Abstract
Numerous attributes render the domestic dog a highly pertinent model for cancer-associated gene discovery. We performed microarray-based comparative genomic hybridization analysis of 60 spontaneous canine intracranial tumors to examine the degree to which dog and human patients exhibit aberrations of ancestrally related chromosome regions, consistent with a shared pathogenesis. Canine gliomas and meningiomas both demonstrated chromosome copy number aberrations (CNAs) that share evolutionarily conserved synteny with those previously reported in their human counterpart. Interestingly, however, genomic imbalances orthologous to some of the hallmark aberrations of human intracranial tumors, including chromosome 22/NF2 deletions in meningiomas and chromosome 1p/19q deletions in oligodendrogliomas, were not major events in the dog. Furthermore, and perhaps most significantly, we identified highly recurrent CNAs in canine intracranial tumors for which the human orthologue has been reported previously at low frequency but which have not, thus far, been associated intimately with the pathogenesis of the tumor. The presence of orthologous CNAs in canine and human intracranial cancers is strongly suggestive of their biological significance in tumor development and/or progression. Moreover, the limited genetic heterogenity within purebred dog populations, coupled with the contrasting organization of the dog and human karyotypes, offers tremendous opportunities for refining evolutionarily conserved regions of tumor-associated genomic imbalance that may harbor novel candidate genes involved in their pathogenesis. A comparative approach to the study of canine and human intracranial tumors may therefore provide new insights into their genetic etiology, towards development of more sophisticated molecular subclassification and tailored therapies in both species.
Keywords: Comparative genomic hybridization, Canine, Brain tumor, Chromosome, Microarray
Introduction
It is estimated that in 2008, 22,000 Americans will be diagnosed with cancer of the nervous system, equivalent to 6.5 people per 100,000 individuals in the US population. Over 13,000 people will succumb to the disease [1]. While advances have been made in the development of improved diagnostic and prognostic tools and therapeutic modalities for brain tumors (see [2] for a review), progress has generally been slow. More recently, focus has shifted towards a better understanding of the molecular basis of cancer. Numerous studies of human brain tumors have revealed specific chromosome aberrations that appear to be associated with tumor histology, clinical grade, therapeutic response and/or metastatic potential (see [3-5] for a review). Other aberrations appear to be random consequences of uncontrolled cell proliferation, and yet more are present at such low frequency that their significance has not yet been established. Despite the spectrum of resources and techniques now available for human molecular studies, identification of the key genes involved in these cytogenetic aberrations has met with varying degrees of success. Moreover the successful translation of molecular findings into meaningful clinical enhancements has been limited [6], with relatively little known about the specific genetic factors that define risk and response to therapy.
The traditional rodent model of brain neoplasia presents with several crucial limitations, among which are the reduced genetic homogeneity and invasiveness of xenograft tumors and the requisite immune-deficient nature of the host, which hamper the ability to make direct and meaningful comparisons with spontaneous tumors of human patients (for example [7]). We, and others, have proposed that the domestic dog represents a compelling additional model, perhaps most significantly because unlike the rodent system, canine brain tumors occur spontaneously and with an incidence similar to (if not higher than) that of human populations [8, 9]. Human cancers of the nervous system are primarily diseases of middle and old age, with 67% of cases diagnosed in individuals aged 45 years or older [1]. Similarly, over 70% of such tumors in the dog occur in ‘middle-aged’ and older animals (age six or above), at a rate equating in both species to approximately 1–3% of adult deaths [9]. The clinical and histologic presentation of human and canine brain tumors is almost identical and they are classified according to similar diagnostic criteria in both species [9–12]. Interestingly, dog and human counterparts of certain brain tumor subtypes share key histopathological features that appear to be absent in rodent models [13]. Given the anatomical and physiological similarities between dogs and people, surgical procedures are highly translatable. Moreover, the increasing profile of the veterinary neurosurgery field means that sophisticated diagnostic and therapeutic capabilities for canine brain tumors are now widely available in specialist veterinary care establishments worldwide, alongside numerous exciting clinical and basic science research studies. While there are certain logistical challenges associated with performing clinical trials in companion animals (for example, rate of patient accrual, elevated expense over rodent systems; see [7] for a review), there is immense potential to exploit the dog as a preclinical model for development and evaluation of novel brain tumor therapies (for example [14]). The many attributes of spontaneous canine brain tumors as a model system may also help to expand the increasingly attractive stem cell hypothesis for cancer development (for example [15, 16]), which represents one of the most crucial and pressing goals of molecular medicine.
As with many other cancers, clinical management of intracranial tumors in both human and veterinary medicine is challenged by the variation in outcome that exists between different morphological subtypes and grades of tumor cell differentiation. There is an increasing need to develop more clinically relevant systems for disease classification, with the ultimate goal of tailoring therapy to the individual patient. It is likely that novel indicators of diagnosis and prognosis will emerge through endeavors within the emerging field of molecular oncology. The many attributes of the domestic dog as a model for gene discovery, and the wealth of resources available for genomic studies in this species, suggests that the dog has great potential to contribute towards advancements in our understanding of tumor pathogenesis at the molecular level. In particular the restricted genetic variation that exists within purebred dog populations [17] provides an opportunity to scan the genome for tumor-associated genomic lesions that may not be readily detectable in the more heterogeneous genetic background of human populations. Interestingly, in the dog, predisposition to intracranial tumors is highly breed-related and demonstrates a particularly strong association with cranio-facial morphology. Brachycephalic (short-nosed) breeds have an elevated incidence of tumors of glial cell origin as compared to the general population (reviewed in [9, 18]). In contrast, mesocephalic (moderate length nose) and doliochocephalic (long-nosed) breeds are more highly predisposed to tumors of the meninges. Dog breeds with elongated cranio-facial morphology are particularly highly represented among family-owned pets in the US, which correlates with a relative inversion in the incidence of meningiomas versus gliomas in dogs, as compared to human populations. This suggests that there may be a strong genetic component to intracranial tumor development and progression that may be characterized through genome-wide evaluation of spontaneous dog malignancies. Consequently we propose that by studying canine brain tumors we may more readily identify underlying genetic factors that have to date been hidden amongst the background ‘noise’ associated with more heterogeneous human populations.
Consideration of the dog as a model for intracranial tumors offers a unique and exciting opportunity to gain new insight into the underlying genetic etiology of this devastating range of neoplasms, with potential for significant clinical benefit to both species through the identification of specific molecular factors that define disease risk and prognosis. Towards this goal, we present data from the first reported application of microarray-based comparative genomic hybridization (aCGH) analysis to the study of spontaneous dog intracranial tumors, using a 1 Mb resolution, genome assembly-integrated microarray platform. We describe and compare their cytogenetic profiles, and draw comparisons with evolutionarily-related regions of the human genome, to assess evidence for conserved genomic changes associated with intracranial neoplasia in both species. We highlight imbalances of chromosome regions that are significantly associated with disease subtype that have not previously been identified as highly significant in human populations. These regions warrant further investigation to determine whether they harbor factors involved in the biology of brain tumors that may reveal novel targets for therapeutic design. The compilation of these data will also aid development of additional diagnostic and prognostic markers for canine brain tumors, which may equally prove informative in the human counterpart.
Materials and methods
Case materials
Tumor specimens (n = 60) were recruited from client-owned dogs admitted to the College of Veterinary Medicine at North Carolina State University and the University of California at Davis from 2001 to 2006. All specimens were acquired prior to initiation of chemotherapy or radiotherapy, and under approved protocols with informed client consent. A total of 54 cases underwent routine diagnostic evaluation that included magnetic resonance imaging (MRI) or computed tomography (CT) of the brain. The remaining six cases did not undergo detailed diagnostic evaluation since the owners opted for euthanasia (based on the severity of neurological signs) and tumor tissue specimens were acquired at necropsy.
Representative brain tumor tissue specimens (retrieved either as surgical biopsies, n=34 cases or at necropsy, n=26 cases) were (1) used to initiate primary cell cultures in RPMI-1640 medium supplemented with 10% fetal bovine serum, (2) snap-frozen in liquid nitrogen for subsequent DNA extraction, and (3) formalin-fixed and paraffin-embedded for histological evaluation of hematoxylin and eosin (H&E)-stained sections. Standard morphological features were used to define the histological subtype and grade of each tumor [9, 12]. Tumor specimens were reviewed independently by two veterinary pathologists (RH, KL). Fixed tumor specimens were also stained for glial fibrillary acidic protein (GFAP) and vimentin using standard immunohistochemical techniques [19].
aCGH analysis
DNA was isolated from snap-frozen tumor tissue by conventional methods. aCGH analysis was carried out using a genomic microarray comprising 2097 cytogenetically-mapped clones from the CHORI-82 dog BAC library (http://bacpac.chori.org, BACPAC Resources, Children’s Hospital Oakland Research Institute, Oakland, CA), distributed at approximately 1 Mb intervals throughout each dog autosome and the X chromosome (mean interval 1.10 Mb, range 0.28–3.28 Mb), in addition to clones representing canine orthologues of 53 human genes associated with a range of cancers [20]. aCGH analysis was performed as described previously [20, 21] using either the patient’s own blood-derived DNA, if available, or equimolar pools of DNA isolated from 10 or more healthy, sex-matched individuals of the same breed, as the reference sample. Locations of BAC clones are denoted herein according to their chromosome of origin and then their Mb position on that chromosome, according to the dog genome assembly (for example, CFA 1; 3.2 Mb). Cytogenetic assignments are reported according to the DAPI-banded dog karyotype nomenclature of Breen et al. [22]. Regions of conserved synteny between the dog (CanFam v2.0) and human (build 36.1) genomes are based on comparative genome sequence assembly data located at http://genome.ucsc.edu [23].
aCGH data were assessed using Fisher’s exact test to identify significant associations between tumor type and the copy number status of whole chromosomes and chromosome regions. Tumor cases were first scored according to their copy number status for each chromosome (balanced, gain or loss), based on the aCGH Smooth algorithm [24]. For those chromosome aberrations present in more than 40% of the cohort, a binary categorization was then used where each tumor case was scored as one when that aberration was present, and as zero when not present. The pairwise phi correlation coefficient between any two aberrations (A and B) was defined as where PAB = P(A = 1, B = 1), PA = P(A = 1), and PB = P(B = 1). The sigsignificance of association between all pairwise combinations of these chromosome aberrations was then assessed using Fisher’s exact test. Hierarchical cluster analysis of all tumor cases was performed using the function hclust in software R with the Euclidean distance measure and the complete linkage clustering algorithm. All statistical assessments were performed using software R 2.7.0.
Evaluation of genomic imbalances by single-locus probe FISH analysis
Interphase nuclei were prepared for FISH analysis by direct harvest of primary tumor cells using conventional techniques of hypotonic treatment and methanol/glacial acetic acid fixation. For consistency, these cells were prepared from the same biopsy specimen from which the tumor DNA was isolated for aCGH analysis. Multicolor FISH analyses were carried out as described elsewhere [25] using BAC clones from the 1 Mb array as single locus probes (SLPs) to investigate regions showing a range of normal and aberrant copy number ratios in aCGH. The same probes were hybridized to dog chromosome preparations from clinical healthy dogs to confirm their normal copy number and expected chromosomal location. Images were acquired from a minimum of 30 representative cells in each instance, and the copy number status of each probe was scored by two independent investigators with no prior knowledge of the corresponding aCGH data.
Results
Diagnostic evaluation of canine intracranial tumors
A total of 60 canine intracranial tumors were analyzed by aCGH, comprising 35 meningiomas and 25 gliomas (Table 1). Fifteen different breeds of dog were represented, in addition to five dogs of mixed breed. Within the panel of gliomas, 64% (16/25 cases) represented brachycephalic breeds, 24% (6/25 cases) were mesocephalic breeds and the three remaining glioma cases were in mixed breed dogs. In contrast, brachycephalic breeds represented only 23% of meningiomas (8/35 cases, all Boxers). The majority of meningioma cases (69%, 24/35 cases) occurred in mesocephalic breeds, in addition to one case in a doliochocephalic breed and two in dogs of mixed breed. Among meningiomas, 63% of cases (22/35) were classified as grade I tumors, 23% of cases (8/35) were grade II and 9% (3/35) were grade III. Two cases could not be graded conclusively. Of the gliomas, 40% of cases (10/25) were classified as oligodendrogliomas, 36% (9/25) were classified as astrocytomas, 16% (4/25) as oligoastrocytomas (mixed gliomas) and 8% (2/25) as ependymomas. The distribution of tumor types and subtypes was consistent with other published case series [26–28].
Table 1.
Signalment, clinical and histopathological data for dog intracranial tumors
| Case # | Breed | Gender | Age at diagnosis (years) |
Location of tumor | Histological subtype | Grade |
|---|---|---|---|---|---|---|
| G01 | Mix | Fs | 7 | Metencephalon | Astrocytoma | 2 |
| G02 | Boxerb | M | 6 | Diencephalon | Astrocytoma | 2 |
| G03 | Boxerb | M | 9 | Telencephalon | Astrocytoma | 2 |
| G04 | Boston Terrierb | Mc | 12 | Telencephalon | Astrocytoma | 2 |
| G05 | Boxerb | M | 8 | Telencephalon | Astrocytoma | 2 |
| G06 | Boston Terrierb | Mc | 7 | Diencephalon | Astrocytoma | 2 |
| G07 | German Shepherd Dogm | Mc | 8 | Cerebellum | Astrocytoma | 3 |
| G08 | English Bulldogb | M | 12 | Telencephalon | Astrocytoma | 4 |
| G09 | Mix | F | 10 | Telencephalon | Astrocytoma | 4 |
| G10 | Mix | F | 12 | Metencephalon | Ependymoma | Not done |
| G11 | German Shorthaired Pointerm | M | 7 | Diencephalon | Ependymoma | 2 |
| G12 | English Bulldogb | Mc | 3 | Diencephalon | Oligoastrocytoma | 2 |
| G13 | Boxerb | F | 9 | Telencephalon | Oligoastrocytoma | 2 |
| G14 | Labrador Retrieverm | Mc | 10 | Telencephalon | Oligoastrocytoma | 2 |
| G15 | Boxerb | Fs | 7 | Telencephalon | Oligoastrocytoma | 2 |
| G16 | English Bulldogb | Mc | 11 | Telencephalon | Oligodendroglioma | 2 |
| G17 | Boston Terrierb | Mc | 6 | Telencephalon | Oligodendroglioma | 2 |
| G18 | Boxerb | Fs | 6 | Diencephalon | Oligodendroglioma | 3 |
| G19 | Boxerb | Fs | 3.5 | Diencephalon | Oligodendroglioma | 3 |
| G20 | German Shorthaired Pointerm | Mc | 9 | Telencephalon | Oligodendroglioma | 3 |
| G21 | Bullmastiffb | Mc | 5 | Diencephalon | Oligodendroglioma | 3 |
| G22 | Boxerb | Fs | 9 | Telencephalon | Oligodendroglioma | 3 |
| G23 | Labrador Retrieverm | F | 10 | Telencephalon | Oligodendroglioma | 3 |
| G24 | Labrador Retrieverm | Mc | 8 | Telencephalon | Oligodendroglioma | 3 |
| G25 | Boxerb | Mc | 8 | Telencephalon | Oligodendroglioma | Not done |
| M01 | Rhodesian Ridgebackm | M | 4 | Telencephalon | Meningothelial | 1 |
| M02 | Golden Retrieverm | Mc | 12 | Cerebellum | Meningothelial | 1 |
| M03 | Boxerb | Fs | 10 | Telencephalon | Meningothelial | 1 |
| M04 | Boxerb | Mc | 7 | Cerebellopontine angle | Meningothelial | 1 |
| M05 | Boxerb | Mc | 6.5 | Cerebellum | Meningothelial | 1 |
| M06 | Golden Retrieverm | Fs | 10 | Telencephalon | Meningothelial | 1 |
| M07 | Labrador Retrieverm | Fs | 10 | Telencephalon | Meningothelial | 1 |
| M08 | Labrador Retrieverm | Fs | 12 | Telencephalon | Meningothelial | 1 |
| M09 | Boxerb | Mc | 7 | Telencephalon | Meningothelial | 1 |
| M10 | Yorkshire Terrierm | Fs | 14 | Telencephalon | Psammomatous | 1 |
| M11 | German Shepherd Dogm | Fs | 10.6 | Telencephalon | Psammomatous | 1 |
| M12 | Beaglem | Mc | 7 | Cerebellopontine angle | Transitional | 1 |
| M13 | Golden Retrieverm | Mc | 11.75 | Telencephalon | Transitional | 1 |
| M14 | Golden Retrieverm | Mc | 10 | Telencephalon | Transitional | 1 |
| M15 | German Shepherd Dogm | Fs | 10.5 | Telencephalon | Transitional | 1 |
| M16 | Labrador Retrieverm | Fs | 9.5 | Telencephalon | Transitional | 1 |
| M17 | Labrador Retrieverm | Fs | 12 | Telencephalon | Transitional | 1 |
| M18 | Mix | Mc | 3 | Cervical spinal cord (C1) | Transitional | 1 |
| M19 | West Highland White Terrierm | Fs | 10.5 | Telencephalon | Transitional | 1 |
| M20 | Golden Retrieverm | Mc | 14 | Telencephalon | Transitional | 1 |
| M21 | Labrador Retrieverm | Mc | 6 | Telencephalon | Transitional with metaplastic change |
1 |
| M22 | Basset Houndd | Fs | 6 | Cerebellum | Transitional with microcystic change |
1 |
| M23 | Golden Retrieverm | M | 8 | Telencephalon | Atypical | 2 |
| M24 | Golden Retrieverm | F | 11 | Telencephalon | Atypical | 2 |
| M25 | Boxerb | F | 5 | Telencephalon | Atypical | 2 |
| M26 | Mix | Mc | 8 | Telencephalon | Atypical | 2 |
| M27 | Boxerb | Fs | 5 | Thoracic spinal cord (T12/13) | Atypical | 2 |
| M28 | Golden Retrieverm | Mc | 9 | Telencephalon | Atypical | 2 |
| M29 | Golden Retrieverm | Mc | 11 | Telencephalon | Atypical | 2 |
| M30 | Boxerb | Fs | 11 | Cervical spinal cord (C1) | Chordoid | 2 |
| M31 | Boxerb | Fs | 10 | Cerebellum | Anaplastic | 3 |
| M32 | Golden Retrieverm | Fs | 12 | Telencephalon | Transitional center with papillary regions invading the brain, and extensive necrosis |
3 |
| M33 | Old English Sheepdogm | Mc | 16 | Telencephalon | Transitional with early papillary change and some necrosis |
3 |
| M34 | Labrador Retrieverm | M | 2 | Telencephalon | Not done | Not done |
| M35 | Dalmationm | Fs | 12 | Telencephalon | Not done | Not done |
The cranial morphology of each purebred case is also indicated (bbrachycephalic, mmesocephalic) For gender: M = intact male; Mc = neutered male; F = intactfemale; Fs = spayedfemale
Global incidence of aneuploidy in dog intracranial tumors
aCGH analysis revealed a wide range of recurrent genomic imbalances both within and between tumor cases (see Fig. 1 for an example). Six cases (one glioma and five meningiomas) showed no evidence of aneuploidy, and were excluded from downstream analysis. The remaining 54 cases were scored initially according to their copy number status for each chromosome (balanced, gain or loss), based on the aCGH Smooth algorithm [24] in order to provide a global overview of recurrent aneuploidy (Fig. 2a). Genomic gains and losses in meningiomas were comparable in number (6.8% and 3.9% respectively). In contrast, in gliomas, chromosome gains were four times as common as losses (25.5% gain vs. 4.8% loss, Fig. 2). aCGH analysis demonstrated that 19 dog chromosomes showed significant (p < 0.05) differences in copy number status between meningiomas and gliomas, consistent with their unrelated cell origin. Of these, 17 were changes that were specifically associated with gliomas, reflecting the relative simplicity of the typical dog meningioma genome in comparison to tumors of glial cell origin. Deletions of dog chromosome 17 (CFA 17) and CFA 27 comprised just greater than one-fourth (26.4%) of all copy number changes in meningiomas, and represented 72.7% of all losses. Four chromosomes (CFA 5, 20, 29 and 37) exhibited a normal copy number in all meningiomas. In contrast, with the exception of CFA 6, each dog autosome was aneuploid in at least one glioma case.
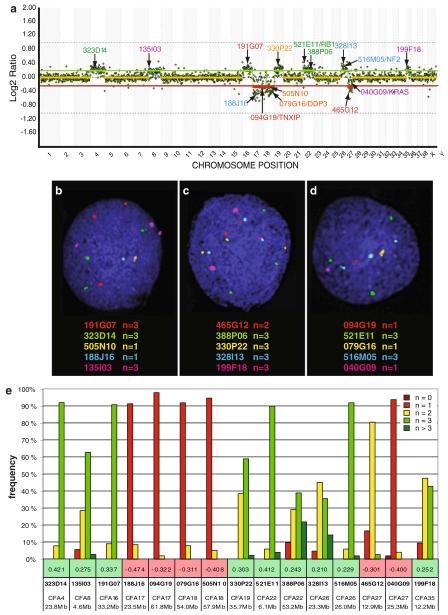
Fig. 1.
a Example whole genome aCGH profile from analysis of a grade I meningioma from a 10 year old male Golden Retriever, co-hybridized with male reference DNA. Data are plotted as the median, block-normalized and background-subtracted log2 tumor DNA:reference DNA ratio of the replicate spots for each arrayed BAC clone. Log2 ratios representing genomic gain and loss are indicated by horizontal bars above (green line) and below (red line) the dashed midline (orange line) that represents normal copy number. The chromosome copy-number status line for the tumor appears as an orange overlay of the center-line when there is a normal copy number, and as either red (loss) or green (gain) in regions where genomic imbalances were apparent, as determined by the aCGH Smooth algorithm [24]. Here, chromosome gain is apparent for CFA 4, 8, 16, 19, 26 and 35. This case also shows the characteristic losses of CFA 17 and 27 that were highly recurrent in our dog meningioma panel, as well as loss of CFA 18. The aCGH profile is annotated with the clone address of 15 BAC clones from the 1 Mb array that were used in subsequent FISH analysis of this case. Five of these 15 clones have previously been shown to contain the full coding sequence of a key cancer-associated gene (TXNIP, DPP3, RB1, NF2, KRAS) [20]. The color of the text denotes the fluorochrome with which the BAC clone was labeled. b–d Targeted FISH analysis of tumor interphase nuclei from the same case using 15 differentially labeled BAC clones (highlighted in a) combined in three separate groups. The modal copy number for each clone is indicated. e Summary of compiled copy number data based on FISH analysis of at least 30 tumor interphase nuclei for each of the 15 BAC clones. The number immediately above each BAC address represents the log2 tumor DNA:reference DNA ratio of the corresponding clone derived from the aCGH analysis. It is evident that the SLP data and the aCGH data are mutually confirmatory
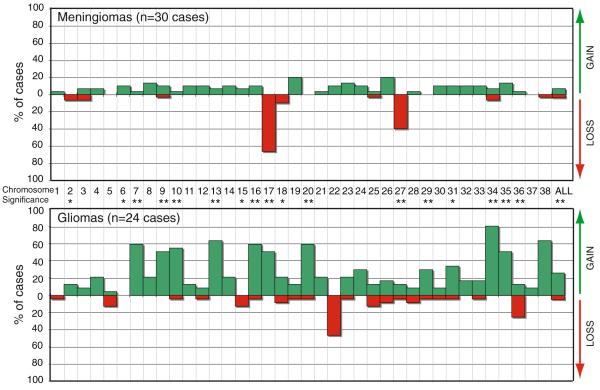
Fig. 2.
Incidence of recurrent chromosome copy number changes in a meningiomas (n = 30 cases) and b gliomas (n = 24 cases). Tumors displaying a normal copy number throughout the genome were excluded from analysis. Each dog autosome is listed along the x-axis, and the y-axis indicates the percentage of the corresponding tumor population that showed copy number gain (green bar above the x-axis) or loss (red bar below the x-axis). The rightmost bars (x = ‘ALL’) show the mean incidence of copy number gain and loss across all chromosomes. Asterisks along the x-axis indicate those chromosomes for which the incidence of recurrent copy number gain or loss differed significantly between the meningiomas and glioma cases analyzed in the present study (* indicates p < 0.05, ** indicates p < 0.01)
Recurrent chromosome copy number aberrations in dog meningiomas
Among 30 cases of canine meningioma that showed evidence of gross aneuploidy, the characteristic cytogenetic features (Fig. 2a) were losses of CFA 17 (20/30 cases, 66.7%) and CFA 27 (12/30 cases, 40.0%). Copy number increases were infrequent among meningiomas, of which gains of CFA 19 and CFA 26 were the most common (6/24 cases, 20%). No other whole chromosome aberrations were observed in ≥20% of meningioma cases. Of the 12 cases with loss of CFA 27, 10 also showed loss of CFA 17, indicating a strong positive association between these aberrations (p = 0.83). In contrast, gain of CFA 19 and loss of CFA 27 were mutually exclusive events in meningiomas, since there were no instances in which both these aberrations occurred within the same case. There was no significant association between any other pairwise combinations of these four most common aberrations. The distribution of these four aberrations was not significantly different when meningiomas were classified according to their clinical grade.
Recurrent chromosome copy number aberrations in dog gliomas
In contrast to meningiomas, dog gliomas showed extensive genomic complexity, with a high incidence of genomic gain accompanied by a lower background of genomic loss (Fig. 2b). Ten different chromosomes (CFA 7, 9, 10, 13, 16, 17, 20, 34, 35 and 38) were each gained in ≥50% of the glioma cases analyzed. The most frequent of these was gain of CFA 34 (19/24 cases, 79%), followed by gains of CFA 13 and 38 (both evident in 15/24 cases, 62.5%). With the exception of CFA 22 and CFA 36 (deleted in 11/24 cases, 45.8% and 6/24 cases, 25.0%, respectively), no copy number losses were common to >20% of the glioma cohort.
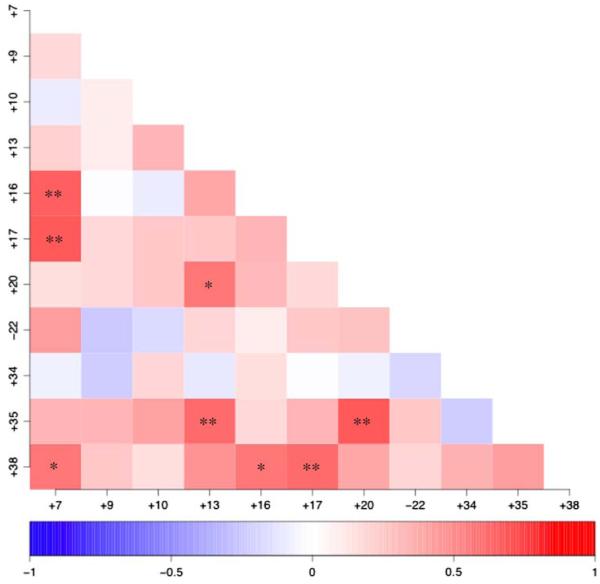
Among those aberrations present in ≥40% of gliomas (gains of CFA 7, 9, 10, 13, 16, 17, 20, 34, 35 and 38, and loss of CFA 22), certain pairs of aberrations showed significant positive correlation (Fig. 3). This indicated that both aberrations occurred within the same case more frequently than would be predicted on the basis of their independent incidence in the population. A particularly strong association (p < 0.01) was identified between five pairs of chromosome aberrations, all of which were genomic gains, namely +7/+16, +7/+17, +13/+35, +17/+38, and +20/+35 (Fig. 3). No significant negative association was observed between any pairs of chromosome aberrations. Comparison of grade II and III dog astrocytomas (n = 7) with grade II and III oligodendrogliomas (n = 9) did not reveal any significant difference in the distribution of their copy number changes. In contrast, comparison of all grade II dog gliomas (n = 12) with grade III and IV cases (n = 10) revealed that grade II tumors are significantly associated with copy number gain of CFA 20 (p = 0.006) and gain of CFA 35 (p = 0.03). The number of other glioma subtypes was insufficient to make these comparisons.
Fig. 3.
Correlation analysis of recurrent chromosome aberration in gliomas. Cases were scored according to the presence or absence of each pairwise combination of the 11 chromosome aberrations that were observed in ≥40% of the glioma population. The degree of correlation between aberrations is indicated on a scale of red (positive correlation) ↔ blue (negative correlation). Asterisks indicate pairwise combinations of chromosome aberrations that show significantly strong association i.e. they occur at a frequency that is significantly different from that predicted on the basis of their individual frequency in the population (* indicates p < 0.02, ** indicates p < 0.01)
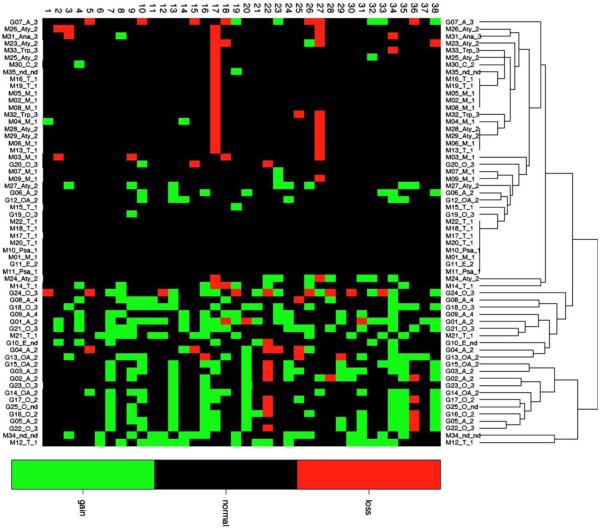
Hierarchical cluster analysis of intracranial tumors
Evaluation of whole chromosome copy number changes in all 60 tumor cases resulted in global segregation of meningiomas and gliomas into distinct clusters (Fig. 4), with a further grouping of cases with no detectable genomic imbalances. Tumors did not cluster tightly according to histological subtype or clinical grade; it is likely that analysis of a larger case series will be required in order for such trends to become apparent. Several meningioma cases clustered more closely with gliomas than with other meningiomas (including M12, M14, M21, M24 and, M34), displaying many of the chromosome gains that were recurrent within gliomas. Of these cases, M12, M21 and M34 showed neither the recurrent loss of CFA 17 nor of CFA 27, which in the present study was characteristic of meningiomas, while M24 and M14 exhibited one or both of these aberrations. With one exception there was no clinical or histological indication that these tumors were not typical of their subtype. In the case of tumor M34, re-evaluation of histological specimens revealed features of cell morphology and immunohistochemical staining that were inconsistent with classical meningiomas, but the case could not be classified unequivocally into a specific tumor subtype. Similarly, four gliomas clustered among meningiomas, namely G06, G07, G12 and G20, primarily due to the small number of genomic aberrations present as compared to other gliomas, for which again there was no apparent clinical or histological explanation. More detailed diagnostic evaluation of these outliers will be performed in downstream studies.
Fig. 4.
Unsupervised cluster analysis of all 60 tumors by chromosome copy number status. All cases are scored according to copy number status (gain = green, black = balanced, red = loss) for all 38 autosomes (horizonatl axis). Each case is denoted according to the tumor type along the vertical axis (G glioma, M meningioma), the histological subtype of the tumor (A astrocytoma, E ependymoma, OA oligoastrocytoma, O oligodendroglioma, Ang angiomatous, M meningothelial, Psa psammomatous, T transitional, C chordoid, Ana anaplastic, Aty atypical, Pap papillary, TrP transitional/papillary, nd not determined) and the clinical grade of the tumor. The panel of cases grossly subdivides into meningiomas and gliomas, with those cases showing a grossly normal karyotype clustered in the center of the figure
Discussion
We have shown previously that evolutionarily conserved chromosome aberrations exist in human and canine lymphoma, osteosarcoma, leukemia and soft-tissue sarcomas [21, 29–31], indicative of a common underlying genetic etiology. We have established that certain tumor-associated genomic aberrations are highly associated with specific dog breeds [32], suggestive of a heritable risk for development of these malignancies. Importantly, these studies have begun to reveal dog tumor-associated chromosome abnormalities for which aberrations of the orthologous regions of the human genome have not been reported widely in the human counterpart. We suggest that this finding is a consequence of the relative homogeneity within purebred dog population and that detailed evaluation of these genomic regions, particularly when targeted to specific breeds, may unearth genes whose key role in defining the risk and progression of tumorigenesis has not yet been recognized in human patients. We propose that a comparative cytogenetic analysis of canine and human intracranial tumors will reveal the subset of evolutionarily-conserved genomic changes that are intimately associated with the disease process among the multitude that are secondary effects of tumor-associated genomic instability. Towards this goal we present microarray-based cytogenetic profiling data from 60 spontaneous intracranial dog tumors, comprising 25 gliomas and 35 meningiomas.
Identification of two characteristic genomic imbalances in canine meningiomas
Meningiomas are frequently benign, slow-growing mesenchymal tumors arising from the arachnoid cells forming a layer of the meninges, the membranes that surround the brain and spinal cord. Since they do not arise from brain parenchyma itself, they are more correctly referred to as intercranial neoplasms. While meningiomas represent approximately 25% of adult primary brain tumors in human patients [12], they comprise up to 40% of all canine intracranial neoplasms [9], which largely reflects the enormous popularity of medium/long-nosed dog breeds. The elevated incidence of meningiomas in the dog, as compared to human populations therefore renders them of particular clinical concern in veterinary medicine. The clinical presentation and histopathology of human and dog meningiomas are highly comparable and they have been shown to share similar immunophenotypes and gene expression profiles [9,12, 33]. Our results showed reduced genomic instability in dog meningiomas as compared to gliomas (Fig. 2), which is consistent with the typically less-aggressive nature of meningioma in both human and veterinary medicine. The most commonly encountered and least aggressive grade I meningiomas of human patients often present with no gross genomic imbalances [9, 12, 34, 35]. Similarly, in our study, grade I tumors comprised 79% of the dog meningioma population (26 of the 33 cases with confirmed clinical grade) and included five of the six dog tumor cases that presented with no evidence of recurrent genomic imbalance. There was no evidence for significant differences in the patterns of chromosome aberrations of the two most numerous of our dog grade I meningioma subtypes, the meningothelial and transitional forms. This is consistent with reports that cytogenetic profiles of human meningioma subtypes that fall within the same histological grade are typically highly conserved (for example [35]), but that they vary more extensively between tumors of different grades. Consistent with their relative infrequency in dog populations (for example [28]), the number of grade III meningiomas that was available in the present study was insufficient to determine whether there was evidence for increasing genomic instability in higher grade tumors, as has been reported in the human counterpart (for example [34, 35]).
The hallmark genomic imbalance in human meningioma, present in approximately 50–75% of cases of all grades, is monosomy of human chromosome 22 (HSA 22) ([35, 36] and others), which frequently (but not always) results in loss of the tumor suppressor gene NF2. In our dog meningioma panel we did not detect recurrent deletions of the regions of CFA 10 and CFA 26 that share conserved synteny with HSA 22 and there were no instances of deletion of the region of the canine genome specifically encoding NF2 (CFA 26; 26 Mb). Moreover, expression array analyses of an overlapping meningioma case series (39 cases, of which 19 cases in common with the present study) have revealed no evidence for global under-representation of NF2 gene expression (Thomson et al., manuscript in preparation). This suggests that genomic deletion of sequences orthologous to HSA 22, and particularly the NF2 region, may not constitute as integral a role in dog meningiomas as is the case for its human counterpart. The small segment of sequence conservation shared by HSA 22qprox and CFA 27qtel, deletions of which are highly recurrent in human and canine meningiomas, respectively, now raises the intriguing possibility that this region may harbor as yet unidentified genes that are fundamental to the development of these tumors in both species.
In human patients the transition from a benign grade I meningioma to a more aggressive, higher grade malignancy has been associated with stepwise accumulation of several key molecular events, including a small subset of recurrent chromosome losses and a wide range of gains [37, 38]; reviewed in [3]. Among these, transition from grade I to grade II frequently involves gains of HSA 1q, 9q, 12q, 15q, 17q and 20, and losses of HSA 1p, 6q, 10, 14q and 18q [35]. Loss of HSA 9p (including the p16/CDKN2A gene region) and amplification of HSA 17q have been proposed as additional events that accumulate in the progression of a grade II to grade III meningioma [35]. There were no instances of p16/CDKN2A (CFA 11; 44 Mb) deletion in the present dog meningioma study panel, nor was there recurrent loss of the dog chromosome regions orthologous to HSA 9 (CFA 9qdist and 11) or gain of the regions orthologous to HSA 17q (CFA 9) in any of the grade III dog meningiomas. Thus, we did not find evidence that these changes are conserved in high-grade dog meningiomas, although this may simply reflect the limited number of such cases available for analysis.
Deletion of HSA 1p has long been linked with initiation and progression of meningioma (for example [35]), and is associated with poor prognosis (for example [39]), in direct contrast with gliomas ([40–46] and others). In the present study loss of CFA 17 was observed in 20/30 cases [67%] of meningiomas that showed evidence of aneuploidy. CFA 17q22-q24 shares evolutionarily-conserved synteny with HSA 1p13.2-q21.3, thus it would be interesting to establish whether this region of HSA 1p has particular biological significance in the pathogenesis of human meningioma. Similarly, loss of CFA 27, which shares conserved synteny with HSA 12p-qprox, was highly recurrent in dog meningiomas, occurring in 40% of the cases that showed evidence of aneuploidy (12/30 cases). Recurrent loss of HSA 12p has indeed been reported in human meningioma patients (for example [35]) but has received little attention due to its relatively low incidence compared to other genomic imbalances. In light of the findings of the present report it will be interesting to monitor the relative incidence and significance of these orthologous chromosome regions in more extensive studies of canine and human meningiomas, since this may further refine the site of the key sequences in both genomes that are intimately associated with meningioma development. Moreover, the highly non-random nature of loss of both CFA 17 and 27 in meningiomas suggests a fundamental biological basis for their manifestation, which warrants detailed evaluation in future studies.
Detection of diverse and extensive genomic instability in canine gliomas
As with their human counterparts, our analyses revealed extensive complexity and diversity in the cytogenetic profiles of canine gliomas, the second most common intracranial tumors of dogs ([26] and others). Gliomas, a diverse group of neuroepithelial tumors originating from the glial cell lineage, represent approximately 50–70% of primary human adult brain tumors [11]. Unequivocal histological distinction between gliomas of astrocytic, oligodendrocytic and ependymal cell origin can be highly challenging [47] and there is considerable interest in developing a molecular means to extend existing diagnostic modalities, through identification of subtype-specific genomic aberrations. Several studies have, however, shown that tumors of glial cell origin, particularly astrocytomas and oligodendrogliomas, as well as the mixed gliomas (oligoastrocytomas) that comprise both these cell lineages, share many common chromosome aberrations, although their relative frequency may differ between subtypes (for example [3, 4]). The present study showed that there was no significant difference in the distribution of copy number changes when comparing dog astrocytomas and oligodendrogliomas of the same grade, consistent with the outcome of clustering analysis (Fig. 3). The number of ependymomas was too small to make this comparison. Ependymomas are relatively infrequent in both human and canine populations; consequently relatively little is known about their cytogenetic basis, and typically they too present with cytogenetic profiles highly similar to those of other glioma subtypes, (reviewed in [3]). Of the two dog ependymomas in the present study, one showed a grossly normal karyotype, whilst the other presented with chromosome aberrations that overlapped fully with those found among the rest of the glioma case panel.
It has been suggested that cytogenetic aberrations may in fact be more highly associated with glial tumors of the same histological grade rather than tumors of the same histological subtype [48]. In the present study of dog gliomas we identified two chromosomes (CFA 20 and 35) whose copy number status was significantly associated with grade. This suggests that as in human patients, the histological grade of dog gliomas is a greater determining factor in their cytogenetic profiles than is the histological subtype. In human patients, gliomas are associated with step-wise accumulation of characteristic molecular defects in their progression from a low-grade to a high-grade tumor, which is associated with decreased median survival (reviewed in [3]). The present study shows that certain of these characteristic aberrations also accumulate as evolutionarily-conserved genomic imbalances in the dog counterpart. Gain of HSA 7 is a characteristic event in early development of gliomas [4]. HSA 7 shares extensive conserved synteny with CFA 14, and also with smaller regions of CFA 6, 16 and 18. Gains of CFA 14 and CFA 18 were not highly recurrent in our dog gliomas (each present in only 5/24, 21% of cases) and there were no gains of CFA 6; however gain of CFA 16 was among the most common copy number changes we observed (15/24, 63% of cases, including 5/9 astrocytomas and 7/10 oligodendrogliomas). HSA 7p gain, consistent with amplification of the EGFR gene (HSA 7; 55.1 Mb), is a particular hallmark of human high-grade astrocytomas [4]. Elevated EGFR expression has been identified previously in several studies of canine astrocytomas (for example [26, 27, 49, 50]). Consistent with these findings, in the present study, the dog orthologue of EGFR (CFA 18, 9.0 Mb) showed increased genomic copy number in both grade IV astrocytoma cases, as well as two grade II mixed gliomas. Human gliomas also share a high incidence of gain of HSA 8q, including the MYC oncogene (HSA 8; 128.8 Mb). This is consistent with gain of CFA 13 (including MYC, CFA 13; 28.2 Mb) in 63% (15/24) dog gliomas, an aberration that we have shown previously also to be highly recurrent in a wide range of other dog tumor types [21, 30, 31]. HSA 8q also shares conserved synteny with CFA 29, gained in 29% (7/24) gliomas, five of which were astrocytomas. Losses of HSA 9 (including p16/INK4A), HSA 10q (including PTEN) and HSA 19q are common to both astrocytomas and oligodendrogliomas in human patients; however we did not detect recurrent aberrations consistent with these aberrations in dog gliomas, as deletions of CFA 9/11, CFA 4/28 and CFA 1, respectively. Losses of HSA 10q and 19q are particularly associated with progression from low to high-grade tumors, and thus a focused analysis of an extensive panel of high-grade dog gliomas will be necessary to determine their incidence in canine patients.
Deletion of HSA 1p is a key hallmark aberration shared by astrocytomas and oligodendrogliomas, but with opposing prognostic significance. Approximately 60–80% of patients with oligodendrogliomas demonstrate a combined deletion of HSA 1p and HSA 19q through an unbalanced chromosome translocation [51] that is rarely encountered in astrocytomas (for example [47, 52]). Particularly in the absence of any other gross aberrations, this aberration confers increased chemosensitivity and in turn an increased survival time, as compared to glioma patients that present without these genomic imbalances ([53–57] and others). Molecular studies of human oligodendrogliomas therefore represent one success story in which this approach has been translated into a meaningful improvement in predicting prognosis for cancer patients. Interestingly, none of our dog glioma cohort demonstrated extensive recurrent genomic losses of regions corresponding to HSA 1p/19q, namely CFA 2qtel, 5qmid, 6qdist, 15qprox, 17qtel and 1qdist. If this observation is upheld in additional studies, the HSA 1p/19q prognostic factor may therefore not be translatable to the canine counterpart. This would have immense significance within the concept of a comparative approach to molecular medicine. In turn, if this genetic aberration is not evolutionarily conserved, detailed evaluation of its precise role in the initiation and evolution of human oligodendro-gliomas is warranted. It is possible that the related HSA 1p/19q translocation event may occur in the dog without incurring any genomic imbalance, which would be undetectable by aCGH analysis. Investigation of this hypothesis will require targeted cytogenetic analysis of a larger case series of canine oligodendroglioma interphase nuclei or chromosome preparations with dog markers selected from these chromosome regions.
While these key hallmarks of human gliomas did not appear to be highly conserved in canine gliomas, several recurrent chromosome aberrations were shown to be characteristic of the dog counterpart but with no apparent conservation in human patients. The most striking of these was gain of CFA 34, evident in 79% of gliomas, which shares conserved synteny with HSA 3qtel. While gain of the entire long arm of HSA 3 is not a common event in astrocytoma, the subregion HSA 3q26-q27 has been highlighted as a small scale aberration associated with recurrent genomic gain in human patients [4] that may harbor an as-yet unidentified oncogene. This comparative correlation thus invites more detailed assessment. There is no obvious evolutionary-based explanation for the majority of the gross chromosome aberrations that were observed in 40% of dog cases; however, as with the characteristic losses of CFA 17 and CFA 27 in meningiomas, their non-random nature indicates a strong likelihood that they have biological relevance, and clearly merits further investigation.
Genome architecture of the dog may aid determination of minimal regions of significance
It was evident in the present study that despite profiling at ~1 Mb intervals, the vast majority of genomic imbalances in both canine meningiomas and gliomas involved whole chromosome gains and losses. The dog genome comprises 38 pairs of acrocentric/telocentric autosomes, ranging in size from 125 to 26 Mb, plus a pair of bi-armed sex chromosomes [58]. The human genome is packaged into 22 autosomal pairs ranging from 247 to 47 Mb in size (NCBI Build 36.1, http://genome.ucsc.edu), with a combination of metacentric, submetacentric and acrocentric morphologies, plus bi-armed sex chromosomes. The largest dog chromosome (CFA 1) is smaller than HSA 12, and with the exception of CFA 1–5, all dog autosomes are smaller than HSA 18 [59]. Considering that intracranial tumors in human patients frequently demonstrate extensive regions of genomic DNA copy number imbalance (whole chromosomes and chromosome arms), it seems reasonable to expect similarly large regions to be detected in comparable studies of canine patients. With the dog genome packaged into a higher number of smaller, single-armed chromosomes, such changes would be predicted to manifest predominantly as whole chromosome gains and losses, as was observed in the present study. Thus the contrasting structural organization of the dog and human genomes offers tremendous opportunities for refining conserved regions of tumor-associated genomic imbalance, identifying new candidate genes and developing novel diagnostic, prognostic and therapeutic targets. It is interesting to note that in contrast to our findings in dog intracranial tumors, our ongoing studies of other dog cancers, including lymphomas, leukemias, soft tissue sarcomas and osteosarcomas, show that, as in their human counterparts, partial chromosome gains and losses, and amplifications and deletions of genomic regions <10 Mb in size (particularly surrounding sites of known oncogenes and tumor-suppressor genes) are frequent events. Although this may in part reflect the greater number of higher-resolution human cytogenetics studies of these tumor types as compared to intracranial neoplasms, it will be interesting to establish whether these observations persist in future studies.
The dog as a cytogenetic model for gene discovery in intracranial tumors
In recent years the increasing resolution and throughput of microarray-based CGH analyses have generated extensive data regarding recurrent genomic copy number changes in human intracranial tumors. We now recognize certain key molecular events involved in the progression of low to high-grade tumors, and have some ability to provide more accurate and refined diagnoses on the basis of presence or absence of specific genomic aberrations. A molecular approach to classification now enables us to categorise many tumors into major groups that correlate with histological and prognostic factors, based on the presence or absence of a small number of specific and in part, mutually exclusive, genomic changes (for example [60]). However, it is clear that our understanding is far from complete. We have yet to develop fully comprehensive means for molecular subclassification, to establish the precise cause of non-random genomic changes, and to understand fully their effect on cell cycle regulation. Although by no means without exception, there is evidence for correlation between chromosomal copy number gains and increased expression of key regulatory genes encoded within those genomic regions (for example [61, 62]); however the relationship between tumor-associated genomic copy number status and gene expression levels also remains to be fully understood, in part due to the paucity of studies in which both parameters have been evaluated on the same case series.
Towards addressing these key questions, we have shown that as with human patients, dog intracranial tumors present with a diverse range of recurrent chromosome aberrations, a proportion of which show evidence for association with specific tumor subtypes. Our data show that a subset of these highly recurrent aberrations appear as orthologous counterparts in both species, suggestive of an evolutionarily conserved genetic etiology that merits further evaluation. Interestingly, however, several of the key hallmark chromosome aberrations encountered in human populations, particularly deletion of HSA 22 in meningiomas, gain of HSA 7p in astrocytomas and loss of HSA 1p/19q in oligodendrogliomas, were not highly conserved as gross genomic imbalances in the corresponding canine tumors. Conversely, we identified highly recurrent CNAs in dog tumors that have not been reported as characteristic of the corresponding human tumor. Analysis of a greater number of cases will indicate whether these patterns are upheld in a more extensive case series. Using the results of the present study, we may now embark upon aCGH-directed cytogenetic analysis of larger cohort, utilizing strategically-selected genomic loci representing non-random CNAs worthy of special attention. The application of these genomic markers to retrospective case series with known clinical outcome will enable assessment of the potential prognostic significance of the corresponding chromosome aberration, while continued assessment of prospective cases will be necessary in order to evaluate in detail the role of structural chromosome defects in tumorigenesis.
It is possible that there is greater evolutionary conservation in the structural rather than numerical changes that occur in both dog and human tumors; however in the absence of associated genomic imbalance these aberrations are intractable to high-throughput aCGH methods and are thus less readily characterized. It is evident from the literature that independent studies of the same cancer frequently reveal highly discordant cytogenetic profiles, which can be attributed in part to differences in study design and application. Two hypotheses have been proposed recently to explain these findings [63]; firstly that a vast multitude of different genomic regions may be involved in tumorigenesis and that each case series will identify a varying combination of these regions. A contrasting hypothesis suggests that the majority of recurrent chromosome aberrations reported thus far are effectively random events, occurring secondary to a small number of (largely already known) key causative changes that may become masked by this vast excess of genomic ‘noise’. Recent work, taking into account the incidence, physical extent and the amplitude of copy number aberrations, points to the latter being the more likely explanation, at least for the glioma genome. It is therefore possible that in both human and dog tumors, the more extensive genomic imbalances mask smaller, more subtle aberrations that, while neither widely reported nor well characterized, may be more intimately associated with the biological behavior of the tumor. Based on the findings of the present study, we propose that comparative cytogenetic profiling of dog and human intracranial (and other) tumors will enable this hypothesis to be examined in greater detail, by revealing the evolutionarily-conserved subset of genomic changes that are common to both species. The association of these changes with tumor histopathology and patient survival will then be crucial in translating these findings into more sophisticated means for disease diagnosis and prognosis. Extension of these studies to the targeted analysis of canine cases based on their cranial morphology has enormous potential to identify genetic features associated with heritable risk of tumor development. The many attributes of spontaneous dog brain tumors as a model system, combined with their immense impact on the welfare of our canine companions in their own right, suggests that such an endeavour would represent a classic example of the synergy afforded by the ‘one medicine’ concept of biomedical research.
Acknowledgements
We would like to acknowledge Pragna Mehta and the Veterinary Neurology and Pathology residents and clinicians from North Carolina State University and the University of California at Davis for assistance with tissue procurement. We thank Eric Seiser for assistance with data analysis. This work was supported by grants from the National Institutes of Health (NS051190) and the American Kennel Club Canine Health Foundation (CHF-403) awarded to MB and NJO. CFL and PE are supported by funds from the Wellcome Trust. HJW is supported by National Science Foundation Award (DMS-0706963). PD and RJH are supported by the Paul C and Borghild T Petersen Foundation.
Contributor Information
Rachael Thomas, Department of Molecular Biomedical Sciences, College of Veterinary Medicine, North Carolina State University, 4700 Hillsborough Street, Raleigh, NC 27606, USA; Center for Comparative Medicine and Translational Research, North Carolina State University, Raleigh, NC 27606, USA.
Shannon E. Duke, Department of Molecular Biomedical Sciences, College of Veterinary Medicine, North Carolina State University, 4700 Hillsborough Street, Raleigh, NC 27606, USA;
Huixia J. Wang, Department of Statistics, College of Agriculture and Life Sciences, North Carolina State University, Patterson Hall, 2501 Founders Drive, Raleigh, NC 27695, USA
Tessa E. Breen, Department of Clinical Sciences, College of Veterinary Medicine, North Carolina State University, 4700 Hillsborough Street, Raleigh, NC 27606, USA
Robert J. Higgins, Department of Pathology, Microbiology and Immunology, School of Veterinary Medicine, University of California-Davis, Davis, CA 95616, USA
Keith E. Linder, Center for Comparative Medicine and Translational Research, North Carolina State University, Raleigh, NC 27606, USA; Department of Population Health and Pathobiology, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, USA
Peter Ellis, Microarray Facility, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK.
Cordelia F. Langford, Microarray Facility, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK
Peter J. Dickinson, Department of Surgical and Radiological Sciences, School of Veterinary Medicine, University of California-Davis, Davis, CA 95616, USA
Natasha J. Olby, Center for Comparative Medicine and Translational Research, North Carolina State University, Raleigh, NC 27606, USA; Department of Clinical Sciences, College of Veterinary Medicine, North Carolina State University, 4700 Hillsborough Street, Raleigh, NC 27606, USA
Matthew Breen, Department of Molecular Biomedical Sciences, College of Veterinary Medicine, North Carolina State University, 4700 Hillsborough Street, Raleigh, NC 27606, USA; Center for Comparative Medicine and Translational Research, North Carolina State University, Raleigh, NC 27606, USA; Cancer Genetics Program, UNC Lineberger Comprehensive Cancer Center, Chapel Hill, NC 27599, USA.
References
- 1.Ries L, Melbert D, Krapcho M, Stinchcomb D, Howlader N, Horner M, Mariotto A, Miller B, Feuer E, Altekruse S, Lewis D, Clegg L, Eisner M, Reichman M, Edwards BK. SEER cancer statistics review, 1975–2005. National Cancer Institute; Bethesda, MD: 2008. http://seer.cancer.gov/csr/1975_2005/, based on November 2007 SEER data submission, posted to the SEER web site. [Google Scholar]
- 2.Soffietti R, Ruda R. Neuro-oncology: new insights and advances in treatment. Lancet Neurol. 2008;7:14–16. doi: 10.1016/S1474-4422(07)70305-8. doi:10.1016/S1474-4422(07)70305-8. [DOI] [PubMed] [Google Scholar]
- 3.Collins VP. Brain tumours: classification and genes. J Neurol Neurosurg Psychiatry. 2004;75(Suppl 2):ii2–ii11. doi: 10.1136/jnnp.2004.040337. doi:10.1136/jnnp.2004.040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koschny R, Koschny T, Froster UG, Krupp W, Zuber MA. Comparative genomic hybridization in glioma: a meta-analysis of 509 cases. Cancer Genet Cytogenet. 2002;135:147–159. doi: 10.1016/s0165-4608(01)00650-1. doi:10.1016/S0165-4608(01)00650-1. [DOI] [PubMed] [Google Scholar]
- 5.van Tilborg AA, Al Allak B, Velthuizen SC, de Vries A, Kros JM, Avezaat CJ, de Klein A, Beverloo HB, Zwarthoff EC. Chromosomal instability in meningiomas. J Neuropathol Exp Neurol. 2005;64:312–322. doi: 10.1093/jnen/64.4.312. [DOI] [PubMed] [Google Scholar]
- 6.Puget S, Rutka J. Malignant brain tumors: two steps for-ward. Clin Neurosurg. 2007;54:4–9. [PubMed] [Google Scholar]
- 7.Kimmelman J, Nalbantoglu J. Faithful companions: a proposal for neurooncology trials in pet dogs. Cancer Res. 2007;67:4541–4544. doi: 10.1158/0008-5472.CAN-06-3792. doi:10.1158/0008-5472.CAN-06-3792. [DOI] [PubMed] [Google Scholar]
- 8.Dobson JM, Samuel S, Milstein H, Rogers K, Wood JL. Canine neoplasia in the UK: estimates of incidence rates from a population of insured dogs. J Small Anim Pract. 2002;43:240–246. doi: 10.1111/j.1748-5827.2002.tb00066.x. doi:10.1111/j.1748-5827.2002.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 9.Koestner A, Higgins R. Tumors of the nervous system. In: Meuten D, editor. Tumors in domestic animals. Blackwell Publishing; 2002. p. 697. [Google Scholar]
- 10.Kleihues P, Cavenee WK. World Health Organization Classification of Tumors. IARC Press; Lyon: 2000. Tumors of the nervous system: pathology and genetics; pp. 9–70. [Google Scholar]
- 11.Kleihues P, Louis DN, Scheithauer BW, Rorke LB, Reifenberger G, Burger PC, Cavenee WK. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol. 2002;61:215–225. doi: 10.1093/jnen/61.3.215. (discussion 226–219) [DOI] [PubMed] [Google Scholar]
- 12.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. doi:10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candolfi M, Curtin JF, Nichols WS, Muhammad AG, King GD, Pluhar GE, McNiel EA, Ohlfest JR, Freese AB, Moore PF, Lerner J, Lowenstein PR, Castro MG. Intracranial glioblastoma models in preclinical neuro-oncology: neuropathological characterization and tumor progression. J Neurooncol. 2007;85:133–148. doi: 10.1007/s11060-007-9400-9. doi:10.1007/s11060-007-9400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Candolfi M, Pluhar GE, Kroeger K, Puntel M, Curtin J, Barcia C, Muhammad AK, Xiong W, Liu C, Mondkar S, Kuoy W, Kang T, McNeil EA, Freese AB, Ohlfest JR, Moore P, Palmer D, Ng P, Young JD, Lowenstein PR, Castro MG. Optimization of adenoviral vector-mediated transgene expression in the canine brain in vivo, and in canine glioma cells in vitro. Neuro Oncol. 2007;9:245–258. doi: 10.1215/15228517-2007-012. doi:10.1215/15228517-2007-012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 16.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. doi:10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 17.Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, Johnson GS, DeFrance HB, Ostrander EA, Kruglyak L. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. doi:10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 18.Summers B, Cummings J, de Lahunta A. Veterinary neuropathology. Mosby; St Louis: 1995. Tumors of the central nervous system. [Google Scholar]
- 19.Dickinson PJ, Keel MK, Higgins RJ, Koblik PD, LeCouteur RA, Naydan DK, Bollen AW, Vernau W. Clinical and pathologic features of oligodendrogliomas in two cats. Vet Pathol. 2000;37:160–167. doi: 10.1354/vp.37-2-160. doi:10.1354/vp.37-2-160. [DOI] [PubMed] [Google Scholar]
- 20.Thomas R, Duke SE, Karlsson EK, Evans A, Ellis P, Lindblad-Toh K, Langford CF, Breen M. A genome assembly-integrated dog 1 Mb BAC microarray: a cytogenetic resource for canine cancer studies and comparative genomic analysis. Cytogenet Genome Res. 2008;122:110–121. doi: 10.1159/000163088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas R, Duke SE, Bloom SK, Breen TE, Young AC, Feiste E, Seiser EL, Tsai PC, Langford CF, Ellis P, Karlsson EK, Lindblad-Toh K, Breen M. A cytogenetically characterized, genome-anchored 10-Mb BAC set and CGH array for the domestic dog. J Hered. 2007;98:474–484. doi: 10.1093/jhered/esm053. doi:10.1093/jhered/esm053. [DOI] [PubMed] [Google Scholar]
- 22.Breen M, Bullerdiek J, Langford CF. The DAPI banded karyotype of the domestic dog (Canis familiaris) generated using chromosome-specific paint probes. Chromosome Res. 1999;7:401–406. doi: 10.1023/a:1009224232134. doi:10.1023/A:1009224232134. [DOI] [PubMed] [Google Scholar]
- 23.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jong K, Marchiori E, Meijer G, Vaart AV, Ylstra B. Breakpoint identification and smoothing of array comparative genomic hybridization data. Bioinformatics. 2004;20:3636–3637. doi: 10.1093/bioinformatics/bth355. doi:10.1093/bioinformatics/bth355. [DOI] [PubMed] [Google Scholar]
- 25.Breen M, Hitte C, Lorentzen TD, Thomas R, Cadieu E, Sabacan L, Scott A, Evanno G, Parker HG, Kirkness EF, Hudson R, Guyon R, Mahairas GG, Gelfenbeyn B, Fraser CM, Andre C, Galibert F, Ostrander EA. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics. 2004;5:65–75. doi: 10.1186/1471-2164-5-65. doi:10.1186/1471-2164-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder JM, Shofer FS, Van Winkle TJ, Massicotte C. Canine intracranial primary neoplasia: 173 cases (1986–2003) J Vet Intern Med. 2006;20:669–675. doi: 10.1892/0891-6640(2006)20[669:cipnc]2.0.co;2. doi:10.1892/0891-6640(2006)20[669:CIPNC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 27.Stoica G, Kim HT, Hall DG, Coates JR. Morphology, immunohistochemistry, and genetic alterations in dog astrocytomas. Vet Pathol. 2004;41:10–19. doi: 10.1354/vp.41-1-10. doi:10.1354/vp.41-1-10. [DOI] [PubMed] [Google Scholar]
- 28.Sturges BK, Dickinson PJ, Bollen AW, Koblik PD, Kass PH, Kortz GD, Vernau KM, Knipe MF, Lecouteur RA, Higgins RJ. Magnetic resonance imaging and histological classification of intracranial meningiomas in 112 dogs. J Vet Intern Med. 2008;22:586–595. doi: 10.1111/j.1939-1676.2008.00042.x. doi:10.1111/j.1939-1676.2008.00042.x. [DOI] [PubMed] [Google Scholar]
- 29.Breen M, Modiano JF. Evolutionarily conserved cytogenetic changes in hematological malignancies of dogs and humans—man and his best friend share more than companionship. Chromosome Res. 2008;16:145–154. doi: 10.1007/s10577-007-1212-4. doi:10.1007/s10577-007-1212-4. [DOI] [PubMed] [Google Scholar]
- 30.Thomas R, Scott A, Langford CF, Fosmire SP, Jubala CM, Lorentzen TD, Hitte C, Karlsson EK, Kirkness E, Ostrander EA, Galibert F, Lindblad-Toh K, Modiano JF, Breen M. Construction of a 2-Mb resolution BAC microarray for CGH analysis of canine tumors. Genome Res. 2005;15:1831–1837. doi: 10.1101/gr.3825705. doi:10.1101/gr.3825705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas R, Smith KC, Ostrander EA, Galibert F, Breen M. Chromosome aberrations in canine multicentric lymphomas detected with comparative genomic hybridisation and a panel of single locus probes. Br J Cancer. 2003;89:1530–1537. doi: 10.1038/sj.bjc.6601275. doi:10.1038/sj.bjc.6601275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modiano JF, Breen M, Burnett RC, Parker HG, Inusah S, Thomas R, Avery PR, Lindblad-Toh K, Ostrander EA, Cutter GC, Avery AC. Distinct B-cell and T-cell lymphoproliferative disease prevalence among dog breeds indicates heritable risk. Cancer Res. 2005;65:5654–5661. doi: 10.1158/0008-5472.CAN-04-4613. doi:10.1158/0008-5472.CAN-04-4613. [DOI] [PubMed] [Google Scholar]
- 33.Thomson SA, Kennerly E, Olby N, Mickelson JR, Hoffmann DE, Dickinson PJ, Gibson G, Breen M. Microarray analysis of differentially expressed genes of primary tumors in the canine central nervous system. Vet Pathol. 2005;42:550–558. doi: 10.1354/vp.42-5-550. doi:10.1354/vp.42-5-550. [DOI] [PubMed] [Google Scholar]
- 34.Arslantas A, Artan S, Oner U, Durmaz R, Muslumanoglu H, Atasoy MA, Basaran N, Tel E. Comparative genomic hybridization analysis of genomic alterations in benign, atypical and anaplastic meningiomas. Acta Neurol Belg. 2002;102:53–62. [PubMed] [Google Scholar]
- 35.Weber RG, Bostrom J, Wolter M, Baudis M, Collins VP, Reifenberger G, Lichter P. Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci USA. 1997;94:14719–14724. doi: 10.1073/pnas.94.26.14719. doi:10.1073/pnas.94.26.14719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mark J, Levan G, Mitelman F. Identification by fluorescence of the G chromosome lost in human meningomas. Hereditas. 1972;71:163–168. doi: 10.1111/j.1601-5223.1972.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 37.Sawyer JR, Husain M, Pravdenkova S, Krisht A, Al-Mefty O. A role for telomeric and centromeric instability in the progression of chromosome aberrations in meningioma patients. Cancer. 2000;88:440–453. doi:10.1002/(SICI)1097-0142(20000115)88:2<440::AID-CNCR27>3.0.CO;2-5. [PubMed] [Google Scholar]
- 38.Al-Mefty O, Kadri PA, Pravdenkova S, Sawyer JR, Stangeby C, Husain M. Malignant progression in meningioma: documentation of a series and analysis of cytogenetic findings. J Neurosurg. 2004;101:210–218. doi: 10.3171/jns.2004.101.2.0210. [DOI] [PubMed] [Google Scholar]
- 39.Ishino S, Hashimoto N, Fushiki S, Date K, Mori T, Fujimoto M, Nakagawa Y, Ueda S, Abe T, Inazawa J. Loss of material from chromosome arm 1p during malignant progression of meningioma revealed by fluorescent in situ hybridization. Cancer. 1998;83:360–366. doi:10.1002/(SICI)1097-0142(19980715)83:2<360::AID-CNCR21>3.0.CO;2-Q. [PubMed] [Google Scholar]
- 40.Caren H, Fransson S, Ejeskar K, Kogner P, Martinsson T. Genetic and epigenetic changes in the common 1p36 deletion in neuroblastoma tumours. Br J Cancer. 2007;97:1416–1424. doi: 10.1038/sj.bjc.6604032. doi:10.1038/sj.bjc.6604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okawa ER, Gotoh T, Manne J, Igarashi J, Fujita T, Silverman KA, Xhao H, Mosse YP, White PS, Brodeur GM. Expression and sequence analysis of candidates for the 1p36.31 tumor suppressor gene deleted in neuroblastomas. Oncogene. 2008;27:803–810. doi: 10.1038/sj.onc.1210675. doi:10.1038/sj.onc.1210675. [DOI] [PubMed] [Google Scholar]
- 42.Tamimi Y, Ziebart K, Desaulniers N, Dietrich K, Grundy P. Identification of a minimal region of loss on the short arm of chromosome 1 in Wilms tumor. Genes Chromosomes Cancer. 2007;46:327–335. doi: 10.1002/gcc.20413. doi:10.1002/gcc.20413. [DOI] [PubMed] [Google Scholar]
- 43.Thelander E Flordal, Ichimura K, Collins VP, Walsh SH, Barbany G, Hagberg A, Laurell A, Rosenquist R, Larsson C, Lagercrantz S. Detailed assessment of copy number alterations revealing homozygous deletions in 1p and 13q in mantle cell lymphoma. Leuk Res. 2007;31:1219–1230. doi: 10.1016/j.leukres.2006.10.022. doi:10.1016/j.leukres.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 44.Zahn S, Sievers S, Alemazkour K, Orb S, Harms D, Schulz WA, Calaminus G, Gobel U, Schneider DT. Imbalances of chromosome arm 1p in pediatric and adult germ cell tumors are caused by true allelic loss: a combined comparative genomic hybridization and microsatellite analysis. Genes Chromosomes Cancer. 2006;45:995–1006. doi: 10.1002/gcc.20363. doi:10.1002/gcc.20363. [DOI] [PubMed] [Google Scholar]
- 45.Idbaih A, Marie Y, Pierron G, Brennetot C, Hoang-Xuan K, Kujas M, Mokhtari K, Sanson M, Lejeune J, Aurias A, Delattre O, Delattre JY. Two types of chromosome 1p losses with opposite significance in gliomas. Ann Neurol. 2005;58:483–487. doi: 10.1002/ana.20607. doi:10.1002/ana.20607. [DOI] [PubMed] [Google Scholar]
- 46.Kilic E, Naus NC, van Gils W, Klaver CC, van Til ME, Verbiest MM, Stijnen T, Mooy CM, Paridaens D, Beverloo HB, Luyten GP, de Klein A. Concurrent loss of chromosome arm 1p and chromosome 3 predicts a decreased disease-free survival in uveal melanoma patients. Invest Ophthalmol Vis Sci. 2005;46:2253–2257. doi: 10.1167/iovs.04-1460. doi:10.1167/iovs.04-1460. [DOI] [PubMed] [Google Scholar]
- 47.Giannini C, Scheithauer BW, Weaver AL, Burger PC, Kros JM, Mork S, Graeber MB, Bauserman S, Buckner JC, Burton J, Riepe R, Tazelaar HD, Nascimento AG, Crotty T, Keeney GL, Pernicone P, Altermatt H. Oligodendrogliomas: reproducibility and prognostic value of histologic diagnosis and grading. J Neuropathol Exp Neurol. 2001;60:248–262. doi: 10.1093/jnen/60.3.248. [DOI] [PubMed] [Google Scholar]
- 48.Trost D, Ehrler M, Fimmers R, Felsberg J, Sabel MC, Kirsch L, Schramm J, Wiestler OD, Reifenberger G, Weber RG. Identification of genomic aberrations associated with shorter overall survival in patients with oligodendroglial tumors. Int J Cancer. 2007;120:2368–2376. doi: 10.1002/ijc.22574. doi:10.1002/ijc.22574. [DOI] [PubMed] [Google Scholar]
- 49.Dickinson PJ, Roberts B, Higgins R, Leutenegger C, Bollen AW, Kass PH, LeCouteur RA. Expression of receptor tyrosine kinases VEGFR-1 (FLT-1), VEGFR-2 (KDR), EGFR-1, PDG-FRa and c-Met in canine primary brain tumours. Vet Comp Oncol. 2006;4:132–140. doi: 10.1111/j.1476-5829.2006.00101.x. doi:10.1111/j.1476-5829.2006.00101.x. [DOI] [PubMed] [Google Scholar]
- 50.Lipsitz D, Higgins RJ, Kortz GD, Dickinson PJ, Bollen AW, Naydan DK, Le Couteur RA. Glioblastoma multiforme: clinical findings, magnetic resonance imaging, and pathology in five dogs. Vet Pathol. 2003;40:659–669. doi: 10.1354/vp.40-6-659. doi:10.1354/vp.40-6-659. [DOI] [PubMed] [Google Scholar]
- 51.Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, Flynn H, Passe S, Felten S, Brown PD, Shaw EG, Buckner JC. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. doi:10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 52.Iwamoto FM, Nicolardi L, Demopoulos A, Barbashina V, Salazar P, Rosenblum M, Hormigo A. Clinical relevance of 1p and 19q deletion for patients with WHO grade 2 and 3 gliomas. J Neurooncol. 2008;88:293–298. doi: 10.1007/s11060-008-9563-z. doi:10.1007/s11060-008-9563-z. [DOI] [PubMed] [Google Scholar]
- 53.Cairncross JG, Macdonald DR. Oligodendroglioma: a new chemosensitive tumor. J Clin Oncol. 1990;8:2090–2091. doi: 10.1200/JCO.1990.8.12.2090. [DOI] [PubMed] [Google Scholar]
- 54.Smith JS, Perry A, Borell TJ, Lee HK, O’Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, Jenkins RB. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 55.Ino Y, Betensky RA, Zlatescu MC, Sasaki H, Macdonald DR, Stemmer-Rachamimov AO, Ramsay DA, Cairncross JG, Louis DN. Molecular subtypes of anaplastic oligodendroglioma: implications for patient management at diagnosis. Clin Cancer Res. 2001;7:839–845. [PubMed] [Google Scholar]
- 56.van den Bent MJ, Looijenga LH, Langenberg K, Dinjens W, Graveland W, Uytdewilligen L, Smitt PA Sillevis, Jenkins RB, Kros JM. Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer. 2003;97:1276–1284. doi: 10.1002/cncr.11187. doi:10.1002/cncr.11187. [DOI] [PubMed] [Google Scholar]
- 57.Felsberg J, Erkwoh A, Sabel MC, Kirsch L, Fimmers R, Blaschke B, Schlegel U, Schramm J, Wiestler OD, Reifenberger G. Oligodendroglial tumors: refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol. 2004;14:121–130. doi: 10.1111/j.1750-3639.2004.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, III, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli KP, Parker HG, Pollinger JP, Searle SM, Sutter NB, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait-Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang Y, Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, De-Gray S, Dhargay N, Dooley K, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M, Foley K, Foley C, Franke A, Friedrich D, Gage D, Garber M, Gearin G, Giannoukos G, Goode T, Goyette A, Graham J, Grandbois E, Gyaltsen K, Hafez N, Hagopian D, Hagos B, Hall J, Healy C, Hegarty R, Honan T, Horn A, Houde N, Hughes L, Hunnicutt L, Husby M, Jester B, Jones C, Kamat A, Kanga B, Kells C, Khazanovich D, Kieu AC, Kisner P, Kumar M, Lance K, Landers T, Lara M, Lee W, Leger JP, Lennon N, Leuper L, LeVine S, Liu J, Liu X, Lokyitsang Y, Lokyitsang T, Lui A, Macdonald J, Major J, Marabella R, Maru K, Matthews C, McDonough S, Mehta T, Meldrim J, Melnikov A, Meneus L, Mihalev A, Mihova T, Miller K, Mittelman R, Mlenga V, Mulrain L, Munson G, Navidi A, Naylor J, Nguyen T, Nguyen N, Nguyen C, Nguyen T, Nicol R, Norbu N, Norbu C, Novod N, Nyima T, Olandt P, O’Neill B, O’Neill K, Osman S, Oyono L, Patti C, Perrin D, Phunkhang P, Pierre F, Priest M, Rachupka A, Raghuraman S, Rameau R, Ray V, Raymond C, Rege F, Rise C, Rogers J, Rogov P, Sahalie J, Settipalli S, Sharpe T, Shea T, Sheehan M, Sherpa N, Shi J, Shih D, Sloan J, Smith C, Sparrow T, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Stone S, Sykes S, Tchuinga P, Tenzing P, Tesfaye S, Thoulutsang D, Thoulutsang Y, Topham K, Topping I, Tsamla T, Vassiliev H, Venkataraman V, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Yang S, Yang X, Young G, Yu Q, Zainoun J, Zembek L, Zimmer A, Lander ES. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. doi:10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 59.Breen M. Canine cytogenetics—from band to basepair. Cytogenet Genome Res. 2008;120:50–60. doi: 10.1159/000118740. doi:10.1159/000118740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Idbaih A, Marie Y, Lucchesi C, Pierron G, Manie E, Raynal V, Mosseri V, Hoang-Xuan K, Kujas M, Brito I, Mokhtari K, Sanson M, Barillot E, Aurias A, Delattre JY, Delattre O. BAC array CGH distinguishes mutually exclusive alterations that define clinicogenetic subtypes of gliomas. Int J Cancer. 2008;122:1778–1786. doi: 10.1002/ijc.23270. doi:10.1002/ijc.23270. [DOI] [PubMed] [Google Scholar]
- 61.Lo KC, Rossi MR, LaDuca J, Hicks DG, Turpaz Y, Hawthorn L, Cowell JK. Candidate glioblastoma development gene identification using concordance between copy number abnormalities and gene expression level changes. Genes Chromosomes Cancer. 2007;46:875–894. doi: 10.1002/gcc.20474. doi:10.1002/gcc.20474. [DOI] [PubMed] [Google Scholar]
- 62.Ruano Y, Mollejo M, Ribalta T, Fiano C, Camacho FI, Gomez E, de Lope AR, Hernandez-Moneo JL, Martinez P, Melendez B. Identification of novel candidate target genes in amplicons of Glioblastoma multiforme tumors detected by expression and CGH microarray profiling. Mol Cancer. 2006;5:39. doi: 10.1186/1476-4598-5-39. doi:10.1186/1476-4598-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. doi:10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]