Abstract
Adeno-associated viral (AAV) vectors preferentially integrate into the genome of cells that are defective in DNA repair, such as occurs with DNA-PKcs deficiency or poly(ADP-ribose) polymerase-1 down-regulation. As the tumor suppressor protein p53 regulates the transcription of many genes involved in DNA repair, we sought to determine whether functional p53 affects the efficiency of AAV integration. p53 is mutated in more than 50% of cancers, and site-specific integration of AAV into the AAVS1 site of human chromosome 19 has frequently been observed in transformed cancer cell lines, but rarely seen in primary cells or in vivo. We therefore hypothesized that p53-negative cells would be more permissive to AAV integration than p53-positive cells. The integration of a rep- and green fluorescent protein–encoding recombinant AAV vector was quantified in p53-expressing and p53-deficient HCT116 colon cancer cells. Our results show that there is a higher efficiency of AAV integration in p53-negative cells compared with p53-positive cells, indicating that p53 does indeed inhibit AAV integration. Further experiments suggest that this p53-mediated block to AAV integration is likely to be through binding of p53 to the AAV Rep protein and the consequent inhibition of Rep activity during AAV integration.
As the tumor suppressor p53 is known to regulate transcription of many DNA repair genes, Zacharias and colleagues sought to determine whether functional p53 affects AAV integration. They show AAV integration efficiency is higher in p53-deficient colon cancer cells, and suggest this inhibitory role for p53 in AAV integration likely occurs through p53 binding to the AAV Rep protein.
Introduction
Adeno-associated virus (AAV) is a nonpathogenic human parvovirus with a 4.7-kb genome (Berns and Linden, 1995). The AAV genome contains two open reading frames, rep and cap, which are flanked by two inverted terminal repeats (Srivastava et al., 1983; Srivastava, 1987). AAV2 has been considered unique among other human viruses due to its ability to site-specifically integrate into the human genome. AAV-targeted integration occurs at the AAVS1 site, and this event is mediated by the viral Rep protein (Kotin et al., 1990; Samulski et al., 1991). The AAVS1 site is located on chromosome 19 at position q13.4, within the first exon of the myosin-binding protein subunit p85 gene (Tan et al., 2001). AAV site-specific integration is initiated by Rep proteins binding to the Rep-binding elements situated both in the AAV genome and at the AAVS1 site (Giraud et al., 1994; Weitzman et al., 1994; Chiorini et al., 1995). The endonuclease activity of Rep then mediates a nicking event at the terminal resolution site within the AAVS1 (Linden et al., 1996b; Young and Samulski, 2001). Finally, the Rep proteins that are bound to the AAV DNA and to the AAVS1 site interact, and a nonhomologous recombination event occurs resulting in integration of the AAV genome (Walsh et al., 1992; Urcelay et al., 1995; Linden et al., 1996a; Xiao et al., 1996; Surosky et al., 1997; Yang et al., 1997; Dyall and Berns, 1998; Lamartina et al., 1998; Dyall et al., 1999; Young et al., 2000).
Numerous studies have observed AAV site-specific integration in transformed cell lines, but this event has rarely been shown in vivo or in primary cells (Kotin et al., 1990; Halbert et al., 1995; Linden et al., 1996a; Schnepp et al., 2005). We speculated that the transformed nature of cell lines could render them more permissive to AAV site-specific integration due to the lack of functional DNA repair machinery. In primary cells, it is possible that endogenous DNA repair pathways compete with Rep-mediated integration events, thus blocking AAV site-specific integration. In fact, previous studies have shown that cellular proteins involved in DNA repair, such as DNA-PKcs, Ku70, and poly(ADP-ribose) polymerase-1 (PARP1), inhibit AAV integration (Song et al., 2004; Fattah et al., 2008; Romanova, unpublished observations).
The cellular tumor suppressor p53 regulates the transcription of many different genes, including those involved in DNA repair (Riley et al., 2008). The p53 protein is encoded by the TP53 gene, which is frequently mutated or inactivated in human cancer and transformed cell lines. A lack of functional p53 then leads to inadequate DNA repair mechanisms within the cell. Furthermore, AAV Rep and p53 have been shown to interact during the lytic phase of the AAV life cycle, during which AAV integration does not occur (Batchu et al., 1999). We hypothesized that functional p53 would block Rep-mediated AAV integration, including targeted insertion into the AAVS1 site of human chromosome 19.
Here we show that AAV vector integration is significantly more efficient in the absence of p53 than in its presence. Southern blot experiments then reveal that site-specific integration of a Rep-expressing AAV vector into the AAVS1 site occurs more frequently in p53-negative cells than in cells with functional p53. Further analysis indicates that Rep does not increase either the total level of p53 expression or the level of activated p53 within a cell. Therefore, the p53-mediated block to Rep-induced integration is most likely due to p53 associating with Rep and inhibiting its function. Results from this study suggest that AAV Rep-mediated integration could be used as a tool to specifically target p53-negative transformed cells as part of a therapeutic regimen for treatment of malignant disease.
Materials and Methods
Plasmid
AAV plasmid pRepGFP was constructed as previously described (Philpott et al., 2002a). The AAV packaging construct pDG is well-established for AAV vector production (Grimm et al., 1998). Plasmid construct pcDNA-Rep was generated by PCR amplifying the AAV Rep gene using primers 5’-ctggctaggcccaccatgccggggttttacgagattgtg-3’ and 5’-cgcctcgag ttattgttcaaagatgcagt-3’ and cloning into pcDNA3.1(+) (Invitrogen, Carlsbad, CA). The plasmid peGFP-N1 was used as a control plasmid (Clontech, Mountain View, CA).
Cell lines
HeLa and HEK 293T cells were cultured in Dulbecco's modified Eagle's medium. p53+/+ and p53–/– HCT116 cells were obtained from Bert Vogelstein (Bunz et al., 1998) and were maintained in McCoy's 5A medium. All cells were supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Western blotting
Proteins were separated by SDS-PAGE and transferred onto an Immobilon P membrane (Millipore, Billerica, MA). The membrane was incubated in a blocking solution containing 5% skim milk and 0.05% Tween 20 in PBS. Incubation with primary antibodies was 1 hr or overnight. The following antibodies were used: mouse anti-p53 at a 1:1,000 dilution and rabbit anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH) at a 1:2,500 dilution (Santa Cruz Biotechnology, Santa Cruz, CA). After incubation with primary antibodies, membrane was washed and incubated with the corresponding horseradish peroxidase–conjugated secondary antibody of the appropriate species at a 1:1,000 dilution (Santa Cruz Biotechnology) for 1 hr. Chemiluminescence signal was detected using either the standard Chemiluminescent Western Blot substrate or SuperSignal West Dura substrate (ThermoFisher Scientific, Rockford, IL).
AAV vector production and titration
The preparation of recombinant AAV (rAAV) vector involved the cotransfection of 293T cells with pDG and pAAV-Rep-GFP plasmids. At 2 days post transfection, cells and media were collected and the cells disrupted by three cycles of freeze-thawing. Cellular DNA and RNA were degraded with benzonase digestion, and cell debris was pelleted. AAV2 virions were purified from the supernatant by binding to a heparin column; the column was washed and virions were eluted in a high-salt buffer. Salt was removed and the vector sample concentrated by centrifugation through Centricon protein concentrators. Viral DNA of the rAAV vector preparations was quantified by dot blot using the NEBlot Phototope Kit and Phototope-Star Detection Kit (New England BioLabs, Ipswich, MA).
Integration assays
Our standard protocol to accurately quantify the rate of AAV integration involved the following: HCT116 cells were infected with rAAV-Rep-GFP, and at 2 days post infection, the cells were sorted by flow cytometry to isolate green fluorescent protein (GFP)–expressing transduced cells. The cells were immediately plated into 96-well plates with 1 cell per well, and clonal cell lines were generated. After expansion of the clonal cell lines, genomic DNA was harvested from these clones at 1 month post infection using a standard salting-out protocol (Miller et al., 1988). Clones that contained AAV DNA were identified by Rep PCR using primers 5’-gatcgaagcttccgcgtctgacgtcgatgg-3’ and 5’-ggaccaggcctcatacatctccttcaatgc-3’. Previous experience has shown us that the persistence of Rep DNA in actively dividing cells for more than 3 weeks occurs due to integration of the AAV DNA (Philpott et al., 2002a,b, 2004). For PARP inhibition, cells were treated with 20 μM 1,5-isoquinolinediol (DIQ) in 0.1 M NaOH. For vehicle control experiments, cells were incubated with 0.06 mM NaOH, which corresponded to the final NaOH vehicle concentration in DIQ-treated cell culture.
Statistical analysis of AAV integration experiments
Four 2×2 tables, stratified by experiment, comparing AAV DNA retention for p53-negative and p53-positive cells are presented in Table 1. Odds ratios for each experiment were computed, as well as the Mantel-Haenszel common odds ratio for the four experiments. Exact tests were computed to test the null hypothesis of conditional independence between retaining AAV DNA and cell type (p53+/+ or p53–/–) conditioned on the experiment. Exact confidence intervals for the common odds ratio were also computed along with Zelen's exact test for equal odds ratios. Zelen's exact test was computed to test the null hypothesis that the odds ratios for the experiments were equal. A conservative two-sided p value was computed by doubling the exact one-sided p values. Odds ratios and exact confidence limits for retaining AAV DNA and the presence or absence of PARP inhibitor were computed separately for p53-negative and p53-positive cells. All analyses were performed using SAS 9.2 software (SAS Institute Inc., Cary, NC).
Table 1.
Quantification of the Number of Clones Retaining AAV DNA
| |
|
Retained AAV DNA |
|
|
|---|---|---|---|---|
| Experiment | Cells | Yes | No | Odds ratio and 95% CI for retaining AAV DNA in p53–/– vs. p53+/+ cells |
| p53–/– | 4 | 4 | ||
| 1 | p53+/+ | 2 | 9 | 4.50 [0.40, 64.41] |
| p53–/– | 5 | 8 | ||
| 2 | p53+/+ | 2 | 17 | 5.31 [0.65, 63.62] |
| p53–/– | 5 | 23 | ||
| 3 | p53+/+ | 3 | 22 | 1.59 [0.27, 11.41] |
| p53–/– | 2 | 25 | ||
| 4 | p53+/+ | 1 | 29 | 2.32 [0.12, 147.40] |
The number of clones retaining AAV DNA is significantly higher in p53-negative cells compared with p53-positive cells (p<0.05).
CI, confidence interval.
Quantification of integration into the AAVS1 site
The rate of targeted integration into the AAVS1 site was quantified by Southern blot analysis (Philpott et al., 2002a,b, 2004). Genomic DNA from the rAAV-Rep-GFP–transduced clonal lines was digested using EcoRI restriction enzyme. The DNA was separated on two agarose gels and transferred to nylon membranes. The first membrane was hybridized to an 800-bp rep probe that was generated by PCR using oligos 5’-gatcgaagcttccgcgtctgacgtcgatgg-3’ and 5’-ggaccaggcctcatacatctccttcaatgc-3’. The second membrane was hybridized to a 2-kb AAVS1 probe that was generated from a PCR product using oligos 5’-gcgccgtgacgtcagcacgc-3’ and 5’-caccagataaggaatctgcc-3’. Disruption to the AAVS1 site was detected by the presence of bands in addition to the band representing the 8-kb wild-type AAVS1 site. Hybridization to the rep probe indicated the presence of the rAAV-Rep-GFP genome, and comigration of the rep band with the disrupted AAVS1 band signified a targeted integration event at the AAVS1 site.
Transient transfection and cell viability assays
p53+/+ HCT116 cells were transfected with pcDNA-Rep or peGFP-N1. Adherent and floating cells were isolated and counted at 48 hr post transfection. Cell viability was measured on a Countess Cell Counter (Invitrogen).
Nuclear extraction and p53 activity assay
293T cells were plated into 10-cm dishes with 1.5×106 cells per dish and, the following day, were transfected with 24 μg of pcDNA-Rep or peGFP-N1 using 0.5 μM polyethylenimine. The transfections were carried out in triplicate along with untransfected controls. Nuclear extracts were prepared at 48 hr post transfection using a Nuclear Extraction Kit (Cayman Chemical Company, Ann Arbor, MI) according to the manufacturer's instructions. The nuclear extracts were used for p53 transcription factor assays using the p53 Transcription Factor Assay Kit (Cayman Chemical Company) following the manufacturer's instructions.
Results and Discussion
Previous studies show that the AAV genome is not active in primary cells and such cells are not permissive to AAV integration (Halbert et al., 1995). Furthermore, a screen of 175 human tissue samples by Schnepp and colleagues detected wild-type AAV DNA in only nine samples. Eight of these samples harbored episomal DNA, and only one sample contained integrated DNA (into chromosome 1) (Schnepp et al., 2005). Site-specific integration was not observed. The discrepancy in AAV integration efficiency between primary cells and transformed cell lines, which are permissive to AAV-targeted integration (Kotin et al., 1990; Halbert et al., 1995; Linden et al., 1996a; Schnepp et al., 2005), led us to consider whether tumor suppressor genes influence the efficiency of AAV Rep-mediated integration. As Rep has been shown to interact with the tumor suppressor p53 (Batchu et al., 1999), we hypothesized that p53 would have an inhibitory effect on AAV integration.
p53 blocks AAV integration
To define the influence of p53 on AAV-mediated insertion events, integration assays were carried out using p53-positive and p53-deficient HCT116 cells (Bunz et al., 1998). The western blot in Fig. 1A confirms the presence of p53 in the wild-type HCT116 cells and the absence of p53 expression in the p53-deficient HCT116 cells. Our goal was to evaluate the effect of p53 on the rate of AAV integration and to determine whether the absence of p53 pushes integration toward a site-specific or random integration pathway. Therefore, we initially assessed integration efficiency within the context of the whole human genome by generating clonal cell lines and quantifying AAV DNA retention by PCR. We followed this with analysis of site-specific integration into the AAVS1 site by Southern blot.
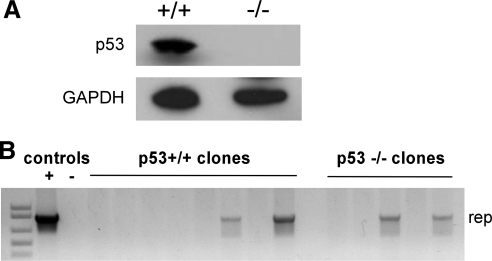
FIG. 1.
p53-positive and p53-negative HCT116 cells. (A) Western blot showing the presence or absence of p53 in HCT116 cell lysates. GAPDH western blot shows equal loading. (B) HCT116 cells were infected with rAAV-Rep-GFP, and clonal cell lines were generated. Clones were screened for AAV DNA by Rep PCR, and a representative gel is shown. HCT116(p53+/+) genomic DNA mixed with pRepGFP plasmid DNA was used as a positive control (+), and HCT116(p53+/+) genomic DNA from uninfected cells was used as a negative control (−).
HCT116 p53+/+ and HCT116 p53–/– cells were infected with the GFP-expressing AAV vector (rAAV-Rep-GFP); at 2 days post infection, GFP-expressing cells were selected by sorting and clonal cell lines were grown from single-cell isolates. The GFP selection process ensured that all of the clones that were maintained had originally been transduced by the rep-containing AAV vector. The clones were grown for at least 1 month, and the genomic DNA was isolated and screened for the presence of rep DNA by PCR. A sample PCR screen gel is shown in Fig. 1B. The infections were carried out using four different vector preparations at a range of multiplicities of infection, and at least eight clones from each infection were analyzed. Results from the four experiments are presented in Table 1. A total of 160 clonal lines, consisting of 85 p53+/+ clones and 75 p53–/– clones, were tested. The odds ratio for each experiment indicates a positive association of retaining AAV DNA in p53–/– cells compared with p53+/+ cells, because significantly more p53–/– clones retained AAV rep DNA compared with the p53+/+ clones. The estimated common odds ratio for retaining AAV DNA for p53–/– cells compared with p53+/+ cells was 2.89 (95% confidence interval: [1.05, 8.72], p=0.0394). Taken together, the four experiments show that rep DNA was retained in 21% of p53-deficient HCT116 cells and in only 9% of p53-positive HCT116 cells. These data indicate that p53-deficient cells more efficiently retain the AAV genome than p53-positive cells, therefore suggesting that p53 inhibits AAV integration.
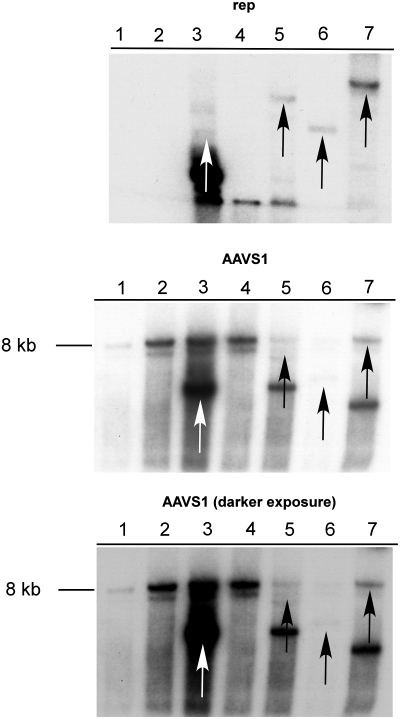
To analyze further the integrity of the integration events, site-specific integration of the AAV genome into the AAVS1 site was quantified by Southern blot. Sample blots are shown in Fig. 2. Out of eight rep-positive, p53-deficient clones analyzed, five contained site-specifically integrated AAV rep DNA (i.e., 62%±12%). In contrast, only 20% (one out of five) of the rep-positive, p53-expressing clones contained site-specifically integrated rep DNA. Although this is a small sample size with only 13 clones tested, the data suggest that p53 specifically blocks Rep-mediated integration into the AAVS1 site of human chromosome 19.
FIG. 2.
Southern blot analysis showing site-specific integration into the AAVS1 site. A representative image of a Southern blot hybridization of rep-positive HCT116 clones is shown. HCT116(p53+/+) and HCT116(p53–/–) cells were infected with rAAV-Rep-GFP. Clones containing rep DNA were selected by PCR, and genomic DNA was analyzed by Southern hybridization with probes specific for rep or AAVS1 sequence. Genomic DNA from uninfected p53–/– and p53+/+ HCT116 cells are shown in lanes 1 and 2, respectively. Lanes 3 and 4 show rep-containing p53+/+ clones, and lanes 5, 6, and 7 show rep-containing p53–/–clones. The arrows indicate the overlapping AAVS1 and rep bands in clones where rAAV genome site-specifically integrated.
p53 and PARP inhibit AAV integration independently
We have previously shown that PARP1 inhibits AAV integration (Romanova, unpublished observations), and as PARP1 and p53 are known to interact in mammalian cells (Wesierska-Gadek et al., 1996; Vaziri et al., 1997; Malanga et al., 1998), we sought to determine whether p53 and PARP1 inhibit integration as part of the same pathway or independently of each other. We therefore performed integration assays in p53-negative cells in the presence of a PARP inhibitor (DIQ) or vehicle control (NaOH). Integration assays were carried out by infecting p53-deficient HCT116 cells with the rAAV-Rep-GFP vector, in the presence of DIQ or NaOH control. Clonal cell lines were generated and grown for at least a month, and then the genomic DNA was isolated. Using the genomic DNA as a template, rep PCR was used to quantify the rate of AAV integration. Results for testing the association between DIQ and NaOH vehicle for p53-negative cells are presented in Table 2. A total of 51 p53-negative clones were included in the analysis. An odds ratio of 2.98 indicates a positive association between retention of AAV DNA and PARP inhibition. As AAV integration increases in the presence of DIQ in p53-negative cells, our results indicate that p53 and PARP function independently to inhibit AAV integration.
Table 2.
Quantification of the Number of Clones Retaining AAV DNA in the Presence or Absence of PARP Inhibitor
| |
|
Retained AAV Rep DNA |
|
|
|---|---|---|---|---|
| Cells | Chemical | Yes | No | Odds ratio and 95% CI for presence of Rep DNA in DIQ-treated cells vs. controls |
| DIQ PARP inhibitor | 7 | 18 | ||
| p53-negative cells | NaOH vehicle | 3 | 23 | 2.98 [0.57, 19.98] |
| DIQ PARP inhibitor | 4 | 23 | ||
| p53-positive cells | NaOH vehicle | 2 | 29 | 2.52 [0.36, 33.75] |
Quantification of the number of clones retaining AAV DNA in the presence or absence of PARP inhibitor shows that p53 and PARP inhibit AAV integration independently.
PARP inhibition enhances AAV integration in p53-positive cells
To determine whether inhibition of PARP affects AAV integration efficiency in p53-positive cells, we performed integration assays in HCT116(p53+/+) cells in the presence of DIQ or NaOH vehicle control. Again, the cells were infected with the rAAV-Rep-GFP vector, and genomic DNA was isolated from clonal cell lines as described above. Results from rep PCR analysis of the clonal DNA are shown in Table 2. A total of 58 p53-postive cells were tested, and an odds ratio of 2.52 indicates a positive association between retention of AAV DNA and PARP inhibition. The data in Table 2 show that PARP inhibition enhances AAV integration in both p53-positive and p53-negative cells, again supporting our conclusion that PARP and p53 block AAV integration independently.
p53-mediated inhibition of AAV integration is not through Rep-dependent toxicity
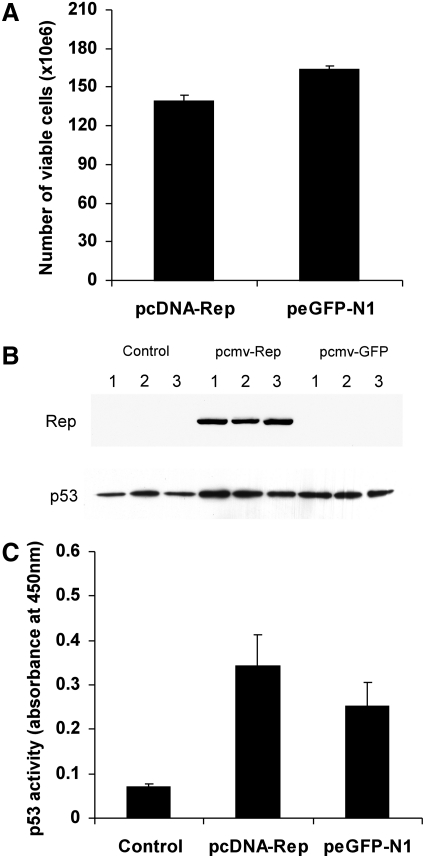
To confirm that p53-mediated inhibition of Rep-dependent integration was not caused by p53-induced apoptosis in response to the presence of Rep, we carried out viability assays in p53-positive Rep-transfected cells. HCT116(p53+/+) cells were seeded into six-well plates at 200,000 cells per well and transfected the following day with a Rep-expressing or GFP-expressing plasmid. At 48 hr post transfection, adherent and floating cells were isolated and the numbers of viable cells were quantified. The results in Fig. 3A show that Rep expression did not cause a significant increase in toxicity in p53-containing cells (Student's independent t test, p=0.248965).
FIG. 3.
Rep does not activate p53. (A) Viability of Rep-transfected HCT p53+/+ cells. p53+/+ HCT116 cells were transfected with pRep or pGFP plasmids (pcDNA-Rep and peGFP-N1, respectively). Viable cells were counted at 48 hr post transfection. Rep does not cause significant toxicity to p53+/+ cells (Student's independent t test, p=0.248965). Error bars represent standard deviations. (B) Western blots of triplicate nuclear lysates from pRep- and pGFP-transfected 293T cells showing that Rep does not affect expression of p53. (C) p53 activity assay using nuclear lysates from untransfected (control), pcDNA-Rep-, and peGFP-N1-transfected 293T cells showing that Rep does not significantly increase the activity of p53 (Student's independent t test, p=0.133542). Error bars represent standard deviations.
Rep does not increase the expression of total p53 or the level of activated p53
To understand the mechanism of p53-mediated integration inhibition, we assessed whether Rep would cause an increase in either p53 expression or the level of activated p53, either of which could result in growth arrest of the cell, and not necessarily cell death. As plasmid transfection would achieve a higher level of Rep expression than transduction of cells using a rep-containing AAV vector or wild-type AAV, HEK293T cells were transfected with the Rep-expressing plasmid “pcDNA-Rep” or Rep-negative plasmid “peGFP-N1.” Untransfected cells were included as a negative control. Nuclear lysates were then harvested at 48 hr post transfection and examined for changes in p53 expression by western blot (Fig. 3B) and for levels of activated p53 by transcription factor assay (Fig. 3C). The western blot analysis and quantification of activated p53 levels in Fig. 3 show no significant Rep-mediated changes in p53 expression or activation, as compared with GFP-transfected control cells (Student's independent t test, p=0.133542). These data suggest that Rep and Rep-mediated nicking of the genome has no affect on p53 activity within a cell. However, previous studies by Batchu and co-workers have shown that p53 does bind to Rep 78 (Batchu et al., 1999). As we have found that p53 is not up-regulated or activated by Rep either directly or by introducing single-stranded breaks in the genome, we suggest that p53 functions to inhibit Rep-mediated site-specific integration by associating with Rep 78 and blocking its activity. This would result in p53 preventing Rep-mediated integration of AAV vector DNA.
Conclusions
The results presented in this work show that p53-deficient cells are more permissive to AAV integration than cells that express functional p53. The mechanism of AAV integration inhibition by p53 remains to be elucidated, but our results indicate that Rep does not induce p53-mediated cell death. Of note, this is in agreement with observations by Schmidt and colleagues (Schmidt et al., 2000). We also found that p53-mediated inhibition of AAV integration is independent of the PARP1-mediated inhibition that we have previously shown (Romanova, unpublished observations). We believe that the most likely mechanism for p53-mediated inhibition of integration is through the binding of p53 to Rep (Batchu et al., 1999). This interaction could interfere with the functional activity of Rep, thus preventing efficient insertion of AAV DNA into the human genome. The mechanistic details warrant further investigation.
Acknowledgments
We would like to thank Tyler Enright for technical support. Funding was provided by the Minnesota Medical Foundation award no. 3876-9221-08, the American Cancer Society Institutional Research Grant no. IRG-58-001-49-IRG34, and the University of Minnesota Academic Health Center Seed Grant no. 2009-17 to N.J.P. We acknowledge the assistance of the Flow Cytometry Core Facility of the Masonic Cancer Center, a comprehensive cancer center designated by the National Cancer Institute, supported by award no. P30CA77598.
Author Disclosure Statement
The authors have no commercial associations and no conflicts of interest. No competing financial interests exist.
References
- Batchu R.B. Shammas M.A. Wang J.Y. Munshi N.C. Interaction of adeno-associated virus Rep78 with p53: implications in growth inhibition. Cancer Res. 1999;59:3592–3595. [PubMed] [Google Scholar]
- Berns K.I. Linden R.M. The cryptic life style of adeno-associated virus. Bioessays. 1995;17:237–245. doi: 10.1002/bies.950170310. [DOI] [PubMed] [Google Scholar]
- Bunz F. Dutriaux A. Lengauer C., et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Chiorini J.A. Wendtner C.M. Urcelay E., et al. High-efficiency transfer of the T cell co-stimulatory molecule B7-2 to lymphoid cells using high-titer recombinant adeno-associated virus vectors. Hum. Gene Ther. 1995;6:1531–1541. doi: 10.1089/hum.1995.6.12-1531. [DOI] [PubMed] [Google Scholar]
- Dyall J. Berns K.I. Site-specific integration of adeno-associated virus into an episome with the target locus via a deletion-substitution mechanism. J. Virol. 1998;72:6195–6198. doi: 10.1128/jvi.72.7.6195-6198.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall J. Szabo P. Berns K.I. Adeno-associated virus (AAV) site-specific integration: formation of AAV-AAVS1 junctions in an in vitro system. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12849–12854. doi: 10.1073/pnas.96.22.12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattah F.J. Lichter N.F. Fattah K.R., et al. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8703–8708. doi: 10.1073/pnas.0712060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud C. Winocour E. Berns K.I. Site-specific integration by adeno-associated virus is directed by a cellular DNA sequence. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10039–10043. doi: 10.1073/pnas.91.21.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D. Kern A. Rittner K. Kleinschmidt J.A. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 1998;9:2745–2760. doi: 10.1089/hum.1998.9.18-2745. [DOI] [PubMed] [Google Scholar]
- Halbert C.L. Alexander I.E. Wolgamot G.M. Miller A.D. Adeno-associated virus vectors transduce primary cells much less efficiently than immortalized cells. J. Virol. 1995;69:1473–1479. doi: 10.1128/jvi.69.3.1473-1479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotin R.M. Siniscalco M. Samulski R.J., et al. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U.S.A. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamartina S. Roscilli G. Rinaudo D., et al. Lipofection of purified adeno-associated virus Rep68 protein: toward a chromosome-targeting nonviral particle. J. Virol. 1998;72:7653–7658. doi: 10.1128/jvi.72.9.7653-7658.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden R.M. Ward P. Giraud C., et al. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U.S.A. 1996a;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden R.M. Winocour E. Berns K.I. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. U.S.A. 1996b;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanga M. Pleschke J.M. Kleczkowska H.E. Althaus F.R. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J. Biol. Chem. 1998;273:11839–11843. doi: 10.1074/jbc.273.19.11839. [DOI] [PubMed] [Google Scholar]
- Miller S.A. Dykes D.D. Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott N.J. Giraud-Wali C. Dupuis C., et al. Efficient integration of recombinant adeno-associated virus DNA vectors requires a p5-rep sequence in cis. J. Virol. 2002a;76:5411–5421. doi: 10.1128/JVI.76.11.5411-5421.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott N.J. Gomos J. Berns K.I. Falck-Pedersen E. A p5 integration efficiency element mediates Rep-dependent integration into AAVS1 at chromosome 19. Proc. Natl. Acad. Sci. U.S.A. 2002b;99:12381–12385. doi: 10.1073/pnas.182430299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott N.J. Gomos J. Falck-Pedersen E. Transgene expression after rep-mediated site-specific integration into chromosome 19. Hum. Gene Ther. 2004;15:47–61. doi: 10.1089/10430340460732454. [DOI] [PubMed] [Google Scholar]
- Riley T. Sontag E. Chen P. Levine A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Samulski R.J. Zhu X. Xiao X., et al. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. Afione S. Kotin R.M. Adeno-associated virus type 2 Rep78 induces apoptosis through caspase activation independently of p53. J. Virol. 2000;74:9441–9450. doi: 10.1128/jvi.74.20.9441-9450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepp B.C. Jensen R.L. Chen C.L., et al. Characterization of adeno-associated virus genomes isolated from human tissues. J. Virol. 2005;79:14793–14803. doi: 10.1128/JVI.79.23.14793-14803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S. Lu Y. Choi Y.K., et al. DNA-dependent PK inhibits adeno-associated virus DNA integration. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2112–2116. doi: 10.1073/pnas.0307833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. Replication of the adeno-associated virus DNA termini in vitro. Intervirology. 1987;27:138–147. doi: 10.1159/000149732. [DOI] [PubMed] [Google Scholar]
- Srivastava A. Lusby E.W. Berns K.I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surosky R.T. Urabe M. Godwin S.G., et al. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J. Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan I. Ng C.H. Lim L. Leung T. Phosphorylation of a novel myosin binding subunit of protein phosphatase 1 reveals a conserved mechanism in the regulation of actin cytoskeleton. J. Biol. Chem. 2001;276:21209–21216. doi: 10.1074/jbc.M102615200. [DOI] [PubMed] [Google Scholar]
- Urcelay E. Ward P. Wiener S.M., et al. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J. Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H. West M.D. Allsopp R.C., et al. ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the post-translational activation of p53 protein involving poly(ADP-ribose) polymerase. EMBO J. 1997;16:6018–6033. doi: 10.1093/emboj/16.19.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.E. Liu J.M. Xiao X., et al. Regulated high level expression of a human gamma-globin gene introduced into erythroid cells by an adeno-associated virus vector. Proc. Natl. Acad. Sci. U.S.A. 1992;89:7257–7261. doi: 10.1073/pnas.89.15.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman M.D. Kyostio S.R. Kotin R.M. Owens R.A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc. Natl. Acad. Sci. U.S.A. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesierska-Gadek J. Bugajska-Schretter A. Cerni C. ADP-ribosylation of p53 tumor suppressor protein: mutant but not wild-type p53 is modified. J. Cell. Biochem. 1996;62:90–101. doi: 10.1002/(sici)1097-4644(199607)62:1<90::aid-jcb10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Xiao X. Li J. Samulski R.J. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J. Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.C. Xiao X. Zhu X., et al. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S.M., Jr. Samulski R.J. Adeno-associated virus (AAV) site-specific recombination does not require a Rep-dependent origin of replication within the AAV terminal repeat. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13525–13530. doi: 10.1073/pnas.241508998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S.M., Jr. McCarty D.M. Degtyareva N. Samulski R.J. Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J. Virol. 2000;74:3953–3966. doi: 10.1128/jvi.74.9.3953-3966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]