SUMMARY
Androgen receptor (AR) is reactivated in castration resistant prostate cancer (CRPC) through mechanisms including marked increases in AR gene expression. We identify an enhancer in the AR second intron contributing to increased AR expression at low androgen levels in CRPC. Moreover, at increased androgen levels the AR binds this site and represses AR gene expression through recruitment of lysine specific demethylase 1 (LSD1) and H3K4me1,2 demethylation. AR similarly represses expression of multiple genes mediating androgen synthesis, DNA synthesis and proliferation, while stimulating genes mediating lipid and protein biosynthesis. Androgen levels in CRPC appear adequate to stimulate AR activity on enhancer elements, but not suppressor elements, resulting in increased expression of AR and AR repressed genes that contribute to cellular proliferation.
Keywords: prostate cancer, androgen receptor, androgen deprivation therapy, H3K4 methylation, LSD1
INTRODUCTION
The standard treatment for metastatic prostate cancer (PCa) is surgical or medical castration to reduce circulating androgens (androgen deprivation therapy, ADT) and suppress activity of the androgen receptor (AR), but patients invariably relapse with more aggressive castration resistant prostate cancer (CRPC). Significantly, early studies showed that AR was highly expressed in CRPC (Ruizeveld de Winter et al., 1994), and further studies in clinical samples and xenograft models have confirmed that AR mRNA is highly expressed and consistently increased in CRPC compared to levels prior to ADT (Taplin et al., 1995; Gregory et al., 2001; Holzbeierlein et al., 2004; Chen et al., 2004; Stanbrough et al., 2006). Multiple androgen regulated-genes, including prostate specific antigen (PSA) and the TMPRSS2:ERG fusion gene, are also highly expressed in CRPC, indicating that AR transcriptional activity has been reactivated despite castrate serum androgen levels (Stanbrough et al., 2006; Cai et al., 2009). Mechanisms that may contribute to restoring AR activity in CRPC include AR mutations or alternative splicing, increased intratumoral androgen synthesis, increased coactivator expression, and activation of several kinases that may directly or indirectly sensitize AR to low levels of androgens (Yuan and Balk, 2009). Moreover, studies in xenograft models indicate that even modest increases in AR protein expression may alone render tumors resistant to castration and to available AR antagonists (Chen et al., 2004).
Despite the critical role AR plays in PCa development and progression to CRPC, the mechanisms that regulate its expression, and contribute to its increased expression in CRPC, are not well understood. AR mRNA levels may be controlled physiologically by a suppressor element in the 5'UTR of the AR gene that regulates transcription (Kumar et al., 1994; Wang et al., 2004; Wang et al., 2008) and by an element in the 3'UTR that regulates mRNA stability (Yeap et al., 2002). Mechanisms contributing to the increased AR mRNA in CRPC include AR gene amplification in about one-third of CRPC patients (Visakorpi et al., 1995) and increased E2F activity in RB deficient tumors (Sharma et al., 2010). Previous studies in androgen sensitive rodent tissues and in LNCaP PCa cells have shown that androgens can negatively regulate AR gene transcription, suggesting that AR mRNA may also increase after ADT due to relief from this negative regulation (Quarmby et al., 1990; Shan et al., 1990; Krongrad et al., 1991; Blok et al., 1992). However, the androgen mediated changes in AR mRNA levels in LNCaP cells are modest and the molecular basis for this negative regulation has not been determined. In contrast to these findings in LNCaP cells, we reported recently that AR mRNA levels in VCaP PCa cells and xenografts were rapidly and substantially increased in response to androgen deprivation, suggesting that relief from AR mediated negative regulation of AR gene expression may make a significant contribution to increasing AR mRNA in CRPC (Cai et al., 2009). This study addresses the molecular basis for this negative regulation of AR gene expression by the androgen liganded AR.
RESULTS
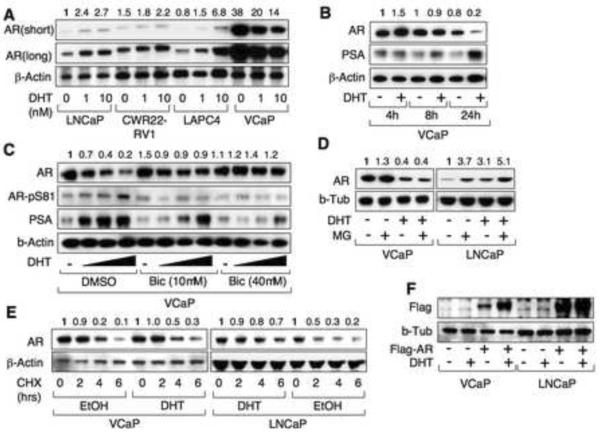
Androgen decreases AR protein in VCaP cells
The VCaP PCa cell line was derived from a vertebral metastasis in a patient with CRPC and it expresses wild-type (WT) AR and AR-regulated genes such as PSA and the TMPRSS2:ERG fusion gene (Korenchuk et al., 2001; Loberg et al., 2006; Cai et al., 2009). In the absence of exogenous androgen, AR protein expression in VCaP cells was higher than in other PCa cell lines including LNCaP, LAPC4, and CWR22Rv1 cells (the latter express a mutant AR with a duplicated exon 3) (Fig. 1A). AR protein was increased by 24 hours of DHT treatment in LNCaP, LAPC4, and CWR22Rv1 cells, consistent with previous data showing that androgen binding increases AR protein stability (Kemppainen et al., 1992). In contrast, although AR protein in VCaP was modestly increased after 4 hours of DHT (Fig. 1B), it was markedly decreased at 24 hours (Fig. 1A) and after 3 days of DHT (Fig. S1). This decrease could be blocked by bicalutamide, an AR antagonist, indicating it was dependent on the agonist liganded AR (Fig. 1C). While AR protein was decreased by DHT, serine 81 phosphorylation (associated with AR transcriptional activity) and PSA expression were markedly increased, indicating that DHT was strongly inducing AR transcriptional activity (Fig. 1B and C).
Figure.1. Androgen decreases AR protein expression in VCaP cells.
(A) LNCaP, CWR22Rv1, LAPC4 or VCaP cells were treated with 0, 1, or 10 nM DHT for 24h and AR or β-actin were immunoblotted. (B) VCaP cells were treated with/out DHT for 4h, 8h, or 24h and AR, PSA, or β-actin were immunoblotted. (C) VCaP cells were treated with 0, 0.1, 1, or 10 nM DHT and with 0, 10, or 40 μM bicalutamide for 24h and immunobloted for AR, Ser 81 phosphorylated AR, PSA, or β-actin. (D) VCaP or LNCaP cells were pre-treated with/out 10 nM DHT for 24h and then treated with MG115/MG132 for 4h. (E) VCaP or LNCaP cells were pre-treated with/out DHT for 2h and then treated with cycloheximide (10 ng/mL) for 0, 2, 4, or 6h. (F) VCaP or LNCaP cells were transiently transfected with empty vector or 3×Flag-AR. After 24h, cells were treated with/out 10 nM DHT for 24h (note: the prostate cancer cells were steroid-depleted by culturing in medium with charcoal/dextran stripped serum, CSS, for 3d before treatments in all experiments). See also Figure S1.
AR protein levels in VCaP and LNCaP cells were increased by proteasome inhibitors (MG115 and MG132, MG) in the absence of DHT, but these inhibitors did not prevent the marked decrease in AR protein in response to DHT in VCaP cells, indicating that the molecular basis for this decline was not increased proteasome mediated AR degradation (Fig. 1D). To directly address whether the DHT liganded AR was less stable in VCaP versus LNCaP cells, we pretreated androgen depleted cells with DHT or vehicle for 2 hours and then added cycloheximide (CHX) to block new protein synthesis. Significantly, AR protein half-life in VCaP cells, similarly to LNCaP cells, was not decreased by DHT, demonstrating that DHT was not directly (through binding to the AR) enhancing AR degradation (Fig. 1E). Finally, DHT in VCaP cells markedly increased expression of transiently transfected Flag-tagged AR regulated by a CMV promoter, further indicating that DHT was not enhancing AR protein degradation (Fig. 1F). Therefore, we next examined effects on AR mRNA.
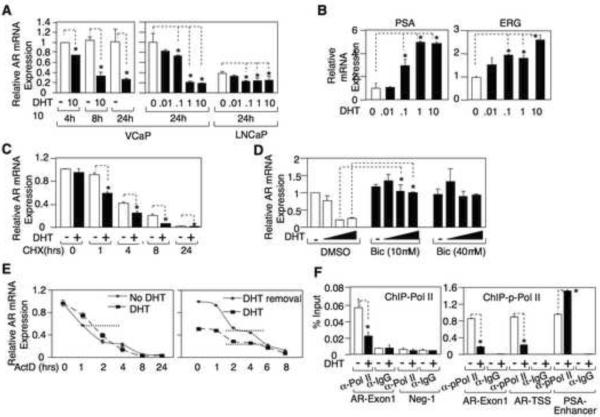
Agonist-liganded AR negatively regulates AR gene transcription
Androgen has been reported to cause a modest decrease in AR mRNA in LNCaP cells (Krongrad et al., 1991), but DHT in VCaP caused a rapid and more dramatic decrease in AR mRNA (Fig. 2A). Interestingly, a higher DHT concentration was required to suppress AR mRNA compared to the levels for induction of PSA and ERG mRNA (the latter from the androgen regulated TMPRSS2:ERG fusion gene), which were half-maximal at <0.1 nM DHT (Fig. 2B). To determine whether this decrease in AR mRNA required new protein synthesis, including the synthesis of ERG that was recently reported to suppress AR gene expression (Yu et al., 2010), we treated androgen starved cells with cycloheximide and DHT, and then measured AR mRNA levels over 24 hours. Significantly, treatment with cycloheximide did not prevent the enhanced decline in AR mRNA, indicating that it was not dependent on the DHT stimulated synthesis of new proteins (Fig. 2C). Bicalutamide blocked the suppression of AR mRNA by DHT (Fig. 2D), consistent with the effect being dependent on the agonist-liganded AR. To determine whether DHT was increasing AR mRNA degradation, we pretreated androgen starved VCaP cells with DHT for 2 hours and then added actinomycin D to block new mRNA synthesis. Significantly, AR mRNA half-life was not decreased by DHT (Fig. 2E, left panel), suggesting that DHT was decreasing AR gene transcription. We also assessed AR mRNA half-life in VCaP cells growing in medium with DHT versus cells where DHT was removed for 16 hours prior to addition of actinomycin D. While AR mRNA was decreased in the presence of DHT, there was no evident decrease in AR half-life (Fig. 2E, right panel). Finally, we found by chromatin immunoprecipitation (ChIP) that DHT decreased the binding of RNA polymerase II to exon 1 in the AR gene (Fig. 2F, left panel), and also decreased binding of active RNA polymerase II as shown by anti-phospho-RNA polymerase II ChiP (Fig. 2F, right panel). Together these results indicated that the DHT liganded AR in VCaP cells was directly repressing AR gene transcription.
Figure.2. Agonist-liganded AR negatively regulates AR gene transcription.
(A) VCaP or LNCaP cells were treated with 0, 0.01, 0.1, 1, or 10 nM DHT for 4h, 8h, or 24h and AR mRNA was measured using qRTPCR. (B) VCaP cells were DHT stimulated for 24h and mRNA for PSA and ERG were measured by qRT-PCR. (C) VCaP cells were treated with cycloheximide (10 ng/mL) and DHT or vehicle, and AR mRNA was then measured by qRT-PCR after.0, 1, 4, 8, or 24h (mRNA expression was normalized to internal control 18S RNA in all the experiments). (D) VCaP cells were treated with 0, 0.1, 1, or 10 nM DHT and with 0, 10, or 40 μM bicalutamide for 24h and AR mRNA was measured by qRT-PCR. (E) Left panel - androgen starved VCaP cells were pretreated with DHT or vehicle for 2 hours followed by addition of actinomycin D (10 μM); right panel - VCaP cells growing in medium with DHT were switched to the same medium with or without DHT for 16 h, followed by addition of actinomycin D. AR mRNA was measured by qRT-PCR at the indicated times after actinomycin D addition. Levels at time 0 were normalized to 1 under both conditions in the left panel and under the DHT removal condition in the right panel. Dotted lines indicate 50% maximal level. (F) VCaP cells were treated with/out DHT for 4h. The DNA bound to RNA polymerase II or active RNA polymerase II (phospho-Ser5) was immunoprecipitated and measured by qPCR. Error bars in each experiment indicate standard deviation (SD).
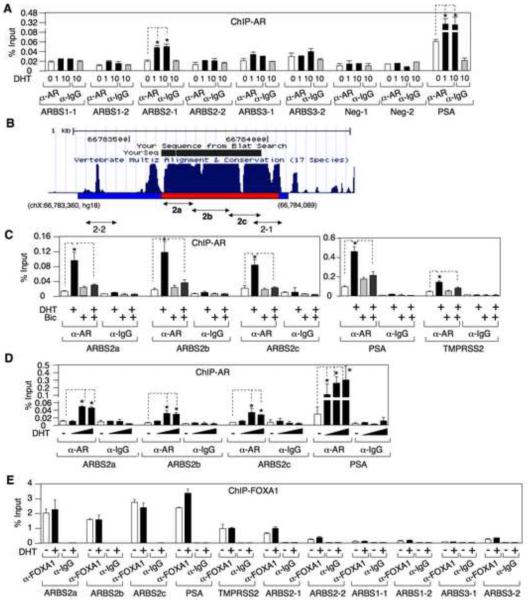
Androgen stimulates AR recruitment to a conserved site in intron 2 of the AR gene
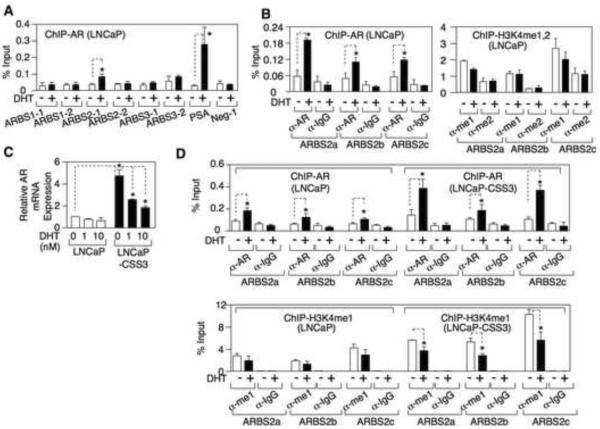
Data from a recent ChIP-chip analysis of AR binding sites (ARBSs) in LNCaP cells identified three sites linked to the AR gene, ARBS1 in the promoter region (10% FDR), ARBS2 in intron 2 (5% FDR), and ARBS3 in the 3' downstream region (5% FDR) (Wang et al., 2009) (Fig. S2A). To assess these binding sites in VCaP cells, we designed two pairs of primers for each ARBS and utilized ChIP coupled with quantitative real-time PCR to measure AR binding. Only the ARBS2 site (ARBS2-1) showed clear DHT induced AR binding, although basal and androgen induced AR binding to the well characterized major ARE upstream of the PSA gene (ARE III) were higher (Fig. 3A). As important regulatory elements may be conserved between species, we compared the human ARBS2 region to the corresponding regions in other species. Interestingly, a fragment of ARBS2 (~400 bp) that overlapped ARBS2-1 was highly conserved among species (100% identical between mouse and rat, 88% identical between mouse/rat and human) and contained multiple binding sites for FOXA1, a pioneer transcription factor that interacts with AR and is generally found at steroid responsive enhancer elements (Fig. 3B and Fig. S2B). Therefore, we synthesized an additional set of primer pairs spanning this conserved region (ARBS2a, 2b, and 2c) and repeated the AR ChIP assays. AR binding to all three sites was substantially increased by DHT and this binding was blocked by the AR antagonist bicalutamide (Fig. 3C). The DHT stimulated increase was comparable to the ~5-fold increase on the AREs in the control PSA and TMPRSS2 enhancers, but basal binding to ARBS2 was again lower (Fig. 3C). As observed on the PSA enhancer, DHT stimulated AR recruitment to ARBS2 was maximal at early times (2 hours), but still persisted after 24 hours (Fig. S2C). As noted for suppression of AR mRNA versus induction of PSA and ERG mRNA (Fig. 2), AR binding to ARBS2 required higher DHT concentrations (Fig. 3D). Finally, anti-FOXA1 ChIP showed that FOXA1 was associated constitutively with ARBS2 (Fig. 3E).
Figure.3. Androgen stimulates AR recruitment to a site in intron 2 of the AR gene.
(A) VCaP cells in steroid depleted medium (CSS mediom) were treated with 0, 1, or 10 nM DHT for 4h and the DNA bound to AR was measured by ChIP followed by qPCR. (B) The conserved region of ARBS2 (intron2) among 17 vertebrate sepcies was plotted using UCSC Genome Browser. (C) VCaP cells were pre-treated with/out 10 μM bicalutamide for 4h followed by treatment with 10 nM DHT for 4h. The DNA bound to AR was measured by ChIP followed by qPCR. (D) VCaP cells were treated for 4h with 0, 0.1, 1, or 10 nM DHT. AR binding to ARBS2 or the PSA enhancer ARE were measured by ChIP followed by qPCR. (E) VCaP cells were treated with/out 10 nM DHT for 4h and the DNA bound to FOXA1 was measured by ChIP and qPCR. Error bars in each experiment indicate SD. See also Figure S2 and see Table S1 for raw qPCR data for experiments shown.
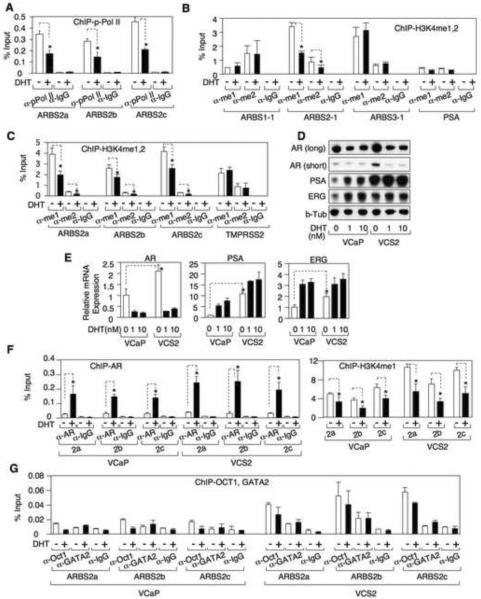
Androgen stimulates demethylation of H3K4 associated with ARBS2
Consistent with ARBS2 functioning as an enhancer, ChIP with an anti-TATA binding protein (TBP) antibody indicated that there was an interaction between this site and AR gene promoter (Fig. S3A). Significantly, we also detected a basal association between activated RNA polymerase II and ARBS2 that was decreased by DHT, suggesting that the agonist liganded AR may be mediating repression through this site (Fig. 4A). Further evidence for an interaction between the AR recruited to ARBS2 and the AR gene promoter was obtained by anti-AR ChIP followed by a chromatin conformation capture (3C) assay, which identified a DHT dependent association between AR, ARBS2, and the AR gene promoter (Fig. S3B).
Figure.4. Androgen stimulates rapid demethylation of H3K4 in VCaP and VCaP-derived VCS2 cells.
(A, B, C) VCaP cells were treated with/out DHT for 4h and the DNA bound to active RNA polymerase II, mono- or di-methylated H3K4 were measured ChIP and qPCR. (D,E) VCaP or VCS2 cells were treated with 0, 1, or 10 nM DHT for 24h and AR, PSA, ERG, and β-tubulin proteins were immunoblotted or mRNA were measured by ChIP followed by qRT-PCR (18S as internal control). (F,G) VCaP or VCS2 cells were treated with/out DHT for 4h and the DNA bound to AR, mono-methylated H3K4, Oct1, or GATA2 were measured by ChIP followed by qPCR. Error bars in each experiment indicate SD. See also Figure S3 and see Table S2 for raw qPCR data for experiments shown.
The agonist liganded AR generally stimulates transcription through recruitment of coactivator proteins and histone acetyltransferases, but can more weakly mediate recruitment of transcriptional corerpessors such as NCoR or SMRT and their associated histone deacetylases (HDACs) (Cheng et al., 2002). Therefore, we next used ChIP to determine whether DHT was directly or indirectly stimulating recruitment of an HDAC to AR binding sites in the AR gene. Interestingly, control experiments indicated that HDAC3 (which forms a complex with NCoR and SMRT) was associated with ARE III in the PSA enhancer, and that this association was decreased by DHT (Fig. S3C). There also appeared to be a very weak association of HDAC3 with each of the ChIP-chip identified AR binding sites (ARBS1, 2, and 3) in the AR gene, but these were not increased by DHT (Fig. S3C). Moreover, ChIP with antibodies against acetylated H3K9/14 did not detect decreases in histone acetylation at any of the sites in response to DHT (Fig. S3D). As a positive control, in the absence of DHT we detected high levels of histone acetylation in AR exon 1 and this decreased in response to DHT, consistent with downregulation of AR gene expression.
As interaction with the promoter and FOXA1 binding suggested that ARBS2 may function as an enhancer, we next assessed changes in histone marks that are associated with active enhancers (H3K4 mono- and dimethylation) at ARBS1, 2, and 3. Substantial H3K4 methylation was detected at each site, but there were no changes in response to DHT at ARBS1 or ARBS3, or at the ARE III site in the PSA enhancer (Fig. 4B). The TMPRSS2 enhancer ARE was similarly unaffected (Fig. 4C). In contrast, DHT caused a decrease in both H3K4me1 and H3K4me2 levels at ARBS2-1 (Fig. 4B), and this was confirmed using the set of ARBS2 primers (ARBS2a, b, and c) spanning the conserved region (Fig. 4C). Taken together, these results suggested that ARBS2 contains an enhancer that is rapidly inactivated by androgen.
VCaP xenografts that relapse after castration have higher levels of AR mRNA and renewed expression of AR-regulated genes, similarly to what is observed in patients who progress to CRPC (Cai et al., 2009). To determine whether the ARBS2 site contributes to the increased AR gene expression in these relapsed tumors, we generated a cell line (VCS2) from a relapsed VCaP xenograft tumor. VCS2 cells in steroid depleted medium had higher levels of AR, PSA, and ERG (from the androgen regulated TMPRSS2:ERG fusion gene) relative to the parental VCaP cells (Fig. 4D), and were less dependent on androgens for cell survival (Figure S3E), but AR protein was still markedly decreased by DHT. An analysis of basal (in steroid depleted medium without exogenous DHT) mRNA levels confirmed that AR, PSA, and ERG mRNA were increased in VCS2 cells compared to VCaP, and showed that AR mRNA was markedly decreased in response to DHT (Fig. 4E). AR ChIP showed that DHT stimulated recruitment of AR to ARBS2 in the VCS2 cells, with the increased binding compared to VCaP being consistent with higher AR levels in the VCS2 cells (Fig. 4F, left panel). Significantly, basal ARBS2 H3K4 methylation was increased in the VCS2 cells compared to VCaP, but was still decreased by DHT (Fig. 4F, right panel). Finally, transcription factors shown previously to interact with AR on enhancers, Oct1 and GATA-2 (Wang et al., 2007), were associated with ARBS2 and were increased in VCS2 (Fig. 4G). Overall these findings further supported the conclusion that ARBS2 contains an enhancer that contributes to increased AR gene expression at low androgen levels in CRPC, and indicated that this enhancer is repressed by the agonist liganded AR.
Androgen deprivation activates the ARBS2 site in LNCaP cells
We next examined the LNCaP PCa cell line, which shows only a small decrease in AR mRNA in response to DHT (see Fig. 2A). Anti-AR ChIP showed DHT stimulated recruitment of AR to ARBS2-1 (Fig. 5A), which was confirmed using the ARBS2a, b, and c primers (Fig. 5B, left panel). However, in contrast to VCaP cells, there was less AR binding to ARBS2 and no marked DHT stimulated decreases in H3K4me1 or me2 (Fig. 5B, right panel). Based on the results above in VCaP versus VCS2 cells, we next examined LNCaP cells that were passaged in vitro in steroid depleted medium (basal medium with 5% charcoal/dextran stripped serum, CSS). As shown in Fig. 5C, after 3 weeks in steroid depleted medium the cells expressed higher levels of AR mRNA, which markedly declined in response to DHT. AR ChIP in these LNCaP-CSS3 cells showed increased DHT stimulated AR recruitment to ARBS2 relative to the parental LNCaP cells (Fig. 5D, upper panel). Most significantly, basal H3K4 methylation of ARBS2 was increased in the LNCaP-CSS3 cells, and it declined in response to DHT (Fig. 5D, lower panel). These results in LNCaP cells further support the conclusion that ARBS2 contains an androgen repressed enhancer that contributes to increased AR gene expression in response to androgen deprivation.
Figure.5. Androgen deprivation activates the ARBS2 site in LNCaP cells.
(A) LNCaP cells were treated with/out 10nM DHT for 4h and the DNA bound to AR was immunoprecipitated and measured by qPCR. (B) LNCaP cells were treated with/out 10nM DHT for 4h and the DNA bound to AR, mono- or di-methylated H3K4 was immunoprecipitated and measured by qPCR. (C) LNCaP or LNCaP-CSS3 (adapted to steroind-depleted medium for >3w) were treated with 0, 1, or 10 nM DHT for 24h and AR mRNA was measured by qRT-PCR (18S as internal control). (D) LNCaP or LNCaP-CSS3 cells were treated with/out 10 nM DHT for 4h and the DNA bound to AR or mono-methylated H3K4 was measured by ChIP and qPCR. Error bars in each experiment indicate SD. See Table S3 for raw qPCR data for experiments shown.
Lysine specific demethylase 1 (LSD1) is recruited to ARBS2 in vitro and in vivo by the DHT liganded AR and mediates repression
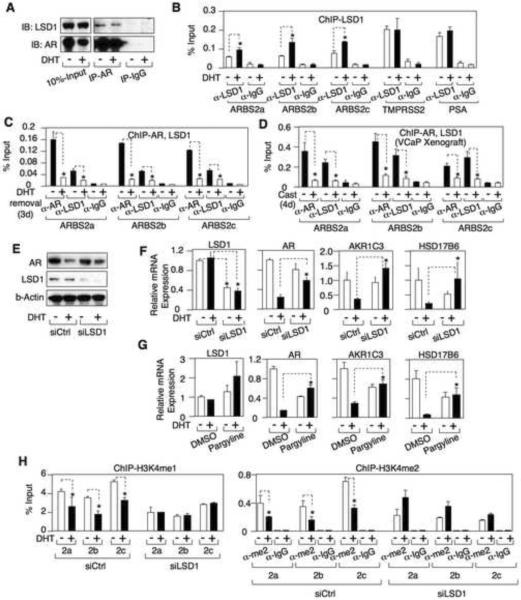
The decrease in H3K4 mono- and dimethylation over the ARBS2 site indicated that AR was either suppressing the activity of a histone methyltransferase or increasing a histone demethylase. Significantly, lysine specific demethylase 1 (LSD1) has been shown to interact with AR (Metzger et al., 2005; Wissmann et al., 2007), and we confirmed this interaction by coimmunoprecipitation of endogenous AR and LSD1 (Fig. 6A). LSD1 is reported to function as an AR coactivator on the PSA gene ARE III enhancer through demethylation of repressive mono- and dimethylated H3K9 (Metzger et al., 2005; Wissmann et al., 2007). However, mono- and dimethylated H3K4 are also substrates for LSD1, and in most contexts LSD1 appears to function as a repressor through H3K4me1 and H3K4me2 demethylation (Shi et al., 2004). Therefore, we next tested the hypothesis that DHT stimulates LSD1 recruitment to ARBS2. An association between LSD1 and ARBS2 was detected by ChIP in VCaP cells (Fig. S4A) and in VCS2 cells (Fig. 6B), and this interaction was increased by DHT. Consistent with previous reports in LNCaP cells (Metzger et al., 2005; Wissmann et al., 2007), LSD1 was constitutively associated with the ARE III in the PSA enhancer and was not clearly increased by DHT (Fig. 6B). LSD1 was similarly constitutively associated with the ARE in the TMPRSS2 enhancer (Fig. 6B). Finally, we confirmed that DHT stimulated the recruitment of LSD1 to ARBS2 in LNCaP cells, and found that LSD1 recruitment to ARBS2 was increased in the LNCaP-CSS3 cells (Fig. S4B).
Figure.6. LSD1 is recruited to ARBS2 by the DHT liganded AR in vitro and in vivo.
(A) VCaP cells were treated with/out 10 nM DHT for 24h and protein was then immunoprecipitated using anti-AR antibody or IgG control, followed by immunoblotting for LSD1 and AR. (B) VCS2 cells were treated with 0 or 10 nM DHT for 4h and the DNA bound to LSD1 was measured by ChIP and qPCR. (C) VCaP cells were grown in steroid-depleted medium supplemented with 10 nM DHT for 3d and then DHT was removed for 3d. The DNA bound to AR or LSD1 was measured by ChIP and qPCR. (D) The tissue of VCaP xenograft tumor (pre-castrated (−) or 4d post-castrated (+) mice) was formalin fixed, lysed and sonicated. The DNA bound to AR or LSD1 was immunoprecipitated and measured by qPCR. (E) VCaP cells were transfected with 20 nM LSD1 siRNA (Dharmacon) for 2d and then treated with/out DHT for 24h. AR, LSD1, and β-actin were immunoblotted. (F) VCaP cells transfected with LSD1 or control siRNA were stimulated with 10 nM DHT and LSD1, AR, AKR1C3, or HSD17B6 mRNA were measured using qRT-PCR. (G) VCaP cells were pre-treated with pargyline (2 mM) for 8h and then treated with/out DHT for 16h. LSD1, AR, AKR1C3, or HSD17B6 mRNA were measured using qRT-PCR (normalized to GAPDH as internal control). (H) VCaP cells were transfected with 20 nM LSD1 siRNA for 2d and then treated with/out 10 nM DHT for 4h. The DNA bound to mono- or di-methylated H3K4 was immunoprecipitated and measured by qPCR. Error bars in each experiment indicate SD. See also Figure S4 and see Table S4 for raw qPCR data for experiments shown.
In the converse experiment we examined VCaP cells cultured in medium with androgen that were then shifted to steroid depleted medium for 3 days. As shown in figure 6C, both AR and LSD1 binding to ARBS2 were decreased in the steroid depleted cells. We showed previously that AR mRNA levels in VCaP xenografts were markedly increased at 4 days after castration (Cai et al., 2009). To determine whether this increase in AR mRNA in vivo correlated with decreased binding of AR and LSD1 to ARBS2, we used ChIP to examine VCaP xenografts prior to castration and at 4 days post castration. As shown in figure 6D, both AR and LSD1 were associated with ARBS2 prior to castration, and these associations were markedly decreased 4 days post-castration.
LSD1 can potentially function as a coactivator or corepressor by demethylating H3K9 or H3K4, respectively, and we found that DHT also stimulated a decline in H3K9 methylation as well as H3K4 methylation across the ARBS2 site (Fig. S4C, left panel). In contrast, DHT did not cause a decrease in H3K4me3, which is associated with both promoters and enhancers but is not a substrate for LSD1 (Fig. S4C, right panel). Therefore, as these changes in methylation would be consistent with LSD1 functioning as a coactivator or corepressor, we next utilized siRNA to address directly whether LSD1 was mediating the downregulation of AR gene expression in response to DHT. Expression of LSD1 protein (Fig. 6E) and mRNA (Fig. 6F) were substantially decreased by the LSD1 siRNA, and the DHT stimulated decrease in AR protein was diminished (Fig. 6E). An analysis of AR mRNA confirmed that the DHT stimulated decrease in AR expression was blunted by LSD1 siRNA (Fig. 6F).
To determine whether this LSD1 dependent suppression was unique to the AR gene, we also examined expression of AKR1C3 and HSD17B6, which are androgen repressed and increased in CRPC. AKR1C3 catalyzes synthesis of testosterone from androstenedione and HSD17B6 oxidizes 5α-androstene-3α, 17β-diol back to DHT (Bauman et al., 2006). Similarly to AR, we reported previously that mRNA expression of AKR1C3 was consistently increased in CRPC (Stanbrough et al., 2006), and both AKR1C3 and HSD17B6 were negatively regulated by androgens in VCaP cells (Cai et al., 2009). As shown in figure 6F, the DHT stimulated declines in AKR1C3 and HSD17B6 mRNA were abrogated by the LSD1 siRNA. Similar results were obtained using a chemical inhibitor of LSD1, pargyline (Fig. 6G), which also prevented the DHT stimulated decline in AR protein (Fig. S4D). Consistent with previous data showing that LSD1 functions as a coactivator on the PSA gene (Metzger et al., 2005; Wissmann et al., 2007), pargyline also blocked the DHT stimulated increase in PSA protein (Fig. S4D).
The LSD1 siRNA did not decrease the DHT stimulated recruitment of AR to ARBS2 (Fig. S4E, left panel). However, the DHT stimulated declines in H3K9 methylation (Fig. S4E, right panel) and H3K4 methylation (Fig. 6H) across ARBS2 were impaired or abrogated by the LSD1 siRNA. Pargyline similarly impaired DHT stimulated H3K4me1 demethylation across ARBS2 (Fig. S4F). Together these data indicated that AR was mediating repression through recruitment of LSD1 and H3K4 demethylation. Finally, we used pargyline to assess whether LSD1 was mediating the DHT stimulated repression of AR gene expression in other PCa cell lines. C4-2 cells were derived from a castration resistant LNCaP xenograft and CWR22Rv1 cells were from a castration resistant CWR22 xenograft. In both cells, pargyline abrogated the DHT stimulated decrease in AR mRNA (Fig. S4G). Moreover, consistent with LSD1 functioning as an AR coactivator on androgen stimulated genes, pargyline suppressed the DHT stimulated increase in FKBP5.
Previous studies have shown that LSD1 functions as a coactivator for AR on the PSA (KLK3) and KLK2 genes due to phosphorylation of H3T6 and H3T11, which suppress LSD1 mediated H3K4 demethylation and enhance H3K9 demethylation, respectively (Metzger et al., 2008; Metzger et al., 2010). Therefore, we next used ChIP to determine whether differences in H3T6 or H3T11 phosphorylation were a basis for the distinct effects of AR and LSD1 on the AR gene versus AR stimulated genes. Significantly, DHT stimulated H3T6 and H3T11 phosphorylation were lower across ARBS2, and were also lower in the androgen suppressed OPRK1 (see Fig. 7) and AKR1C3 genes, compared to AREs in the androgen stimulated PSA, KLK2, and FKBP5 genes (Fig. S4G). However, H3T6 and H3T11 phosphoryation were also low in the strongly androgen stimulated TMPRSS2 gene. These findings are consistent with the conclusion that phosphorylation of H3T6 and H3T11 contribute to the regulation of LSD1 substrate specificity, but additional mechanisms may also contribute to this regulation.
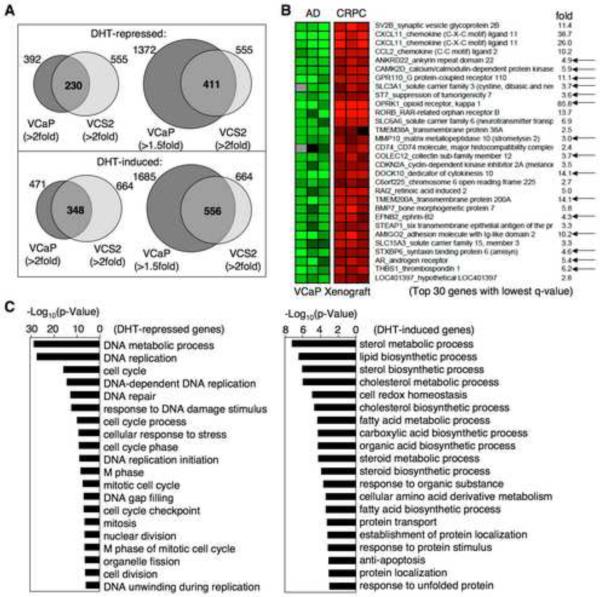
Figure.7. Identification of androgen repressed genes in VCaP cells and xenografts.
(A) VCaP or VCS2 cells were treated with/out 10 nM DHT for 24h and analyzed on Affymetrix U133A microarrays. The numbers of DHT-repressed genes or DHT-induced genes in VCaP and VCS2 cells and their overlaps are shown. (B) VCaP xenografts were established and biopsied at three stages: androgen-dependent tumor (AD), 4d post-castration (CS), and castration-resistant relapsed tumor (CRPC). mRNA were extracted from the biopsies of tumors of AD or CRPC stages and analyzed on Agilient microarrays. The data was analyzed using SAM software (Significance Analysis of Microarrays). The top 30 genes with lowest q-value are shown, with black arrows indicating DHT-repressed genes. (C) GO term analysis of DHT-repressed genes (left panel) versus androgen-induced genes (right panel). See also Figure S5 and Table S5.
Expression of androgen repressed genes is increased in CRPC xenografts
Expression microarrays were used to identify genes that were androgen repressed in both VCaP and VCS2 cells in vitro, and to then assess the expression of these genes in vivo in androgen dependent versus relapsed castration resistant VCaP xenografts. AR, AKR1C3 and HSD17B6 were again found to be androgen repressed in VCaP (4.2, 2.8, and 3.7-fold higher in the absence of androgen, respectively) and were even more highly androgen repressed in VCS2 cells (6.4, 8.5, and 4.7-fold, respectively) (Table S5). In contrast, expression of these genes was highly upregulated in the relapsed VCaP xenografts (5.4, 2.3, and 3.5-fold for AR, AKR1C3, and HSD17B6, respectively). These findings, in conjunction with the low intratumoral androgen levels in these castration resistant tumors (Fig. S5A), support a feedback mechanism that negatively regulates AR signaling at high androgen levels and enhances signaling at the lower androgen levels.
To more systematically assess the significance of additional in vitro identified androgen repressed genes, we next focused on the 411 genes that were repressed by >2 fold in VCS2 and >1.5 fold in VCaP (the lower threshold in VCaP being based on the more robust repression of AR, AKR1C3, and HSD17B6 in VCS2 cells) (Fig. 7A and Table S5). Remarkably, amongst the top 30 genes with most significantly elevated expression in the castration resistant VCaP xenografts, 12 were in this group of 411 androgen-repressed genes (Fig. 7B). In addition, further genes amongst this group of 30 that appeared to be androgen-repressed were ANKRD22 (1.64-fold in VCaP, 1.82-fold in VCS2), MMP10 (1.32-fold in VCaP, 4.2-fold in VCS2), and STXBP6 (1.60-fold in VCaP, 1.93-fold in VCS2).
We next took advantage of recent AR ChIP-seq data in VCaP cells (Yu et al., 2010) to assess the frequency of AR binding sites in androgen repressed versus androgen activated genes in VCaP cells. AR binding sites were found in 20% of AR-activated genes and in 14% of AR repressed genes, with the background being 11% (fraction of total 31,810 genes that contain AR binding sites), indicating that there is enrichment for AR binding sites within the AR repressed genes (Fig. S5B). The lower enrichment versus the AR activated genes could mean that more genes in the AR activated group are directly regulated by AR, but could also be in part technical and reflect somewhat weaker binding of AR to AR repressed genes. To further assess whether suppression of these genes was mediated directly by AR through an LSD1 dependent mechanism, we focused on another androgen repressed gene (OPRK1) that was strongly upregulated in the VCaP CRPC xenografts. Using real time RT-PCR, we first confirmed that DHT markedly decreases OPRK1 mRNA in VCaP cells, similarly to the decreases in AR, AKR1C3, and HSD17B6 (Fig. S5C). Using AR siRNA we also showed that AR downregulation could blunt the DHT mediated repression of these genes, providing further evidence that the repression was AR mediated (Fig. S5C). The AR siRNA also decreased basal, but not DHT stimulated PSA or TMPRSS2 expression, consistent with AR functioning more efficiently on AR stimulated genes. OPRK1 has a single AR binding site in its 3' UTR based on ChIP-chip and ChIP-seq data in both LNCaP and VCaP cells (Wang et al., 2009, Yu et al., 2010) (Fig. S5D). Therefore, we used ChIP with primers covering this site to assess AR and LSD1 binding. Significantly, DHT stimulated AR and LSD1 recruitment to this site, and also decreased H3K4 methylation (Fig. S5E). Together these data indicate that AR is directly negatively regulating a set of genes that are up-regulated in the VCaP CRPC xenografts.
To assess the potential functional consequences of failing to suppress androgen repressed genes after castration, we determined the pathways that were associated with the 411 androgen-repressed genes identified in VCaP and VCS2 cells. Importantly, expression of these genes was most significantly associated with increased DNA replication and cell cycle progression (Fig. 7C, left panel), while genes that were increased in response to DHT in VCaP and VCS2 cells were associated with synthesis of lipids, proteins, and other metabolic processes distinct from DNA replication (Fig. 7C, right panel). Finally, we treated VCaP CRPC xenografts with testosterone to assess effects on AR repressed genes in vivo, and found by RT-PCR that AR, AKR1C3, HSD17B6, and OPRK1 were repressed (Fig. S5F). Testosterone also suppressed expression of BCL11A, another strongly AR repressed gene that was increased in castration resistant VCaP xenografts, but did not clearly suppress PSA or TMPRSS2. Moreover, there was marked regression in the xenografts (Fig. S5G). These findings indicated that a partial restoration of androgen levels and AR transcriptional activity in CRPC cells may drive tumor growth by activating cellular metabolism while failing to suppress DNA replication and proliferation.
Increased expression of androgen repressed genes in CRPC patients
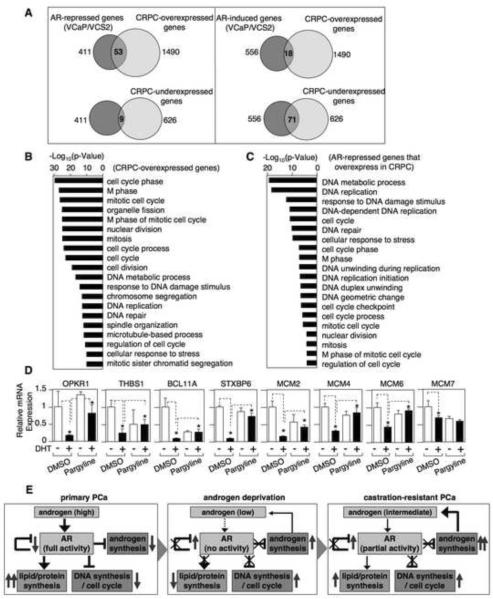
To determine whether increased expression of androgen repressed genes may contribute to CRPC in patients, we used expression data from a set of CRPC bone marrow metastases versus primary prostate cancers that had not received hormonal therapy (Stanbrough et al., 2006; Mendiratta et al., 2009). Consistent with lower androgen levels and reduced AR transcriptional activity in CRPC, only a small fraction of the genes that were androgen induced in VCaP/VCS2 were overexpressed in CRPC (18/556), while a much larger fraction were underexpressed (71/556) (Fig. 8A). Similarly, very few of the AR repressed genes were underexpressed in CRPC (9/411), while many more were overexpressed (53/411) (Table S6). As noted previously, genes that are overexpressed in CRPC are highly associated with proliferation (Stanbrough et al., 2006; Wang et al., 2009) (Fig. 8B), while genes that are underexpressed are more associated with developmental pathways (Fig. S6A). Significantly, the set of 53 androgen repressed genes that were overexpressed in the CRPC biopsy samples were similarly highly associated with DNA replication and proliferation (Fig. 8C).
Figure.8. Expression of androgen repressed genes is increased in human CRPC samples.
(A) Affymetrix microarray expression data showing overlaps between androgen repressed/induced genes and the expression of 1490 genes that were increased and 626 genes that were decreased (p<0.001 and fold-change >1.5) in 34 CRPC bone marrow metastases compared with 27 primary tumors prior to any hormonal therapy. (B) GO term analysis of the group of 1490 CRPC-overexpressed genes and (C) 53 AR-repressed genes that were overexpressed in CRPC. (D) VCaP cells were pre-treated with pargyline (2 mM) for 8h and then treated with/out DHT for 16h. OPKR1, THBS1, BCL11A, STXBP6, MCM2, MCM4, MCM6, or MCM7 mRNA were measured using qRT-PCR (normalized to GAPDH as internal control). Error bars in each experiment indicate SD. (E) Graphical summary showing divergent effects of androgen deprivation on expression of AR stimulated genes, which are decreased, versus AR repressed genes (including the AR gene), which are increased. In castration resistant PCa, mechanisms including further increases in intratumoral androgen synthesis result in partial restoration of AR transcriptional activation function on genes mediating lipid and protein biosynthesis, but do not restore AR repressor function on the AR gene, or on genes mediating androgen synthesis, DNA synthesis and cell cycle progression. See also Figure S6 and Table S6.
To further assess the biological importance of these 53 androgen repressed genes in CRPC, we removed them from the set of 1490 genes that were overexpressed in the CRPC biopsy samples and repeated the Gene Ontology analysis on the remaining 1437 genes. While these 1437 genes were still associated with cell cycle progression and DNA metabolism, the significance of all these associations was markedly decreased, and DNA replication was no longer amongst the most highly associated pathways in the absence of these 53 androgen repressed genes (Fig. S6B). Finally, we selected for further analysis a set of 8 genes that were androgen repressed in VCaP/VCS2 cells and were also overexpressed in the relapsed VCaP xenografts or the clinical CRPC biopsies. Quantitative real time RT-PCR confirmed that they were all DHT repressed in VCaP and VCS2 cells, and that this could be prevented with bicalutamide (Fig. S6C). Moreover, in all cases the androgen stimulated downregulation was decreased or abrogated by treatment with pargyline, indicating that it was mediated by LSD1 (Fig. 8D). Together these findings elucidate a mechanism by which loss of negative regulation by the agonist liganded AR, in association with LSD1, increases the expression of AR and of multiple genes that contribute to increased androgen synthesis, DNA replication and proliferation in CRPC.
DISCUSSION
Studies in clinical samples and xenograft models indicate that increased AR gene expression plays a major role in the progression to CRPC. We observed previously in VCaP cells in vitro and in VCaP xenografts in vivo that AR mRNA levels decline rapidly in response to androgen stimulation and increase rapidly in response to androgen withdrawal (Cai et al., 2009). In this report we have identified a highly conserved site in the second intron of the AR gene that regulates its expression in response to androgen stimulation and withdrawal. RNA polymerase II and FOXA1 are associated with this ARBS2 site, as are OCT1, GATA2 and substantial levels of H3K4 mono- and dimethylation that are further increased in cells adapted to androgen deprivation, consistent with this element functioning as an enhancer that contributes to increased AR gene expression in CRPC. Moreover, we show that the agonist liganded AR decreases AR gene expression by functioning as a transcriptional repressor at this site through recruitment of LSD1 and demethylation of H3K4me1,2. The rapid androgen mediated downregulation of AKR1C3 and HSD17B6 is similarly LSD1 dependent, indicating that the agonist liganded AR directly mediates a physiological intracellular negative feedback loop to regulate AR activity. Taken together, these findings elucidate a mechanism that contributes to increased AR gene expression and restored AR activity in CRPC, and identify a suppressor element and transcriptional repressor function for the agonist liganded AR.
Further analysis of gene expression in androgen starved versus androgen stimulated VCaP and VCS2 cells showed that the agonist liganded AR also suppressed the expression of multiple genes mediating DNA synthesis and cell cycle progression, while it increased the expression of genes mediating synthesis of lipids, amino acids, and other metabolic processes. This profile is consistent with AR function in normal prostate epithelium to drive terminal differentiation and synthesis of seminal fluid, and provides a molecular basis for the biphasic response to androgen stimulation whereby PCa cells proliferate in response to low levels of androgen but are growth arrested at high concentrations (Xu et al., 2006). Significantly, a set of these androgen repressed genes associated with increased DNA synthesis and proliferation were overexpressed in vivo in castration resistant VCaP xenografts and in CRPC patient samples. We suggest that androgen levels in CRPC cells are adequate to stimulate AR activity on enhancer elements of genes mediating certain critical metabolic functions such as lipid synthesis, which are sensitive to lower levels of androgens, but are not adequate to effectively recruit AR and LSD1 to suppressor elements in multiple genes that negatively regulate AR signaling and cellular proliferation. A graphical summary showing divergent effects of AR on expression of AR stimulated versus AR repressed genes after androgen deprivation and in CRPC is shown in figure 8E.
LSD1 was initially identified in corepressor complexes and shown to function by demethylating mono- and dimethylated H3K4 (Shi et al., 2004). However, it was subsequently shown to function as a coactivator through demethylation of repressive mono- and dimethylated H3K9 when associated with AR and possibly other nuclear receptors including estrogen receptor α (Metzger et al., 2005, Garcia-Bassets et al., 2007, Perillo et al., 2008). The results of this study indicate that the association with AR does not determine the coactivator versus corepressor function of LSD1, and that it is instead determined by properties of the element to which it is being recruited. For example, hypoacetylated nucleosomes are more susceptible substrates for LSD1 mediated demethylation (Shi et al., 2005). Moreover, recent data indicate that phosphorylation of H3T11 by an AR associated kinase (PRK1/PKN1) enhances the demethylation of H3K9me3 by JMJD2C and subsequent demethylation of H3K9me1,2 by LSD1 (Metzger et al., 2008), while phosphorylation of H3T6 by a distinct kinase (PKCβ1) can suppress the LSD1 mediated demethylation of H3K4me1,2 (Metzger et al., 2010). Our data indicate that lower H3T6 and H3T11 phosphorylation may contribute to the substrate specificity and corepressor function of LSD1 at AR repressed genes, although LSD1 may be regulated by a distinct mechanism on the TMPRSS2 gene. It will clearly be important to further characterize these and additional AR suppressor elements and determine the extent to which histone modifications or other factors regulate the function of AR and LSD1 on these suppressor versus AR enhancer elements.
It has been well appreciated for many years that AR has both growth promoting and growth suppressing activities, and that androgen deprivation therapies may directly or indirectly stimulate some pathways that contribute to growth and eventual relapse. Indeed, androgens can suppress the growth of some CRPC derived cell lines, and high-dose androgens have been explored as a therapy for CRPC (Umekita et al., 1996, Morris et al., 2009). However, the molecular basis for androgen stimulated growth suppression has not been clear, and there have been no previous studies suggesting that distinct AR transcriptional mechanisms may underlie these functions. Therefore, the results of this study provide a paradigm with implications for both basic molecular mechanisms of steroid action and for AR targeted therapy of prostate cancer. In particular, the distinct mechanisms of AR action on enhancer versus suppressor elements may make it possible to selectively augment AR transcriptional repressor function and thereby prevent or delay the emergence of CRPC.
EXPERIMENTAL PROCEDURES
Cell culture and xenografts
LNCaP or C4-2 cells were cultured in RPMI1640 medium with 10% FBS. VCaP cells were cultured in DMEM medium with 10% FBS and VCS2 cells were cultured in DMEM medium with 8% charcoal/dextran-stripped FBS (CSS) plus 2% FBS. For most immunoblotting, RT-PCR or ChIP assays, cells were grown to 50–60% confluence in 5% (CSS) medium for 3 days and then treated with androgens or drugs. VCaP xenografts were established in the flanks of male scid mice by injecting ~2 million cells in 50% Matrigel. When the tumors reached ~1 cm, biopsies were obtained and then the mice were castrated. Additional biopsies were obtained 4 days after castration, and the tumors were harvested at relapse. Frozen sections were examined to confirm that the samples used for RNA and protein extraction contained predominantly non-necrotic tumor. All animal experiments were approved by the Beth Israel Deaconess Institutional Animal Care and Use Committee and were performed in accordance with institutional and national guidelines.
RT-PCR and immunoblotting
Quantitative real-time RT-PCR amplification was carried out on RNA extracted from tissue samples or cell lines using TRIZOL reagent. 50ng RNA was used for each reaction and the result was normalized by co-amplification of 18S RNA. Reactions were performed on an ABI Prism 7700 Sequence Detection System using Taqman one-step RT-PCR reagents. Primers and probes are listed in supplementary information. PCR data are represented as mean ± STD for repeats. Protein extracts were prepared by boiling for 15 min in 2% SDS. Blots were incubated with anti-PSA (1:3000, polyclonal, BioDesign), anti-AR (1:2000, polyclonal, Upstate), anti-LSD1 (1:1000, Abcam), anti-β-actin (1:5000, monoclonal, Abcom), or anti-β-tubulin (1:2000, Upstate), and then with 1:5000 anti-rabbit or anti-mouse secondary antibodies (Promega).
Co-immunoprecipitation
VCaP cells were harvested in Triton lysis buffer (0.5% Triton X-100, 20 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, and 2 mM dithiothreitol) with protease inhibitors. The protein was immunoprecipitated using monoclonal anti-AR (AR441 from NeoMarkers) or mouse IgG control and then subjected to immunoblotting.
Chromatin-immunoprecipitation (ChIP) assay
Cells were formalin fixed, lysed and sonicated to break the chromatin into 500–800 bp fragments. Anti-AR (Santa Cruz), anti-FOXA1 (Abcam), anti-OCT1 (Santa Cruz), anti-GATA2 (Santa Cruz), anti-RNA Polymerase II (Santa Cruz), anti-RNA Polymerase II CTD repeat (phospho Ser5), anti-TBP (Santa Cruz), anti-LSD1 (Abcam), anti-HDAC1 (Santa Cruz), anti-HDAC2 (Santa Cruz), anti-HDAC3 (Santa Cruz), anti-H3K4me1 (Abcam), anti-H3K4me2 (Upstate), anti-H3K4me3 (Abcam), anti-H3K9me1 (Abcam), anti-H3K9/14ace (Upstate), anti-H3T6pho (Abcam), anti-H3T11pho (Abcam), or rabbit IgG (Santa Cruz) were used to precipitate chromatin fragments from cell extracts. Quantitative real time PCR was used to analyze binding to the ARBS-1, -2, -3, PSA enhancer (ARE3), TMPRSS2 enhancer (−14k upstream), OPRK1 enhancer (3'-UTR), or negative-1 (3' irrelevant region of PSA), -2 (irrelevant region of chromosome 18). The primers are listed in the supplementary information. We used real time quantitative PCR (SYBR green) to amplify the DNA fragment in the antibody precipitated DNA and the un-precipitated input DNA to calculate ΔCT values. The RQ values (RQ=2−ΔCT) are presented and reflect the precipitated DNA as a percentage of the input DNA. Results are represented as mean ± STD for replicate samples. Data are representative of at least three experiments. Significant differences are indicated (*) in the experiments. Raw dat afo rthe real time quantitative PCR are provided in Tables S1–S4.
Gene expression microarray assay
VCaP or VCS2 cells treated with ethanol or 10nM DHT were subjected to microarray assay (Affymetrix) to identify genes whose expression was repressed by DHT in both VCaP and VCS2 cells. Tissue mRNA was extracted and purified from three sets (pre-castrated, 4d-post-castrated, and relapsed) of xenograft tumors (3 mice) and then subjected to microarray assay (Agilent). SAM software was used to perform t-test on these three biological repeats (three mice) to determine the score and q-value. The genes whose expression was significantly elevated in relapsed tumors (q<0.05) were picked for the next screening to determine if they were DHT-repressed in VCaP and VCS2.
Supplementary Material
HIGHLIGHTS
AR gene expression in CRPC is increased by an enhancer in the second intron.
This enhancer is repressed by AR at high androgen levels through LSD1 recruitment.
AR represses genes mediating androgen synthesis, DNA synthesis, and cell cycling.
Decreased androgen in CRPC relieves repression but supports AR dependent growth.
SIGNIFICANCE.
This study shows that AR can function through a suppressor element to repress its own expression and the expression of additional genes including those that mediate androgen synthesis. This negative feedback loop suppresses AR signaling at high androgen levels, but allows increased AR and androgen synthesis in CRPC. Moreover, decreased androgen levels in CRPC, while adequate to stimulate AR on enhancer elements, may relieve AR suppression of genes mediating DNA synthesis/proliferation and thereby contribute to tumor growth. Distinct mechanisms of AR action on enhancer versus suppressor elements may make it possible to selectively augment AR transcriptional repressor function and thereby prevent or delay emergence of CRPC.
ACKNOWLEDGMENTS
We thank J. He for analyzing three AR binding sites among species. We thank E.A. Mostaghel, B. Marck and A.M. Matsumoto for measuring intratumoral level of steroids. This work was supported by grants to S.P.B. from the NIH (R01 CA111803 and Prostate SPORE P50 CA090381), DOD Prostate Cancer Research Program (PC060807), and a Challenge Grant from the Prostate Cancer Foundation. C.C. was supported by a fellowship from the DOD Prostate Cancer Research Program (PC08115) and a Career Development Award from the Prostate SPORE. H.W. was supported by a fellowship from DOD Prostate Cancer Research Program. S.C. was supported by an NIH K99/R00 award. P.N.W. was supported by grants from NIH (Prostate SPORE P50 CA097186 and RC1 CA146849) and from the DOD Prostate Cancer Research Program (PC093509).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS The expression microarray data has been deposited in the Gene Expression Omnibus (GEO) database, available at www.ncbi.nlm.nih.gov/geo, under accession GSE31410.
REFERENCES
- Bauman DR, Steckelbroeck S, Williams MV, Peehl DM, Penning TM. Identification of the major oxidative 3alpha-hydroxysteroid dehydrogenase in human prostate that converts 5alpha-androstane-3alpha, 17beta-diol to 5alpha-dihydrotestosterone: a potential therapeutic target for androgen-dependent disease. Mol. Endocrinol. 2006;20:444–458. doi: 10.1210/me.2005-0287. [DOI] [PubMed] [Google Scholar]
- Blok LJ, Themmen AP, Peters AH, Trapman J, Baarends WM, Hoogerbrugge JW, Grootegoed JA. Transcriptional regulation of androgen receptor gene expression in Sertoli cells and other cell types. Mol. Cell Endocrinol. 1992;88:153–164. doi: 10.1016/0303-7207(92)90020-7. [DOI] [PubMed] [Google Scholar]
- Cai C, Wang H, Xu Y, Chen S, Balk SP. Reactivation of androgen receptor-regulated TMPRSS2:ERG gene expression in castration-resistant prostate cancer. Cancer Res. 2009;69:6027–6032. doi: 10.1158/0008-5472.CAN-09-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- Cheng S, Brzostek S, Lee SR, Hollenberg AN, Balk SP. Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol. Endocrinol. 2002;16:1492–1501. doi: 10.1210/mend.16.7.0870. [DOI] [PubMed] [Google Scholar]
- Garcia-Bassets I, Kwon YS, Telese F, Prefontaine CG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, Fu XD, Rosenfeld MG. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory CW, Johnson RT, Jr., Mohler JL, French FS, Wilson EM. Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Res. 2001;61:2892–2898. [PubMed] [Google Scholar]
- Holzbeierlein J, Lal P, LaTulippe E, Smith A, Satagopan J, Zhang L, Ryan C, Smith S, Scher H, Scardino P, Reuter V, Gerald WL. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am. J. Pathol. 2004;164:217–227. doi: 10.1016/S0002-9440(10)63112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemppainen JA, Lane MV, Sar M, Wilson EM. Androgen receptor phosphorylation, turnover, nuclear transport, and transcriptional activation. Specificity for steroids and antihormones. J. Biol. Chem. 1992;267:968–974. [PubMed] [Google Scholar]
- Korenchuk S, Lehr JE, MClean L, Lee YG, Whitney S, Vessella R, Lin DL, Pienta KJ. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001;15:163–168. [PubMed] [Google Scholar]
- Krongrad A, Wilson CM, Wilson JD, Allman DR, McPhaul MJ. Androgen increases androgen receptor protein while decreasing receptor mRNA in LNCaP cells. Mol. Cell Endocrinol. 1991;76:79–88. doi: 10.1016/0303-7207(91)90262-q. [DOI] [PubMed] [Google Scholar]
- Kumar MV, Jones EA, Grossmann ME, Blexrud MD, Tindall DJ. Identification and characterization of a suppressor element in the 5'-flanking region of the mouse androgen receptor gene. Nucleic Acids Res. 1994;22:3693–3698. doi: 10.1093/nar/22.18.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loberg RD, John LN, Day LL, Neeley CK, Pienta KJ. Development of the VCaP androgen-independent model of prostate cancer. Urol. Oncol. 2006;24:161–168. doi: 10.1016/j.urolonc.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiratta P, Mostaghel E, Guinney J, Tewari AK, Porrello A, Barry WT, Nelson PS, Febbo PG. Genomic strategy for targeting therapy in castration-resistant prostate cancer. J. Clin. Oncol. 2009;27:2022–2029. doi: 10.1200/JCO.2008.17.2882. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Metzger E, Yin N, Wissmann M, Kunowska N, Fischer K, Friedrichs N, Patnaik N, Higgins JM, Potier N, Scheidtmann KH, Buettner R, Schule R. Phosphorylation of histone H3 at threonine 11 establishes a noval chromatin mark for transcriptional regulation. Nat. Cell Biol. 2008;10:53–60. doi: 10.1038/ncb1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Imhof A, Patel D, Hoffmeyer K, Friedrichs N, Muller JM, Greschik H, Kirfel J, Ji S, Kunowska N, Beisenherz-Huss C, Gunther T, Buettner R, Schule R. Phosphorylation of histone H3T6 by PKCbeta1 controls demethylation at histone H3K4. Nature. 2010;464:792–796. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- Morris MJ, Huang D, Kelly WK, Slovin SF, Stephenson RD, Eicher C, Delacruz A, Curley T, Schwartz LH, Scher HI. Phase 1 trial of high-dose exogenous testosterone in patients with castration-resistant metastatic prostate cancer. Eur Urol. 2009;56:237–244. doi: 10.1016/j.eururo.2009.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- Quarmby VE, Yarbrough WG, Lubahn DB, French FS, Wilson EM. Autologous down-regulation of androgen receptor messenger ribonucleic acid. Mol. Endocrinol. 1990;4:22–28. doi: 10.1210/mend-4-1-22. [DOI] [PubMed] [Google Scholar]
- Ruizeveld de Winter JA, Janssen PJ, Sleddens HM, Verleun-Mooijman MC, Trapman J, Brinkmann AO, Santerse AB, Schroder FH, van der Kwast TH. Androgen receptor status in localized and locally progressive hormone refractory human prostate cancer. Am. J. Pathol. 1994;144:735–746. [PMC free article] [PubMed] [Google Scholar]
- Shan LX, Rodriguez MC, Janne OA. Regulation of androgen receptor protein and mRNA concentrations by androgens in rat ventral prostate and seminal vesicles and in human hepatoma cells. Mol. Endocrinol. 1990;4:1636–1646. doi: 10.1210/mend-4-11-1636. [DOI] [PubMed] [Google Scholar]
- Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK, Gomella L, Knudsen ES, Nelson PS, Knudsen KE. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J. Clin. Invest. 2010;120:4478–92. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, Febbo PG, Balk SP. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, Keer HN, Balk SP. Mutation of the androgen-receptor gene in metastatic androgen- independent prostate cancer. N. Engl. J. Med. 1995;332:1393–1398. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- Umekita Y, Hiipakka RA, Kokontis JM, Liao S. Human prostate tumor growth in athymic mice: inhibition by androgens and stimulation by finasteride. Proc. Natl. Acad. Sci. U S A. 1996;93:11802–11807. doi: 10.1073/pnas.93.21.11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi OP. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat. Genet. 1995;9:401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- Wang LG, Johnson EM, Kinoshita Y, Babb JS, Buckley MT, Liebes LF, Melamed J, Liu XM, Kurek R, Ossowski L, Ferrari AC. Androgen receptor overexpression in prostate cancer linked to Pur alpha loss from a novel repressor complex. Cancer Res. 2008;68:2678–2688. doi: 10.1158/0008-5472.CAN-07-6017. [DOI] [PubMed] [Google Scholar]
- Wang LG, Ossowski L, Ferrari AC. Androgen receptor level controlled by a suppressor complex lost in an androgen-independent prostate cancer cell line. Oncogene. 2004;23:5175–5184. doi: 10.1038/sj.onc.1207654. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu S, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors govers androgen receptor-dependent prostate cancer growth. Mol. Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Janne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM, Rubin MA, True L, Fiorentino M, Fiore C, Loda M, Kantoff PW, Liu XS, Brown M. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, Metzger E, Schule R. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- Xu , Chen , Ross KN, Balk SP. Androgens Induce Prostate Cancer Cell Proliferation through Mammalian Target of Rapamycin Activation and Post-transcriptional Increases in Cyclin D Proteins. Cancer Res. 2006;66:7783–7702. doi: 10.1158/0008-5472.CAN-05-4472. [DOI] [PubMed] [Google Scholar]
- Yeap BB, Voon DC, Vivian JP, McCulloch RK, Thomson AM, Giles KM, Czyzyk-Krzeska MF, Furneaux H, Wilce MC, Wilce JA, Leedman PJ. Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3'-untranslated region of the androgen receptor messenger RNA. J. Biol. Chem. 2002;277:27183–27192. doi: 10.1074/jbc.M202883200. [DOI] [PubMed] [Google Scholar]
- Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, Gong Y, Cheng H, Laxman B, Vellaichamy A, Shankar S, Li Y, Dhanasekaran SM, Morey R, Barrette T, Lonigro RJ, Tomlins SA, Varambally S, Qin ZS, Chinnaiyan AM. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Balk SP. Mechanisms mediating androgen receptor reactivation after castration. Urol. Oncol. 2009;27:36–41. doi: 10.1016/j.urolonc.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.