionally and post-transcriptionally. We furthermore confirm the involvement of Tyk2 in the regulation of lipid and carbohydrate metabolism at the level of metabolites. Taken together, our results provide new evidence for important functions of Tyk2 at the molecular interface between innate immunity and cellular metabolism.
Keywords: 2D-DIGE, Mass spectrometry, Poly(I:C), Macrophages, Immunity, Metabolism
Graphical abstract
Highlights
► Comparative 2-DIGE and MS reveals an impact of Tyk2 on the macrophage proteome. ► Tyk2 regulates enzymes involved in lipid and carbohydrate metabolism. ► Tyk2 regulates protein expression transcriptionally and post-transcriptionally. ► Poly(I:C) treatment induces a Tyk2-dependent increase of extracellular lactate. ► We provide evidence for a role of Tyk2 in linking metabolism and innate immunity.
1. Introduction
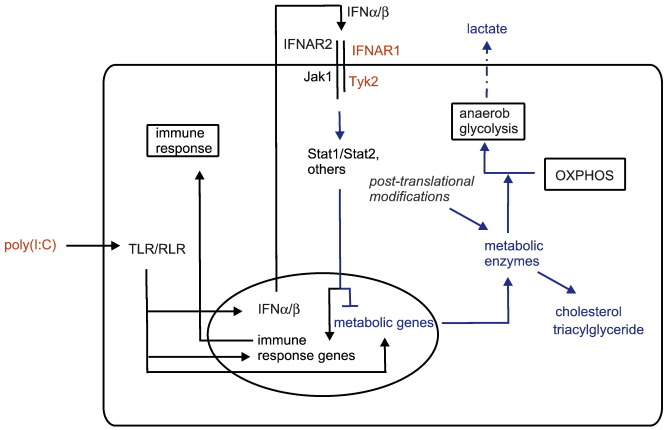
Protein kinases are involved in the regulation of a large number of cellular processes and dysregulated kinase activity is a frequent cause of diseases. Tyrosine kinase 2 (Tyk2) is a member of the Janus kinase (JAK) family of intracellular non-receptor tyrosine kinases, which are an integral part of the JAK-STAT (JAK-signal transducer and activator of transcription) pathway [1,2]. Tyk2 was originally identified in a genetic screen as a crucial component for type I interferon (IFNα/β) signaling in human cell lines [3]. In addition, Tyk2 has a role in signal transduction via a number of other cytokines and some growth factors [4]. We have shown that mice lacking Tyk2 display strongly increased susceptibility to infection with viruses, e.g. lymphocytic choriomeningitis virus (LCMV), vaccinia virus (VV) [5], and murine cytomegalovirus (MCMV) [6]. Consistently, Tyk2-deficient macrophages fail to control MCMV replication in vitro [6]. To further study the contribution of Tyk2 to macrophage activity we used synthetic polyinosinic acid-polycytidylic acid (poly(I:C)) to mimic the presence of double-stranded RNA (dsRNA), a known (by-)product of virus replication. Poly(I:C) is recognized by the pattern recognition receptors (PRRs) Toll-like receptor 3 (TLR3) and by retinoic-acid-inducible gene I (RIG-I)-like receptors (RLRs) [7,8]. Upon ligand binding several distinct intracellular pathways convey the signals from these receptors and result in the activation of a common set of transcription factors. These include NFκB, which is required for the induction of pro-inflammatory genes (e.g. interleukin-1β and tumor necrosis factor α), and members of the IFN-regulatory factor (IRF) family, which induce transcriptional activation of IFNα/β genes [7,8]. Following secretion and binding of IFNα/β to the type I IFN receptor, which is composed of the two subunits IFNAR1 and IFNAR2, Tyk2 and Jak1 are activated. They auto- and/or trans-phosphorylate and subsequently phosphorylate mainly Stat1 and Stat2 heterodimers. The phosphorylated heterodimers then translocate to the nucleus and, in combination with IRF9, induce expression of a large set of IFNα/β-responsive genes [1,9]. The interplay between PRR- and IFNα/β-induced pathways and the contribution of additionally activated signaling cascades determines the complex biological responses to poly(I:C). Although different pathogen-associated patterns, like lipopolysaccharide (LPS) and dsRNA, induce similar and overlapping signaling cascades [10], cellular responses can be fairly different [11].
In a previous study, we used a proteomic approach to investigate the molecular role of Tyk2 in macrophages in response to LPS, a structural membrane component of Gram-negative bacteria recognized by TLR4 [12]. The absence of Tyk2 showed complex consequences on the macrophage proteome and implied regulatory roles of Tyk2 at the mRNA and post-transcriptional level before and after treatment with LPS.
In the current work, we performed a proteome analysis of Tyk2-deficient and wildtype (WT) primary murine macrophages to identify novel molecular functions of Tyk2 in response to poly(I:C). Consistent with our proteomic study using LPS, we show that Tyk2 is also essential for transcriptional and post-transcriptional responses to poly(I:C). More importantly, our data suggest novel roles of Tyk2 in the poly(I:C)-mediated regulation of cellular lipid and carbohydrate metabolism.
2. Material and methods
2.1. Materials
All chemicals were of analytical grade or of higher purity unless stated otherwise. Poly(I:C) (Amersham Biosciences, Piscataway, NJ, USA) was annealed prior to use to ensure a high yield of double-stranded configuration. Purified (≥ 90% by SDS-PAGE) recombinant murine IFNβ (Calbiochem, San Diego, CA, USA) was diluted in PBS containing 0.1% BSA.
2.2. Animals, cells and cell lysates
Tyk2-deficient (Tyk2−/−) [5] and IFNAR1-deficient (IFNAR1−/−) [13] mice have been previously described and were on a C57BL/6 background (B6.129P2-Tyk2tm1 and B6.129P2-IFNAR1tm1). C57BL/6 wildtype (WT) mice were purchased from Charles River Laboratories (Sulzfeld, Germany). Mice were kept under specified pathogen-free (SPF) conditions and were sex- and age-matched (8–12 weeks) for each experiment. Bone marrow-derived macrophages (BMM) were isolated and grown as described previously [14]. Following cultivation for 6–7 days, cells were incubated with 50 μg/ml poly(I:C) or 500 units/ml IFNβ for the times indicated. If not stated otherwise, whole cell lysates were prepared as previously described [12] and used for 2D-DIGE and Western blot analysis. Alternatively, RIPA buffer (20 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% Triton-X-100, 0.5% Na-deoxycholate; 0.1% SDS and protease inhibitors as described [12]) was used. Protein concentration was measured by the Coomassie G-250 (dye reagent, Bio-Rad laboratories, Hercules, CA, USA) protein binding assay [15].
2.3. 2D-DIGE and statistical analysis
2D-DIGE experiments were designed to comprise three biological replicates per genotype (WT vs. Tyk2−/− BMM) and treatment (with or without poly(I:C) for 18 h). DIGE labeling, 2-DE separation and evaluation were performed as previously described [12]. Briefly, samples were minimally labeled with CyDye DIGE™ fluorescent dyes (GE Healthcare Life Sciences, Munich, Germany). Proteins were separated in the first dimension on 24 cm IPG Dry strips with linear pH gradients of pH 4–7 and pH 6–9 using the IPGphor III system (all GE Healthcare). The second dimension was performed on isocratic SDS-PAGE gels (10%T, 25.5 × 20.5 cm) according to Laemmli [16] using an Ettan Dalt Six electrophoresis chamber (GE Healthcare). Low molecular weight (LMW)-SDS marker (GE Healthcare) was used as Mr standard. Fluorescence images of the gels were acquired on a Typhoon 9400 scanner (GE Healthcare). Image analysis including spot detection, matching, normalization and quantification was performed using DeCyder software Version 5.02 (GE Healthcare).
Statistical analysis of all spots was based on spot volume ratios. Differentially regulated spots between genotypes (WT and Tyk2−/−) were selected according to volume ratio and Student's t-test. Spot matching and spot quality of proteins of interest were manually checked to avoid false positives. As we are mainly interested in genotype-specific differences between WT and Tyk2−/− BMM, untreated samples were labeled with Cy3 and treated samples were labeled with Cy5. Differentially regulated spots between WT and Tyk2−/− were crosschecked for dye labeling bias based on reverse labeling experiments (untreated/Cy5, treated/Cy3). A pool of equal amounts of protein from all samples was used as internal standard and labeled with Cy2.
2.4. Protein identification by MALDI-MS
Protein spots were detected by acidic silver nitrate staining as previously described using a mass spectrometry (MS)-compatible protocol [17], manually excised and submitted to MS analysis. In some cases semipreparative 2-DE gels (150 μg protein load) were used to recover sufficient material for MS analysis. Spot destaining, in-gel tryptic digestion and sample preparation using ZipTipμ-C18 (Millipore, Billerica, MA, USA) were performed as described previously [12]. Peptide mass fingerprinting (PMF) and seamless post source decay (PSD) fragment ion analysis experiments were performed on a MALDI-reflectron instrument (AXIMA-CFRplus, Shimadzu Biotech Kratos Analytical, Manchester, UK) equipped with a nitrogen-laser (λ = 337 nm). Dried droplet and thin layer sample preparation techniques were applied using α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich, St. Louis, MO, USA) as matrix [18]. Monoisotopic values of the matrix cluster trimer at [3M + H]+m/z 568.15 and autolytic tryptic products at [M + H]+m/z 805.41, [M + H]+m/z 1153.57 and [M + H]+m/z 2163.05 were used for internal calibration. Autolytic tryptic product ions, matrix cluster ions [19], keratin and gel blank artifacts [20] were sorted out from the obtained PMF mass spectra. The resulting monoisotopic list of m/z values was submitted to search engines MASCOT [21], revision 2.3, and ProFound [22] searching the databases UniProtKB/Swiss-Prot (Release 2011_03 of 08-Mar-11) and NCBI (RefSeq Release 46 of 08-Mar-11) restricting to Mus musculus taxonomy. Search parameters were set for mass accuracy: < 50 ppm, fixed modification: carbamidomethylation, variable modifications: methionine oxidation and acetylation at the protein N-terminal end, and missed cleavages: one. Based on the measured PMF at least two peptides were selected for PSD experiments. Search parameters based on these experiments were identical to PMF experiments, except for precursor ion mass accuracy (< 100 ppm) and product ion tolerance (+/− 1.0 Da). A more stringent search parameter (Da instead of ppm) was chosen for peptide fragmentation to reduce false positive results. A protein was considered as identified, if the scores of database searches clearly exceeded the algorithm's significance threshold (p < 0.05) for PMF data and for sequence tag ion analyses of at least one peptide.
2.5. RNA isolation, reverse transcription (RT) and quantitative PCR (qPCR)
Total RNA was isolated from 106 cells using TRIzol (Invitrogen Life Technologies, Lofer, Austria). RNA (1 μg/20 μl reaction volume) was reverse transcribed using iScript cDNA synthesis kit (Bio-Rad, Vienna, Austria). Ubiquitin-conjugating enzyme E2 D2 (Ube2d2) was used as endogenous control as described previously [6]. qPCR assays for Tgtp (transcript NM_011579), Tim/TpiI (transcript NM_009415), Acsl4 (transcript NM_001033600) and Acly (transcript NM_134037) were carried out with EvaGreen. The reaction mix contained 2 μl cDNA, 0.3 μM of each primer, 2.5 mM MgCl2, 0.2 mM each dNTPs, 1× Hotfire B Puffer, 0.2×EvaGreen (Biotium, Hayward, CA, USA), 1 U Hotfire Polymerase (Solis BioDyne, Tartu, Estonia) in a final volume of 25 μl. For Tgtp assays 5% DMSO was added to the reaction mix. PCR conditions: initial denaturation at 95 °C for 15 min followed by 45 cycles 95 °C 20 s and 63 °C 40 s. Following primers were used:
Tgtp (exons 1 and 2, amplicon length 72 bp): 213.fwd-TTGCCACCAGATCAAGGTCA; 284.rev-TCAAAGCTGGAGGCCCAAG;
Acly (exons 18 and 19, amplicon length 132 bp): 2206.fwd-GCGCTACCAGGACACTCCA, 2337.rev-CCGATACACCAGCAGACCACT;
Acsl4 (exons 9 and 10, amplicon length 145 bp): 1274.fwd-ACTTATGGCCGCTGTTCCG, 1419.rev-GGGCGTCATAGCCTTTCTTG;
Tim/TpiI (exons 6 and 7, amplicon length 92 bp): 860.fwd-GGGATGGCTGAAATCCAATG, 952.rev-AGCTCTTTGCAGGTTGCTCC.
Quantitative PCR (qPCR) was performed on a realplex mastercycler (Eppendorf, Vienna, Austria). Samples (1:4 to 1:6 diluted cDNAs) were amplified in duplicates including a standard curve and samples without reverse transcriptase (RT−) were included. Specificity of EvaGreen signals was controlled with melting curve and agarose gel analyses. qPCR data were analyzed using realplex (Eppendorf, Vienna, Austria) software and the standard curve method was used for the calculation of relative expression levels [23,24]. Statistical analysis was done with the software SPSS 17.0 (Mac OS-X, Chicago, IL, USA). Data were log transformed for approximate normality and analyzed with a linear model (ANOVA) with genotype, treatment and interaction as factors. The experiment was treated as random factor. A design matrix of zeros and ones was used to obtain contrasts between genotypes within a treatment and between treatments for each genotype. Appropriate contrasts were calculated (using SPSS) and resulting p-values are reported. Results are shown as means ± standard error (SE).
2.6. Western blot analysis
Western blots were performed on small-sized SDS-PAGE gels as described previously [6]. PageRuler prestained protein ladder (Fermentas, St. Leon-Rot, Germany) was used as a Mr standard. The following antibodies were used: goat anti-TGTP (A-20), goat anti-ACSL4 (N-18), and rabbit anti-TIM (N-21) (Santa Cruz Biotechnology, Santa Cruz, CA, USA); rabbit anti-ACLY and anti-phospho-S454 ACLY3 (Cell Signalling, Danvers, MA, USA); rabbit anti-phospho-T447/S541 ACLY (Epitomics, Burlingame, CA); mouse anti-extracellular signal regulated kinase (pan-ERK: p42, p44, p56 and p85) (BD Biosciences Transduction Laboratories, Lexington, KY, USA); peroxidase-coupled donkey anti-goat IgG (Jackson ImmunoResearch Laboratories, Bar Harbor, Maine, USA); donkey anti-rabbit IgG-HRP F(ab)'2 fragment; sheep anti-mouse IgG-HRP F(ab)'2 fragment; ECL Western Blotting Detection reagents (all GE Healthcare Life Sciences, Munich, Germany).
2.7. Metabolite analysis: lactate, cholesterol and triacylglycerol
Supernatants from BMM were collected for extracellular lactate measurements using a kit from Greiner Diagnostics (Bahlingen, Germany) following the manufacturer's instructions. Briefly, 10 μl of supernatant were incubated with 1 ml of lactate reagent at room temperature for 5 min and the absorbance was measured at 500 nm.
Lipids were extracted from BMM for total cholesterol (TC) and triacylglycerol (TAG) quantitative analysis. BMM were washed three times with PBS and incubated with hexane/isopropanol (60/40, v/v) for 1 h at 4 °C with gentle mixing. Lipid extracts were transferred to sample vials and evaporated to dryness in a vacuum centrifuge. The residue was dissolved in chloroform/methanol (66/33, v/v) and aliquots were taken for TC and TAG analysis. For TAG quantifications 0.3% Triton X-100 was added for complete solubilization. Samples were vacuum dried before determining cellular TC (Greiner Diagnostics, Bahlingen, Germany) and TAG (Diagnostic Systems, Holzheim, Germany) concentrations by enzymatic assays following the manufacturer's instructions. For total protein content determination (Coomassie G-250 protein binding assay [15]) 0.3 M NaOH was added to the cell culture plates after lipid extraction and incubated for 2 h at room temperature. Cell culture plates were kept at − 20 °C overnight and for 4 h at room temperature before the protein assay was performed. Data of metabolite (lactate, TC and TAG) measurements were normalized to cellular total protein content. Statistical evaluation was performed using the software package SPSS version 14.0 (SPSS, Chicago, IL, USA). ANOVA and linear regression were used for statistical analysis as described in Section 2.5.
3. Results
3.1. Statistical evaluation of protein spot expression patterns obtained from 2D-DIGE analysis
Differences in protein patterns in whole cell lysates were compared between WT and Tyk2−/− bone marrow-derived macrophages (BMM) with and without poly(I:C) treatment. 2D-DIGE was used at two different pH gradients (pH 4–7 and pH 6–9) to cover a broad pH range and representative 2-DE protein patterns are shown in Fig.1. A total number of six analytical gels per pH range was analyzed comprising three biological replicates per genotype and treatment. Only protein spots which were present in all images were included in further data analysis (Table 1; pH 4–7: 505 spots and pH 6–9: 490 spots). Experimental variation was assessed based on spot volume ratios which resulted in a mean SD of ± 0.079 and ± 0.087 for pH 4–7 and pH 6–9, respectively. On average, spot expression differences of more than 29% (pH 4–7) and 31% (pH 6–9) produce significant results (p ≤ 0.05) in 90% of the cases with our experimental set-up (sample size per group = 3). These highly reproducible 2D-DIGE spot patterns from whole cell lysates of primary murine macrophages are in agreement with our previous study [12]. The focus of the present analysis was on the impact of Tyk2 before and/or after poly(I:C) treatment. For further analysis, we only considered spots that were differentially regulated between the genotypes and that showed at least 30% difference in spot intensities. 156 protein spots were significantly changed (p ≤ 0.05) in Tyk2−/− compared to WT macrophages (Table 1): 14 spots were up-regulated and 32 down-regulated in the untreated state, 56 spots were up-regulated and 54 down-regulated after poly(I:C) treatment in the absence of Tyk2. 17 of these 156 spot volume changes were found in more than one state, leaving in summary 139 unique differentially regulated spots. In conclusion, Tyk2 positively and negatively affects protein expression both before and after poly(I:C) treatments.
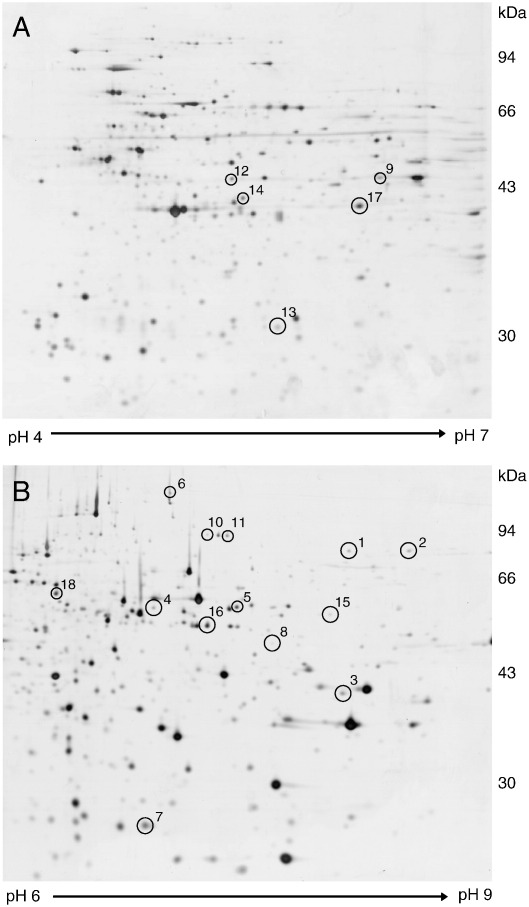
Fig. 1.
Representative silver stained 2-DE protein patterns of macrophages.
Analytical gels visualized by MS-compatible silver stain. Whole cell extracts (75 μg per gel) were separated on pH range (A) 4–7 (sample: WT 18 h poly(I:C) treatment) and (B) 6–9 (sample mix: WT untreated, WT 18 h poly(I:C) and internal standard). Numbered spots indicate proteins which have been identified by PMF and sequence tags applying MALDI-MS (see Table 2 for details).
Table 1.
Numbers of differentially expressed spots and statistical evaluation.
| Spot selection | pH 4–7 | pH 6–9 |
|---|---|---|
| Total number of spots | 1647 | 2036 |
| Number of spots in all images | 505 | 490 |
| Differentially expressed spotsa | ||
| Untreated | 2 (up), 10 (down) | 12 (up), 22 (down) |
| 18 h poly(I:C) | 10 (up), 34 (down) | 46 (up), 20 (down) |
| Selected spots for protein identification | 9 | 17 |
| Spots/proteins identified | 5 | 13 |
| Experimental variation | ||
| Average SD of normalized data | 0.08 | 0.09 |
| Minimal detectable differencesb | 1.29 | 1.31 |
(up) Up-regulated, (down) down-regulated in Tyk2−/− compared to WT macrophages.
Minimal 1.3-fold change in Tyk2−/− compared to WT cells, p < 0.05.
Fold change (power 90%, sample size/group = 3).
3.2. Absence of Tyk2 influences mainly the expression level of proteins functionally assigned to immune response or metabolism
Among the spots showing expression differences, 26 were selected for protein identification based on previously described selection criteria, i.e. spot quality and quantity, spot traceability on silver stained gels and expression patterns [12]. 18 protein spots (Fig. 1) were unambiguously identified by MALDI-MS based on PMF and amino acid sequence tags derived from mass spectrometric fragment ion analysis and results are listed in detail in Table 2. For spots 3 and 18 two highly sequence homologous protein variants were identified showing nearly identical theoretical pI- and Mr-values. In both cases, one unique peptide was assigned to the most probable variant; therefore either only this protein variant or a mixture of both protein variants is contained in the spot. For two proteins multiple spots with similar expression patterns were identified located at similar Mr, but at different pI-positions (Fig. 1, spots 4/5 and spots 10/11). This is likely due to different post-translational modifications, which were not further investigated within the present study. The remaining eight differentially expressed spots could not be successfully identified because database searching with PMFs and sequence tag data did not yield significant results. This can be explained either by insufficient amount of protein and/or incomplete peptide cleavage/extraction (hydrophobic interactions with gel matrix) combined with non-optimal fragmentation behavior.
Table 2.
Summary of differentially expressed protein spots in Tyk2-deficient macrophages +/− poly(I:C).
| Peptide mass fingerprint |
Sequence tag determination |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spot # | Protein name/function | SWISSPROT accession # | WT | Tyk2−/− | WT + poly(I:C) treatment | Tyk2−/− + poly(I:C) treatment | Average ratio Tyk2−/− vs. WT + poly(I:C) treatment | p(gt)a | p(gt × tr)b | MWt (kDa) theor. | p/theor. | Probability based MOWSE score | Sequ. cov. % | Matched/unmatched peptides | Probability based MOWSE score | Precursor Ion(s) m/z (observed) and charge state | Peptide sequence(s) |
| Metabolism | |||||||||||||||||
| 1 | Long-chain-fatty-acid-CoA ligase 4 (ACSL4) | Q9QUJ7 | 1 | 1.28 | 0.76 | 1.45 | 1.90 | nsc | 5.62E-05 | 79.22 | 8.51 | 97 | 18 | 9/7 | 75 | 1002.20 + | FEIPIKVR |
| 1557.62 + | TAEDYCVDENGQR | ||||||||||||||||
| 1628.89 + | YNFPLVTLYATLGR | ||||||||||||||||
| 2 | Peroxisomal multifunctional enzyme type 2 (DHB4) | P51660 | 1 | 1.07 | 0.62 | 0.90 | 1.44 | ns | 7.28E-03 | 79.95 | 8.76 | 218 | 26 | 16/2 | 58 | 1283.52 + | IDVVVNNAGILR |
| 1323.47 + | DATSLNQAALYR | ||||||||||||||||
| 1529.67 + | HVLQQFADNDVSR | ||||||||||||||||
| 3 | 3-ketoacyl-CoA thiolase A, peroxisomal (THIKA)d | Q921H8 | 1 | 1.07 | 0.68 | 1.56 | 2.29 | ns | 8.28E-04 | 43.95 | 8.74 | 157 | 36 | 11/7 | 158 | 1569.73 + | FPQASASDVVVVHGR |
| 2043.37 + | IAQFLSGIPETVPLSTVNR | ||||||||||||||||
| 3-ketoacyl-CoA thiolase B, peroxisomal (THIKB)d | Q8VCH0 | 43.99 | 8.82 | 67 | 20 | 6/12 | 68 | 2043.37 + | IAQFLSGIPETVPLSTVNR | ||||||||
| 4 | Catalase (CATA) | P24270 | 1 | 1.40 | 0.99 | 1.44 | 1.45 | 9.05E-03 | 3.58E-02 | 59.77 | 7.72 | 70 | 13 | 5/2 | 53 | 1277.44 + | LAQEDPDYGLR |
| 1392.53 + | NFTDVHPDYGAR | ||||||||||||||||
| 5 | Catalase (CATA) | P24270 | 1 | 1.42 | 0.58 | 1.22 | 2.12 | 1.34E-02 | 3.97E-03 | 59.77 | 7.72 | 139 | 22 | 10/2 | 77 | 1277.41 + | LAQEDPDYGLR |
| 1392.46 + | NFTDVHPDYGAR | ||||||||||||||||
| 6 | ATP-citrate synthase (ACLY) | Q91V92 | 1 | 1.25 | 0.58 | 0.91 | 1.57 | 2.03E-02 | 7.06E-04 | 119.73 | 7.13 | 185 | 18 | 15/2 | 42 | 875.01 + | EAGVFVPR |
| 1568.84 + | TIAIIAEGIPEALTR | ||||||||||||||||
| 7 | Triosephosphate isomerase (TIM,TPIS) | P17751 | 1 | 1.21 | 0.73 | 1.13 | 1.56 | 8.26E-04 | 7.95E-04 | 26.71 | 6.90 | 244 | 65 | 14/4 | 112 | 1248.34 + | SNVNDGVAQSTR |
| 1540.71 + | DLGATWVVLGHSER | ||||||||||||||||
| 8 | Fructose-bisphosphate aldolase A (ALDOA) | P05064 | 1 | 1.21 | 0.83 | 1.47 | 1.76 | 5.26E-03 | 5.89E-04 | 39.36 | 8.30 | 120 | 29 | 7/1 | 176 | 2108.36 + | IGEHTPSALAIMENANVLAR |
| 2259.38 + | YTPSGQSGAAASESLFISNHAY | ||||||||||||||||
| 9 | α-enolase (ENOA) | P17182 | 1 | 1.33 | 0.86 | 1.20 | 1.40 | 2.72E-02 | 1.63E-02 | 47.14 | 6.37 | 120 | 19 | 8/1 | 63 | 704.80 + | GVPLYR |
| 1440.60 + | YITPDQLADLYK | ||||||||||||||||
| 10 | Aconitate hydratase, mitochondrial (ACON) | Q99KI0 | 1 | 1.11 | 0.48 | 0.78 | 1.63 | 4.11E-02 | 3.21E-03 | 85.46 | 8.08 | 86 | 11 | 8/5 | 84 | 1464.62 + | SQFTITPGSEQIR |
| 1602.73 + | NAVTQEFGPVPDTAR | ||||||||||||||||
| 11 | Aconitate hydratase, mitochondrial (ACON) | Q99KI0 | 1 | 1.23 | 0.82 | 1.19 | 1.46 | 5.40E-04 | 1.08E-03 | 85.46 | 8.08 | 311 | 33 | 23/4 | 136 | 1464.64 + | SQFTITPGSEQIR |
| 1602.77 + | NAVTQEFGPVPDTAR | ||||||||||||||||
| Immune response | |||||||||||||||||
| 12 | T cell-specific GTPase (TGTP) | NP_035709e | 1 | 1.09 | 8.53 | 4.39 | 0.51 | ns | 3.15E-03 | 47.15 | 5.49 | 310 | 54 | 20/0 | 152 | 1679.87 + | FGEYDFFIIISATR |
| 1867.08 + | DIESAPLHIAVTGETGAGK | ||||||||||||||||
| 13 | Proteasome activator complex subunit 1 (PSME1) | P97371 | 1 | 0.70 | 1.53 | 1.37 | 0.89 | 1.07E-04 | ns | 28.67 | 5.73 | 270 | 71 | 18/3 | 146 | 1396.63 + | NAYAVLYDIILK |
| 1873.03 + | QLVHELDEAEYQEIR | ||||||||||||||||
| 14 | Interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) | Q64345 | 1 | 0.66 | 7.15 | 4.78 | 0.67 | 3.61E-03 | 1.36E-03 | 47.93 | 5.51 | 241 | 52 | 22/7 | 237 | 1741.90 + | STVNNSPLYSLVMCR |
| 1891.12 + | KSEDLAALECLLQFPR | ||||||||||||||||
| 15 | Interferon-activable protein 205-B (IFI5B) | Q08619 | 1 | 0.26 | 2.76 | 3.15 | 1.14 | 3.61E-06 | 2.80E-01 | 47.59 | 8.38 | 58 | 11 | 6/5 | 141 | 1361.48 + | QMIEVPNCITR |
| 1772.04 + | GILEINETSSVLEAAPK | ||||||||||||||||
| Others | |||||||||||||||||
| 16 | Immune-responsive protein 1 (IRG-1) | P54987 | 1 | 1.16 | 34.73 | 58.59 | 1.69 | ns | 4.05E-03 | 54.01 | 7.09 | 71 | 13 | 5/2 | 90 | 1169.28 + | ALFPADHIER |
| 1579.68 + | LSSMSSFDHTTLPR | ||||||||||||||||
| 17 | UMP-CMP kinase 2, mitochondrial (CMPK2) (Thymidylate kinase LPS-inducible member - TYKi) | Q3U5Q7 | 1 | 0.84 | 13.96 | 8.03 | 0.58 | ns | 1.26E-03 | 50.03 | 6.89 | 192 | 31 | 13/3 | 142 | 1102.32 + | VLELIQSSGR |
| 1622.73 + | AVLEECTSFIPEAR | ||||||||||||||||
| 18 | EH domain-containing protein 1 (EHD1)d | Q9WVK4 | 1 | 1.34 | 2.03 | 3.18 | 1.57 | ns | 5.00E-04 | 60.60 | v6.35 | 180 | 32 | 15/3 | 140 | 1237.38 + | LNAFGNAFLNR |
| 1675.80 + | LLDTVDDMLANDIAR | ||||||||||||||||
| 2015.27 + | VYIGSFWSHPLLIPDNR | ||||||||||||||||
| EH domain-containing protein 3 (EHD3)d | Q9QXY6 | 60.87 | 6.03 | 69 | 16 | 7/11 | 77 | 1237.38 + | LNAFGNAFLNR | ||||||||
| 2015.27 + | VYIGSFWSHPLLIPDNR | ||||||||||||||||
p-values for the differences between genotypes (gt) under basal conditions.
p-values for the differences between genotypes in response to poly(I:C) treatment (gt × tr).
Not significant (p-value > 0.05).
Proteins with high sequence homology.
Accession # from NCBI nr database.
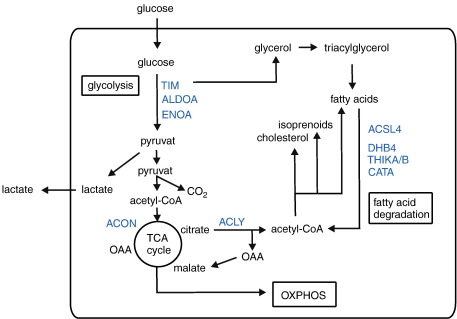
Identified proteins were grouped on the basis of their major cellular function according to the PANTHER classification system [25], gene ontology (GO) [26] and UniProtKB databases [27] (Table 2). In line with the role of Tyk2 in autocrine/paracrine IFNα/β signaling, we identified four proteins known as IFNα/β-regulated proteins: T-cell specific GTPase (TGTP), proteasome activator complex subunit 1 (PSME1), IFN-induced protein with tetratricopeptide repeats 3 (IFIT3) and IFN-activable protein 205-B (IFI5B). All of them showed decreased protein expression in the absence of Tyk2 in the basal state and/or after poly(I:C) stimulation. Remarkably, the majority of differentially expressed protein spots were identified as metabolic enzymes and all showed an increased expression by 1.40- to 2.29-fold in the absence of Tyk2 after poly(I:C) treatment. All of them are involved in cellular lipid or carbohydrate metabolic processes, or both (Fig. 2). Long-chain-fatty-acid-CoA ligase 4 (ACSL4), peroxisomal multifunctional enzyme type 2 (DHB4) and peroxisomal 3-ketoacyl-CoA thiolase A or B (THIKA/B) are enzymes of the fatty acid degradation pathway (β-oxidation) [27,28]. DHB4 and THIKA/B are exclusively allocated to peroxisomal and ACSL4 to mitochondrial and peroxisomal subcellular compartments. In addition, catalase (CATA) degrades H2O2, which is produced as a result of peroxisomal β-oxidation [29]. Regarding carbohydrate metabolism, we found triosephosphate isomerase (TIM), fructose-bisphosphate aldolase A (ALDOA) and α-enolase (ENOA), which are all involved in the glycolytic/gluconeogenesis pathways. Furthermore, ATP-citrate lyase (ACLY), which is required for acetyl-CoA formation, and aconitate hydratase (ACON), which is a part of the tricarboxylic acid cycle (TCA, citrate cycle), were differentially regulated between WT and Tyk2−/− cells. TIM and ACLY are enzymes linking carbohydrate and lipid metabolic processes. TIM participates in the production of glycerol, a precursor for the synthesis of triacylglycerol (TAG), whereas ACLY is involved in the glucose-dependent lipogenesis [27,28]. In summary, the presented results show a substantial impact of Tyk2 on the expression of proteins mainly involved in fatty acid degradation and glycolysis.
Fig. 2.
Overview of identified enzymes within the metabolic pathways.
Scheme showing the identified differentially regulated metabolic enzymes (depicted in blue), metabolic pathways and the connection to specific metabolites (triacylglycerol, cholesterol and lactate). ACLY, ATP-citrate lyase; ACON, aconitate hydratase; ACSL4, long-chain-fatty-acid-CoA ligase 4; ALDOA, fructose-bisphosphate aldolase A; CATA, catalase; DHB4, peroxisomal multifunctional enzyme type 2; ENOA, α-enolase; OAA, oxalacetate; OXPHOS, oxidative phosphorylation; TCA cycle, tricarboxylic acid cycle; THIKA/B, peroxisomal 3-ketoacyl-CoA thiolase A or B; TIM, triosephosphate isomerase.
From the proteins identified, one IFNα/β-regulated protein and three enzymes involved in carbohydrate and/or lipid metabolic pathways were selected for more detailed analysis.
3.3. Transcriptional up-regulation of TGTP in response to poly(I:C) is reduced in the absence of Tyk2
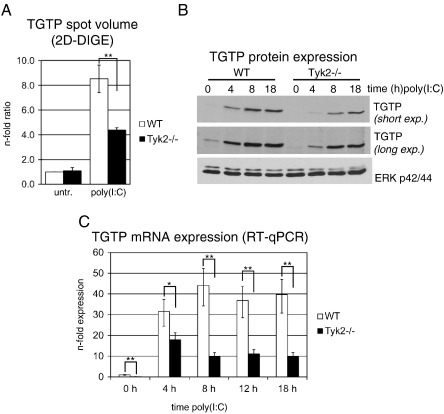
TGTP is a member of the 47 kDa GTPase family [30] and was discovered based on its transcriptional induction by IFNγ in macrophages and T-cells [31]. TGTP is also efficiently induced by transfected poly(I:C) in fibroblast cell lines in an IFNα/β-dependent manner [32]. TGTP possesses specific antiviral activity, is induced upon MCMV infection [33] and is implicated in cell survival after infection with vesicular stomatitis virus, but not herpes simplex virus [32]. Confirming our data from the 2D-DIGE analysis (Table 2, spot 12 and Fig. 3A), we found a strong and partially Tyk2-dependent up-regulation of TGTP by Western blot analysis (Fig. 3B). As expected, induction of TGTP occurred at the transcriptional level (Fig. 3C) and was clearly reduced, but not abolished, in Tyk2-deficient cells. In contrast to the 2D-DIGE data (Fig. 3A), TGTP mRNA expression (Fig. 3C) and TGTP protein expression as determined by Western blot analysis (Fig. 3B, middle panel) were already significantly reduced in untreated Tyk2−/− cells. Despite this discrepancy in the basal state, these data further confirm the requirement of Tyk2 for efficient autocrine/paracrine IFNα/β responses and extend our previous findings to additional IFNα/β-stimulated proteins.
Fig. 3.
Effect of Tyk2 deficiency on TGTP expression levels.
Macrophages were treated with poly(I:C) for the times indicated or left untreated (untr., 0 h) and whole cell lysates were analyzed by (A) 2D-DIGE and (B) Western blotting or (C) total RNA was isolated and mRNA expression determined by RT-qPCR. (A) Expression levels as determined by 2D-DIGE (untr. and 18 h poly(I:C)) are given as fold ratios relative to untreated WT cells. Mean values ± SD of three biological replicates are depicted. (B) 10 μg protein per lane was separated by SDS-PAGE (10%T) and probed with an anti-TGTP antibody (short and long exposures are depicted). ERK p42/p44 was used as loading control. Data are representatives of three independent experiments. (C) Mean values of TGTP mRNA expression ± SE from three independent experiments are depicted. Expression levels are shown relative to untreated WT cells, Ube2d2 was used for normalization. Significance levels of genotype-specific differences for each time point are indicated with * p ≤ 0.05 and ** p ≤ 0.01. Significance levels not indicated in the figures are for WT untreated vs. all treatment time points p ≤ 0.01, Tyk2−/− untreated vs. all treatment time points p ≤ 0.01.
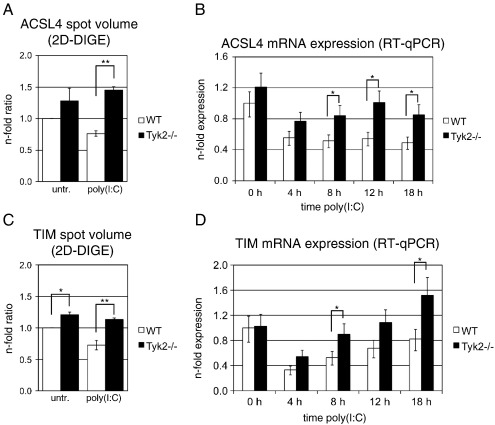
3.4. Tyk2 is required for transcriptional regulation of ACSL4 and TIM
ACSL4 belongs to the large family of long-chain acyl-CoA synthetases (ACSLs) [34], which are essential for de novo lipid synthesis, fatty acid degradation and phospholipid remodeling of membranes. Membrane-bound ACSL proteins catalyze ATP-dependent activation of hydrophobic long-chain fatty acids to hydrophilic acyl-CoA, whereby ACSL4 specifically prefers arachidonate as substrate [35]. According to the presented 2D-DIGE data, ACSL4 was down-regulated by poly(I:C) treatment to 0.76-fold in WT cells (Table 2, spot 1 and Fig. 4A). In the absence of Tyk2, ACSL4 was slightly but not significantly enhanced in untreated cells and, importantly, ACSL4 was not down-regulated after poly(I:C) treatment (Fig. 4A). The mean difference between WT and Tyk2−/− cells in ACSL4 levels after poly(I:C) treatment was 1.9-fold (Table 2, spot 1). We next analyzed whether ACSL4 was also regulated at the transcriptional level. As shown in Fig. 4B, ACSL4 mRNA was down-regulated after 4 h of poly(I:C) treatment and reached levels of 0.5-fold at 18 h after treatment in WT cells. Although ACSL4 mRNA was also slightly down-regulated in Tyk2-deficient cells, mRNA levels were significantly higher than in WT cells from 8 h after poly(I:C) treatment onwards.
Fig. 4.
Effect of Tyk2 deficiency on ACSL4 and TIM expression levels.
Macrophages were treated with poly(I:C) for the times indicated or left untreated (untr., 0 h) and (A, C) whole cell lysates were analyzed by 2D-DIGE or (B, D) total RNA was isolated and mRNA expression determined by RT-qPCR. Expression levels are given as described in the legend to (A, C) Fig. 3A and (B, D) Fig. 3C. Significance levels of genotype-specific differences for each time point are indicated with * p ≤ 0.05 and ** p ≤ 0.01. Significance levels not indicated in the figures are for (B) WT untreated vs. all treatment time points p ≤ 0.01, Tyk2−/− untreated vs. 4 h treatment p ≤ 0.05 and for (D) WT untreated vs. 4 h poly(I:C) p ≤ 0.01 and 8 h treatment p ≤ 0.05, Tyk2-/− untreated vs. 4 h treatment p ≤ 0.05.
TIM, also known as TPI or TPIS, catalyzes the isomerization of dihydroxyacetone phosphate (DHAP) and d-glyceraldehyde-3-phosphate (GADP). DHAP and GADP occur as intermediates in the glycolysis/gluconeogenesis pathways and DHAP can also be metabolized to glycerol, a precursor for the synthesis of TAG [36]. As determined by 2D-DIGE, the TIM spot intensity patterns were very similar to those of ACSL4. TIM was modestly down-regulated by poly(I:C) treatment in WT cells and absence of Tyk2 resulted in significantly higher TIM levels both before and after poly(I:C) treatment (Table 2, spot 7 and Fig. 4C). The differences were again very modest and higher after stimulation (Tyk2−/− vs. WT ratios in the basal and treated state of 1.21-fold and 1.56-fold, respectively). TIM mRNA was transiently down-regulated in WT and, to a lesser extent, in Tyk2−/− macrophages (Fig. 4D). TIM mRNA levels were significantly higher in Tyk2−/− compared to WT cells at 8 h and 18 h after poly(I:C) treatment, however, a trend for enhanced expression was also seen at 4 h and 12 h (p ≤ 0.1). As might be expected based on these relatively small changes in expression levels, we could not detect consistent differences in protein levels of ACSL4 and TIM using Western blot analysis (data not shown). Taken together, both ACSL4 and TIM are clearly, but modestly, transcriptionally down-regulated in response to poly(I:C) treatment and this is at least partially mediated by Tyk2-dependent mechanisms.
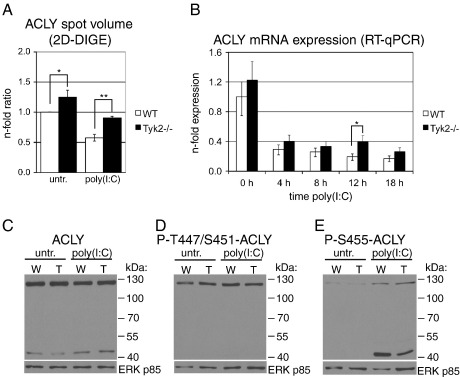
3.5. Tyk2 is involved in the post-translational regulation of ACLY
ACLY is a cytosolic enzyme that catalyzes the conversion of mitochondria-derived citrate to oxalacetate and acetyl-CoA. Acetyl-CoA is further utilized for the synthesis of fatty acids, cholesterol and isoprenoids. Oxalacetate is reduced to malate, which can return to the mitochondria and complete the substrate cycle [28]. ACLY can also positively regulate glycolysis, presumably via the removal of citrate, which acts as a glycolytic inhibitor [37]. ACLY protein spot levels as determined by 2D-DIGE were modestly reduced to 0.58-fold by poly(I:C) treatment in WT cells and 1.57-fold up-regulated in Tyk2−/− as compared to WT cells after poly(I:C) treatment (Table 2, spot 6 and Fig. 5A). Similar to TIM, differences in ACLY spot intensities were already evident in untreated cells. Interestingly, we found only minor differences between WT and Tyk2−/− cells at the mRNA level (Fig. 5B). ACLY mRNA was reduced about 4-fold in both WT and Tyk2−/− cells after poly(I:C) treatment. Significantly enhanced ACLY mRNA expression was only detectable at 12 h after poly(I:C) treatment in Tyk2−/− cells, but mRNA levels were again similar to those in WT cells at the 18 h time point. Surprisingly, despite the clear down-regulation of ACLY mRNA, ACLY protein levels were unchanged in both WT and Tyk2−/− cells in response to poly(I:C) treatment (Fig. 5C). In addition to the full-length ACLY protein (around 125 kDa), we also found an immunoreactive band at a lower Mr. The latter likely corresponds to one of the previously reported proteolytic cleavage products of ACLY with a reported Mr of 50–53 kDa [38–40]. Proteolytically cleaved ACLY has been found in rat liver homogenates [38,39] and in glioblastoma cell lines [37] and, importantly, retains its full enzymatic activity [39]. A 50 kDa ACLY fragment was also identified as a phospho-acceptor protein in rat liver [40]. Murine ACLY can be phosphorylated at serine 455 (S455) and tyrosine 672 (Y672) and multiple additional phosphorylation sites are predicted by similarity to human or rat ACLY, e.g. threonine 447 (T447) and S451 [27,41]. In order to test whether poly(I:C) treatment and/or Tyk2 deficiency affects ACLY phosphorylation, we performed Western blot analysis with the commercially available phospho-ACLY-specific antibodies. As shown in Fig. 5D, phospho(P)-T447/S451 was detectable in untreated cells, unchanged by poly(I:C) treatment and similar between WT and Tyk2−/− cells. Notably, P-T447/S451-ACLY was only detected at the Mr corresponding to full-length ACLY. In contrast, P-S455-ACLY was strongly induced upon poly(I:C) treatment (Fig. 5E). P-S455-ACLY was mainly found at the lower Mr band, indicating that cleaved ACLY is preferentially phosphorylated or, alternatively, P-S455 targets full-length ACLY for proteolysis. As the total amount of the ACLY fragment did not increase upon stimulation (Fig. 5C), the former possibility is more likely. It is possible that proteolytic cleavage occurs during the protein isolation procedure, as activated macrophages express particularly high levels of proteases and ACLY is quite unstable [40]. However, we observed similar levels of the proteolytic product even under more stringent lysis conditions, i.e. using RIPA buffer (data not shown). Importantly, the poly(I:C)-mediated increase of P-S455-ACLY was clearly reduced in Tyk2−/− cells (Fig. 5E). Phosphorylation of human ACLY at S455 has been shown to increase ACLY enzymatic activity around 6-fold, whereas phosphorylation on T447/S541 had no detectable effect [42]. In relation to our 2D-DIGE data, we assume that the poly(I:C)-induced down-regulation of the 125 kDa ACLY spot identified with MS is a result of post-translational modification(s), i.e. shifts in pI caused by phosphorylation and/or changes in Mr by proteolytic cleavage. As indicated by the mRNA data, it is also possible that total ACLY protein levels are decreased in response to poly(I:C), but as we did not find any differences using semi-quantitative Western blots these changes must be rather modest. Taken together, our data suggest that the major effect of poly(I:C) treatment on ACLY is the induced S455 phosphorylation and the likely resulting increase of enzymatic activity. Tyk2 did not grossly impact on total ACLY expression, but was required for maximal poly(I:C)-mediated ACLY S455 phosphorylation.
Fig. 5.
Effect of Tyk2 deficiency on ACLY expression levels.
Macrophages were treated with poly(I:C) for the times indicated or left untreated (untr., 0 h) and whole cell lysates were analyzed by (A) 2D-DIGE, (C–E) Western blotting or (B) total RNA was isolated and mRNA expression determined by RT-qPCR. Expression levels are given as described in the legend to (A) Fig. 3A and (B) Fig. 3C. (C–E) 10 μg protein per lane was separated by SDS-PAGE (8%T) and probed with an (C) anti-ACLY, (D) anti-phospho-T447/S451-ACLY and (E) anti-phospho-S455-ACLY antibody. (C–E) ERK p85 was used as loading control. Data are representatives of three independent experiments. (A, B) Significance levels of genotype-specific differences for each treatment are indicated with * p ≤ 0.05 and ** p ≤ 0.01. Significance levels not indicated in the figures are for both WT and Tyk2−/− cells untreated vs. all treatment time points p ≤ 0.01. (C–E) W, WT; T, Tyk2−/−.
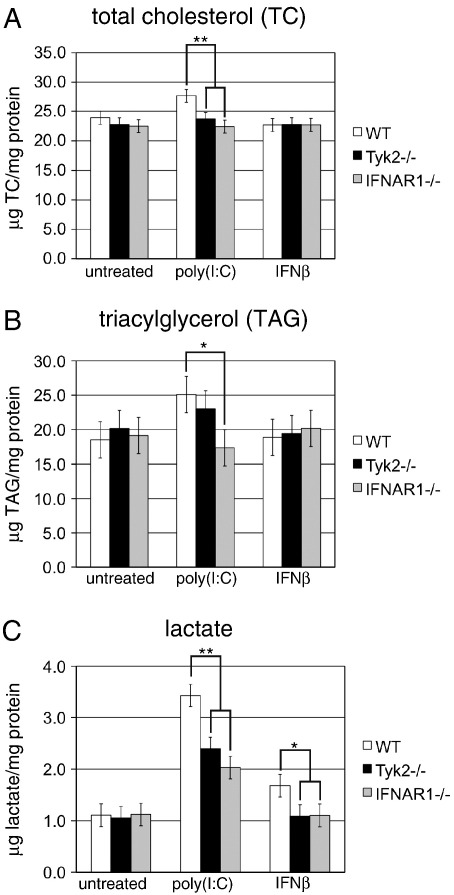
3.6. Tyk2 is required for the up-regulation of cellular total cholesterol and extracellular lactate in macrophages
We next decided to more directly test the influence of poly(I:C) treatment and the consequences of Tyk2 deficiency on cellular metabolism. To this end, we assessed the impact of Tyk2 on the amounts of cellular total cholesterol (TC), TAG and extracellular lactate. As most of the effects of Tyk2 in macrophages were thus far attributable to its role in type I IFN signaling, we included macrophages derived from IFNα/β receptor 1-deficient (IFNAR1−/−) mice in these analyses and, additionally, treated cells with recombinant IFNβ. The cellular TC concentration was slightly (1.16-fold) but significantly increased in WT cells after poly(I:C) treatment (Fig. 6A). TC levels of both Tyk2−/− and IFNAR1−/− macrophages were similar to those of WT cells in the untreated state, but showed no increase in response to poly(I:C). TC levels after IFNβ treatment remained unchanged in all three genotypes (Fig. 6A). Thus, functional IFN signaling is required but not sufficient for the up-regulation of cellular TC. A similar but not identical picture appeared for the TAG concentrations (Fig. 6B). In WT cells, poly(I:C) treatment resulted in a modest but consistent increase in cellular TAG levels (Fig. 6B). Again, IFNAR1 was required for this effect, but treatment with IFNβ alone did not cause any changes in TAG levels. TAG levels in Tyk2−/− cells were comparable to WT cells before and after poly(I:C) treatment (Fig. 6B). The strongest poly(I:C)-mediated changes were observed for extracellular lactate (Fig. 6C). In WT cells, 3.43-fold increased lactate levels were found after 18 h of poly(I:C) treatment. Neither Tyk2 nor IFNAR1 deficiency resulted in changes of extracellular lactate levels in untreated cells, but up-regulation by poly(I:C) was clearly reduced in both genotypes. WT cells treated with exogenous IFNβ showed a modest up-regulation of extracellular lactate and this was, as expected, not observed in cells lacking IFNAR1. In contrast to TC and TAG, IFNβ alone is sufficient to induce a change in lactate release, nevertheless other poly(I:C)-induced factors contribute to the effect. The poly(I:C)- and the IFNβ-mediated increase in lactate release were partially or completely dependent on the presence of Tyk2.
Fig. 6.
Effects of Tyk2 and IFNAR1 deficiency on metabolite concentrations.
Macrophages were cultivated in the absence (untreated) or presence of poly(I:C) or IFNβ for 18 h. Intracellular (A) TC and (B) TAG and (C) extracellular lactate were measured enzymatically and normalized to cellular total protein content. Results are mean values ± SE of the following number of independent experiments: TC, n = 7; TAG, n = 3; lactate, n = 4. Significance levels of genotype-specific differences for each treatment are indicated with * p ≤ 0.05 and ** p ≤ 0.01. Significance levels not indicated in the figures: TC, WT (untreated) vs. WT (poly(I:C)) p ≤ 0.01; TAG, WT (untreated) vs. WT (poly(I:C)) p ≤ 0.05; lactate, WT (untreated) vs. WT (poly(I:C)) p ≤ 0.01, WT (untreated) vs. WT (IFNβ) p ≤ 0.05.
4. Discussion
Tyk2 is mainly associated with its protective role during microbial infections. In macrophages, absence of Tyk2 results in reduced type I IFN signaling, partial defects in LPS responses and increased virus sensitivity [5,12,24]. In the present study, we show that besides the expected effects on immune response proteins, Tyk2 is involved in the regulation of carbohydrate and lipid metabolism in macrophages exposed to poly(I:C). By means of 2D-DIGE and MS we find protein spot expression levels of 9 distinct enzymes known to be involved in carbohydrate and/or fatty acid metabolism up-regulated in the absence of Tyk2. We further analyzed mRNA expression patterns for TIM, ACSL4 and ACLY and found transcriptional and post-transcriptional effects of Tyk2. Importantly, we also correlate our findings to metabolite abundance.
TIM catalyzes the equilibrium of triosephosphates in the gluconeogenesis/glycolysis pathway, which is also connected to fatty acid biosynthesis [36]. We report here that TIM is transcriptionally down-regulated in a Tyk2-dependent manner after poly(I:C) treatment. However, the difference between WT and Tyk2−/− cells was less than 2-fold at the mRNA and protein spot level and could not be detected by Western blot analysis. Similarly, ACSL4, which is involved in fatty acid degradation, showed modest and Tyk2-dependent transcriptional down-regulation by poly(I:C) treatment. This is in accordance with a previous transcriptional study in macrophages treated with LPS, in which genes annotated to lipid and cholesterol metabolism were, as a group, significantly regulated in a Tyk2-dependent manner, but showed a very modest amplitude of regulation at the single gene level [43]. Small changes in multiple enzyme transcript levels that impact on overall cellular metabolic activity have been reported just recently in the context of sterol biosynthesis and viral infection [44]. ACLY is best known for its involvement in glucose-mediated lipogenesis [28]. From a cell-signaling perspective the recent finding that ACLY is also the major source of acetyl-CoA required for histone acetylation and selective gene expression in response to growth factor stimulation and during adipocyte differentiation is of particular interest [45]. We found that ACLY mRNA is down-regulated by poly(I:C) treatment in WT and Tyk2−/− macrophages. Phosphorylation of ACLY at S455 is strongly induced by poly(I:C) treatment and is clearly reduced in the absence of Tyk2. S455 phosphorylation of ACLY substantially enhances its enzymatic activity in vitro [42], arguing for a poly(I:C)-mediated increase in ACLY activity, despite the modest down-regulation of its mRNA level. Nutrients, insulin and interleukin-3 were shown to induce phosphorylation of S455-ACLY and increased P-S455-ACLY was reported in tumor cell lines and tissues [37,46,47]. More related to our study, induction of P-S455-ACLY has recently been demonstrated in human fibroblasts infected with CMV [48]. Protein kinase B (PKB/Akt) and cAMP-dependent protein kinase (PKA) can act as ACLY S455 kinases in vitro and, PKB/Akt has been linked to ACLY S455 phosphorylation in lung adenocarcinoma cell lines and in response to insulin in adipocytes [42,46,49–51]. As type I IFNs can activate PKB/Akt [52] it may be speculated that reduced ACLY phosphorylation in the absence of Tyk2 is due to reduced autocrine/paracrine IFNα/β-mediated PKB/Akt activation.
Interconnections between infection/inflammation and lipid metabolism are becoming increasingly apparent [53–55], but underlying molecular mechanisms are only just beginning to emerge. Because of their anticipated role in atherosclerosis [56], most studies in macrophages focus on their differentiation into lipid-loaden foam cells in the presence of modified low-density lipoprotein. Nevertheless, an increase in intracellular TAG even in the absence of exogenous lipid has been previously reported in RAW264.7 macrophage cells treated with LPS [57,58]. We show a slight but significant increase in TAG levels upon poly(I:C) treatment in WT but not in IFNAR1−/− cells. In contrast, no effect on TAG levels upon poly(I:C) treatment was reported in a previous study [58]. One possible reason for this discrepancy is the difference in the type of macrophages used, i.e. primary macrophages versus a cell line. As we find enzymes involved in fatty acid degradation and synthesis down-regulated by poly(I:C), it is also possible that the fatty acid composition is altered during macrophage activation. Similar to TAG, we found a slight increase in TC concentrations after poly(I:C) treatment. This was dependent on the presence of both Tyk2 and IFNAR1. Liver X receptors (LXRs) are nuclear receptors crucial for the regulation of cholesterol homeostasis and involved in the crosstalk between lipid metabolism and TLR signaling [59,60]. Stimulation with TLR3 or TLR4 ligands blocks macrophage cholesterol efflux by blocking expression of LXR target genes, e.g. the cholesterol efflux mediators ATP-binding cassette sub-family A member 1 (ABCA1) and sub-family G member 1 (ABCG1) [59]. Intracellular cholesterol levels were, however, not determined in this study. The inhibition of LXR target gene expression occurred independent of type I IFN [59], indicating that the IFNAR1-dependent, poly(I:C)-mediated increase in intracellular cholesterol reported herein is caused by a different mechanism. In contrast to our data, MCMV infection and IFNβ treatment resulted in 2-fold decreased cellular free cholesterol levels [44]. This difference might be explained by differences between MCMV- and poly(I:C)-induced cellular responses.
In most cells, including macrophages, glycolysis serves to convert glucose to pyruvate and then generate ATP by oxidative phosphorylation (OXPHOS). At low oxygen level pyruvate is converted to lactate (anaerobic glycolysis) to produce ATP, however with less efficiency [61]. We show that extracellular lactate levels strongly increase after poly(I:C) treatment, indicating an increase in glycolytic rates. In line with this, a strong increase in extracellular lactate was observed in macrophages stimulated with LPS [62,63]. More recently, a shift from OXPHOS to anaerobic glycolysis was reported in dendritic cells activated with LPS and, interestingly, this was dependent on the presence of PKB/Akt [64]. We propose a similar shift in response to poly(I:C) and demonstrate a strong but not absolute requirement for IFNAR1 and Tyk2. Of note, studies in the 1960s already suggested effects of IFNs on OXPHOS in the context of viral infections [65,66]. Our data support this hypothesis and provide evidence that Tyk2 facilitates the transition from OXPHOS to anaerobic glycolysis.
Given the importance of lipid and glucose metabolism for the innate and adaptive immune system [53], it appears likely that the defects in metabolic reprogramming upon immune stimulation in Tyk2−/− macrophages reported herein contribute to immunological phenotypes described in Tyk2−/− mice. In particular the failure to efficiently switch from OXPHOS to anaerobe glycolysis might impair macrophage activation and result in increased sensitivity to death by nutrient limitation. This could broadly inhibit inflammatory processes and contribute to the resistance of Tyk2−/− mice to inflammation-induced pathologies. Control of glucose metabolism is, for example, considered as promising therapeutic approach in sepsis, although it emerged more difficult than initially anticipated [67]. Against this background it seems interesting to determine whether Tyk2−/− mice show reduced hyperglycemia during LPS-induced shock. Our results also raise the question whether the metabolic phenotype extends to other cell types. This is particularly interesting in lymphocytes, as they have a high glucose-dependent energy demand during clonal expansion upon antigen encounter [53], and Tyk2−/− pro-B cells exhibit defects in homeostatic and IFNβ-induced mitochondrial respiration [68].
5. Concluding remarks
The presented data show that the absence of Tyk2 results in significant changes on cellular protein abundance in macrophages in the basal state and upon poly(I:C) treatment, which is in line with our recently published study on LPS responses [12]. We observed an explicit impact of Tyk2 on the abundance of several lipid and carbohydrate metabolic enzymes. We show that Tyk2-dependent regulations occur transcriptionally and/or post-transcriptionally and correlate these data to changes in metabolite levels. Our data emphasize the importance of Tyk2 for the regulation of cellular immune responses and provide new evidence for a novel function of Tyk2 linking metabolic and innate immune networks.
Authors contributions
TG performed the 2D-DIGE, MS and metabolite experiments, participated in the design of the study and wrote the first draft of the manuscript together with MM-D. NL did most of the Western blot analyses, BW the RT-qPCR studies and MR helped with the cell culture and the 2D-DIGE experiments. CV performed and supervised the statistical evaluations and DK supervised the metabolite assays. TK was responsible for the mouse routine and experimental breeding. IM and MG supervised the 2D-DIGE analysis and MM-D and GA the MS experiments. GA, IM and MM-D contributed to the design and coordination of the study. BS and MM co-ordinated and designed the study and wrote the final manuscript. All authors read and approved the final manuscript.
Acknowledgments
We thank Anton Ibovnik (Medical University Graz, Graz, Austria) for help in establishing the lipid extraction procedure in our lab. Mathias Müller and Birgit Strobl are supported by the Austrian Science Fund (FWF, grant SFB-F28) and Dagmar Kratky by the FWF grant SFB-F30. Mathias Müller is also supported by the Austrian Federal Ministry of Science and Research (GEN-AU program project Austromouse).
Footnotes
According to the current amino acid numbering in the UniProtKB database entries, S454 corresponds to S455 in both human and mouse ACLY (S454 in the rat homolog); this is not indicated in the antibody datasheet and therefore mentioned here.
References
- 1.O'Shea J.J., Gadina M., Schreiber R.D. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002;109:S121–S131. doi: 10.1016/s0092-8674(02)00701-8. Suppl. [DOI] [PubMed] [Google Scholar]
- 2.Schindler C., Levy D.E., Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 3.Velazquez L., Fellous M., Stark G.R., Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 4.Rane S.G., Reddy E.P. Janus kinases: components of multiple signaling pathways. Oncogene. 2000;19:5662–5679. doi: 10.1038/sj.onc.1203925. [DOI] [PubMed] [Google Scholar]
- 5.Karaghiosoff M., Neubauer H., Lassnig C., Kovarik P., Schindler H. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity. 2000;13:549–560. doi: 10.1016/s1074-7613(00)00054-6. [DOI] [PubMed] [Google Scholar]
- 6.Strobl B., Bubic I., Bruns U., Steinborn R., Lajko R. Novel functions of tyrosine kinase 2 in the antiviral defense against murine cytomegalovirus. J Immunol. 2005;175:4000–4008. doi: 10.4049/jimmunol.175.6.4000. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto M., Seya T. TLR3: interferon induction by double-stranded RNA including poly(I:C) Adv Drug Deliv Rev. 2008;60:805–812. doi: 10.1016/j.addr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi O., Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Schindler C., Plumlee C. Inteferons pen the JAK-STAT pathway. Semin Cell Dev Biol. 2008;19:311–318. doi: 10.1016/j.semcdb.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitha P.M. Unexpected similarities in cellular responses to bacterial and viral invasion. Proc Natl Acad Sci USA. 2004;101:695–696. doi: 10.1073/pnas.0307303101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doyle S.L., O'Neill L.A. Toll-like receptors: from the discovery of NFkappaB to new insights into transcriptional regulations in innate immunity. Biochem Pharmacol. 2006;72:1102–1113. doi: 10.1016/j.bcp.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Radwan M., Miller I., Grunert T., Marchetti-Deschmann M., Vogl C. The impact of tyrosine kinase 2 (Tyk2) on the proteome of murine macrophages and their response to lipopolysaccharide (LPS) Proteomics. 2008;8:3469–3485. doi: 10.1002/pmic.200800260. [DOI] [PubMed] [Google Scholar]
- 13.Muller U., Steinhoff U., Reis L.F., Hemmi S., Pavlovic J. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 14.Baccarini M., Bistoni F., Lohmann-Matthes M.L. In vitro natural cell-mediated cytotoxicity against Candida albicans: macrophage precursors as effector cells. J Immunol. 1985;134:2658–2665. [PubMed] [Google Scholar]
- 15.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Blum H., Gross H.J., Beier H. The expression of the TMV-specific 30-kDa protein in tobacco protoplasts is strongly and selectively enhanced by actinomycin. Virology. 1989;169:51–61. doi: 10.1016/0042-6822(89)90040-8. [DOI] [PubMed] [Google Scholar]
- 18.Vorm O., Roepstorff P., Mann M. Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal Chem. 1994;66:3281–3287. [Google Scholar]
- 19.Smirnov I.P., Zhu X., Taylor T., Huang Y., Ross P. Suppression of alpha-cyano-4-hydroxycinnamic acid matrix clusters and reduction of chemical noise in MALDI-TOF mass spectrometry. Anal Chem. 2004;76:2958–2965. doi: 10.1021/ac035331j. [DOI] [PubMed] [Google Scholar]
- 20.Mattow J., Schmidt F., Hohenwarter W., Siejak F., Schaible U.E. Protein identification and tracking in two-dimensional electrophoretic gels by minimal protein identifiers. Proteomics. 2004;4:2927–2941. doi: 10.1002/pmic.200400908. [DOI] [PubMed] [Google Scholar]
- 21.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W., Chait B.T. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal Chem. 2000;72:2482–2489. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- 23.Giulietti A., Overbergh L., Valckx D., Decallonne B., Bouillon R. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods. 2001;25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- 24.Karaghiosoff M., Steinborn R., Kovarik P., Kriegshauser G., Baccarini M. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- 25.Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.UniProt-Consortium The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michal G. An atlas of biochemistry and molecular biology. John Wiley & Sons, Inc.; New York, USA: 1999. Biochemical pathways. [Google Scholar]
- 29.Schrader M., Yoon Y. Mitochondria and peroxisomes: are the ‘big brother’ and the ‘little sister’ closer than assumed? Bioessays. 2007;29:1105–1114. doi: 10.1002/bies.20659. [DOI] [PubMed] [Google Scholar]
- 30.Howard J., The I.R.G. Proteins: a function in search of a mechanism. Immunobiology. 2008;213:367–375. doi: 10.1016/j.imbio.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Carlow D.A., Marth J., Clark-Lewis I., Teh H.S. Isolation of a gene encoding a developmentally regulated T cell-specific protein with a guanine nucleotide triphosphate-binding motif. J Immunol. 1995;154:1724–1734. [PubMed] [Google Scholar]
- 32.Carlow D.A., Teh S.J., Teh H.S. Specific antiviral activity demonstrated by TGTP, a member of a new family of interferon-induced GTPases. J Immunol. 1998;161:2348–2355. [PubMed] [Google Scholar]
- 33.van den Pol A.N., Robek M.D., Ghosh P.K., Ozduman K., Bandi P. Cytomegalovirus induces interferon-stimulated gene expression and is attenuated by interferon in the developing brain. J Virol. 2007;81:332–348. doi: 10.1128/JVI.01592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soupene E., Kuypers F.A. Mammalian long-chain acyl-CoA synthetases. Exp Biol Med (Maywood) 2008;233:507–521. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang M.J., Fujino T., Sasano H., Minekura H., Yabuki N. A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proc Natl Acad Sci USA. 1997;94:2880–2884. doi: 10.1073/pnas.94.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wierenga R.K., Kapetaniou E.G., Venkatesan R. Triosephosphate isomerase: a highly evolved biocatalyst. Cell Mol Life Sci. 2010;67:3961–3982. doi: 10.1007/s00018-010-0473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beckner M.E., Fellows-Mayle W., Zhang Z., Agostino N.R., Kant J.A. Identification of ATP citrate lyase as a positive regulator of glycolytic function in glioblastomas. Int J Cancer. 2010;126:2282–2295. doi: 10.1002/ijc.24918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lill U., Schreil A., Eggerer H. Isolation of enzymically active fragments formed by limited proteolysis of ATP citrate lyase. Eur J Biochem. 1982;125:645–650. doi: 10.1111/j.1432-1033.1982.tb06731.x. [DOI] [PubMed] [Google Scholar]
- 39.Singh M., Richards E.G., Mukherjee A., Srere P.A. Structure of ATP citrate lyase from rat liver. Physicochemical studies and proteolytic modification. J Biol Chem. 1976;251:5242–5250. [PubMed] [Google Scholar]
- 40.Linn T.C., Srere P.A. Identification of ATP citrate lyase as a phosphoprotein. J Biol Chem. 1979;254:1691–1698. [PubMed] [Google Scholar]
- 41.Ramakrishna S., D'Angelo G., Benjamin W.B. Sequence of sites on ATP-citrate lyase and phosphatase inhibitor 2 phosphorylated by multifunctional protein kinase (a glycogen synthase kinase 3 like kinase) Biochemistry. 1990;29:7617–7624. doi: 10.1021/bi00485a011. [DOI] [PubMed] [Google Scholar]
- 42.Potapova I.A., El-Maghrabi M.R., Doronin S.V., Benjamin W.B. Phosphorylation of recombinant human ATP:citrate lyase by cAMP-dependent protein kinase abolishes homotropic allosteric regulation of the enzyme by citrate and increases the enzyme activity. Allosteric activation of ATP:citrate lyase by phosphorylated sugars. Biochemistry. 2000;39:1169–1179. doi: 10.1021/bi992159y. [DOI] [PubMed] [Google Scholar]
- 43.Vogl C., Flatt T., Fuhrmann B., Hofmann E., Wallner B. Transcriptome analysis reveals a major impact of JAK protein tyrosine kinase 2 (Tyk2) on the expression of interferon-responsive and metabolic genes. BMC Genomics. 2010;11:199. doi: 10.1186/1471-2164-11-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanc M., Hsieh W.Y., Robertson K.A., Watterson S., Shui G. Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS Biol. 2011;9:e1000598. doi: 10.1371/journal.pbio.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Migita T., Narita T., Nomura K., Miyagi E., Inazuka F. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008;68:8547–8554. doi: 10.1158/0008-5472.CAN-08-1235. [DOI] [PubMed] [Google Scholar]
- 47.Bauer D.E., Hatzivassiliou G., Zhao F., Andreadis C., Thompson C.B. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 48.Yu Y., Maguire T.G., Alwine J.C. Human cytomegalovirus activates glucose transporter 4 expression to increase glucose uptake during infection. J Virol. 2011;85:1573–1580. doi: 10.1128/JVI.01967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berwick D.C., Hers I., Heesom K.J., Moule S.K., Tavare J.M. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem. 2002;277:33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- 50.Pierce M.W., Palmer J.L., Keutmann H.T., Hall T.A., Avruch J. The insulin-directed phosphorylation site on ATP-citrate lyase is identical with the site phosphorylated by the cAMP-dependent protein kinase in vitro. J Biol Chem. 1982;257:10681–10686. [PubMed] [Google Scholar]
- 51.Sale E.M., Hodgkinson C.P., Jones N.P., Sale G.J. A new strategy for studying protein kinase B and its three isoforms. Role of protein kinase B in phosphorylating glycogen synthase kinase-3, tuberin, WNK1, and ATP citrate lyase. Biochemistry. 2006;45:213–223. doi: 10.1021/bi050287i. [DOI] [PubMed] [Google Scholar]
- 52.Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 53.Wolowczuk I., Verwaerde C., Viltart O., Delanoye A., Delacre M. Feeding our immune system: impact on metabolism. Clin Dev Immunol. 2008;2008:639803. doi: 10.1155/2008/639803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maciver N.J., Jacobs S.R., Wieman H.L., Wofford J.A., Coloff J.L. Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J Leukoc Biol. 2008;84:949–957. doi: 10.1189/jlb.0108024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silverstein R.L., Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gordon S. Do macrophage innate immune receptors enhance atherogenesis? Dev Cell. 2003;5:666–668. doi: 10.1016/s1534-5807(03)00329-0. [DOI] [PubMed] [Google Scholar]
- 57.Funk J.L., Feingold K.R., Moser A.H., Grunfeld C. Lipopolysaccharide stimulation of RAW 264.7 macrophages induces lipid accumulation and foam cell formation. Atherosclerosis. 1993;98:67–82. doi: 10.1016/0021-9150(93)90224-i. [DOI] [PubMed] [Google Scholar]
- 58.Kazemi M.R., McDonald C.M., Shigenaga J.K., Grunfeld C., Feingold K.R. Adipocyte fatty acid-binding protein expression and lipid accumulation are increased during activation of murine macrophages by toll-like receptor agonists. Arterioscler Thromb Vasc Biol. 2005;25:1220–1224. doi: 10.1161/01.ATV.0000159163.52632.1b. [DOI] [PubMed] [Google Scholar]
- 59.Castrillo A., Joseph S.B., Vaidya S.A., Haberland M., Fogelman A.M. Crosstalk between LXR and toll-like receptor signaling mediates bacterial and viral antagonism of cholesterol metabolism. Mol Cell. 2003;12:805–816. doi: 10.1016/s1097-2765(03)00384-8. [DOI] [PubMed] [Google Scholar]
- 60.Joseph S.B., Castrillo A., Laffitte B.A., Mangelsdorf D.J., Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 61.Kawaguchi T., Veech R.L., Uyeda K. Regulation of energy metabolism in macrophages during hypoxia. Roles of fructose 2,6-bisphosphate and ribose 1,5-bisphosphate. J Biol Chem. 2001;276:28554–28561. doi: 10.1074/jbc.M101396200. [DOI] [PubMed] [Google Scholar]
- 62.Cramer T., Yamanishi Y., Clausen B.E., Forster I., Pawlinski R. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haji-Michael P.G., Ladriere L., Sener A., Vincent J.L., Malaisse W.J. Leukocyte glycolysis and lactate output in animal sepsis and ex vivo human blood. Metabolism. 1999;48:779–785. doi: 10.1016/s0026-0495(99)90179-8. [DOI] [PubMed] [Google Scholar]
- 64.Krawczyk C.M., Holowka T., Sun J., Blagih J., Amiel E. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115:4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagano Y., Kojima Y., Sawa I., Hagihara B., Kobayashi S. The effect of the virus inhibiting factor on oxidative phosphorylation. Jpn J Exp Med. 1966;36:341–362. [PubMed] [Google Scholar]
- 66.Isaacs A., Klemperer H.G., Hitchock G. Studies on the mechanism of action of interferon. Virology. 1961;13:191–199. doi: 10.1016/0042-6822(61)90053-8. [DOI] [PubMed] [Google Scholar]
- 67.Hirasawa H., Oda S., Nakamura M. Blood glucose control in patients with severe sepsis and septic shock. World J Gastroenterol. 2009;15:4132–4136. doi: 10.3748/wjg.15.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Potla R., Koeck T., Wegrzyn J., Cherukuri S., Shimoda K. Tyk2 tyrosine kinase expression is required for the maintenance of mitochondrial respiration in primary pro-B lymphocytes. Mol Cell Biol. 2006;26:8562–8571. doi: 10.1128/MCB.00497-06. [DOI] [PMC free article] [PubMed] [Google Scholar]