Abstract
In 2010, more than 45 years after the initial discovery of lipoprotein(a) [Lp(a)] by Kare Berg, an European Atherosclerosis Society Consensus Panel recommended screening for elevated Lp(a) in people at moderate to high risk of atherosclerotic cardiovascular disease (CVD). This recommendation was based on extensive epidemiological findings demonstrating a significant association between elevated plasma Lp(a) levels and coronary heart disease, myocardial infarction, and stroke. In addition to those patients considered to be at moderate to high risk of heart disease, statin-treated patients with recurrent heart disease were also identified as targeted for screening of elevated Lp(a) levels. Taken together, recent findings have significantly strengthened the notion of Lp(a) as a causal risk factor for CVD. It is well established that Lp(a) levels are largely determined by the size of the apolipoprotein a [apo(a)] gene; however, recent studies have identified several other LPA gene polymorphisms that have significant associations with an elevated Lp(a) level and a reduced copy number of K4 repeats. In addition, the contribution of other genes in regulating Lp(a) levels has been described. Besides the strong genetic regulation, new evidence has emerged regarding the impact of inflammation as a modulator of Lp(a) risk factor properties. Thus, oxidized phospholipids that possess a strong proinflammatory potential are preferentially carried on Lp(a) particles. Collectively, these findings point to the importance of both phenotypic and genotypic factors in influencing apo(a) proatherogenic properties. Therefore, studies taking both of these factors into account determining the amount of Lp(a) associated with each individual apo(a) size allele are valuable tools when assessing a risk factor role of Lp(a).
Apo(a) Gene Size Polymorphism and Lp(a)
The plasma lipoprotein(a)[Lp(a)] level is one of the most heritable quantitative traits in humans, and the high heritability of this trait has been attributed to variations at LPA locus.1 Lp(a), like low-density lipoprotein (LDL), consists of a cholesteryl ester–rich lipid core and one molecule of apolipoprotein B (apoB), form apoB-100, with an addition of one molecule of the glycoprotein apolipoprotein(a) [apo(a)].2,3 The two apolipoproteins are covalently linked through a single disulfide bond at the carboxy-terminal portion of each respective protein. ApoB is largely hydrophobic, and portions of the protein are embedded in the lipid part of Lp(a). In contrast, apo(a) is hydrophilic, carbohydrate-rich, highly charged, and contains repeated loop structures, so-called kringles (K), of which K4 type 2 is present as multiple copies varying in number from 3 to more than 40 copies.4 Due to the variability in the number of K4 type 2 repeats, the apo(a) gene size is highly polymorphic and is a major predictor of Lp(a) levels.5 Smaller apo(a) sizes tend to associate with higher Lp(a) levels, leading to an increased cardiovascular disease (CVD) risk,6–11 but this association is far from linear.12 In general, two types of variability contribute to this deviation from linearity. First, there are large interindividual differences in Lp(a) levels associated with any given apo(a) allele size. Second, marked differences in Lp(a) levels have been found between ethnic populations. In particular, populations of African descent, including African Americans, have higher Lp(a) levels compared to most other ethnic groups for a range of apo(a) allele sizes.13,14 Due to the extensive apo(a) gene size polymorphism, homozygotes for apo(a) size are rare; a heterozygosity index of approximately 94% was reported among Caucasians.15,16
Previous studies have demonstrated that Lp(a) synthesis strongly predicts plasma levels. Studies in model systems have shown formation of an apo(a)–apoB complex at the hepatocyte surface, resulting in an Lp(a) particle.17 The exact mechanisms involved in Lp(a) synthesis in humans remain unresolved as some studies have reported an intracellular Lp(a) formation as well.18 Results from in vitro studies further suggest that Lp(a) particles with a small-size apo(a) are more efficiently secreted from hepatocytes than Lp(a) particles with a large-size apo(a). However, a small apo(a) allele size in a heterozygous individual does not always correspond to the dominant apo(a) protein isoform.12 In support of this, the size and expression of one apo(a) allele in a genotype influences the Lp(a) level attributable to the other apo(a) allele in both African Americans and Caucasians.19
Apo(a) Gene Non-Size Polymorphism and Lp(a)
Apo(a) size polymorphism is a major predictor of Lp(a) levels contributing between 40% to 70% of the variation in Lp(a) concentrations8,20,21; however, other genetic variants at the LPA locus also contribute with varying effects across ethnicity (Table 1). Distributions of a C/T variation in the promoter region of the apo(a) gene and a pentanucleotide repeat (PNR) (TTTTAn), about 1 kb upstream of the apo(a) gene differed across ethnicity. The T allele was less common and the presence of a small PNR allele was more common in African Americans than Caucasians.19 Controlling for apo(a) sizes, PNR influenced allele-specific apo(a) levels in Caucasians, with a stepwise decrease with increasing PNR number >8. The apo(a) allele size (>24 K4) and PNR >8 were significant predictors for allele-specific apo(a) levels in Caucasians, but not in African Americans. Furthermore, evidence of linkage disequilibrium (LD) to a variable degree for African Americans and Caucasians among the C/T, PNR, and apo(a) size polymorphisms has been found.22 In a biethnic study of subjects with end-stage renal disease (ESRD), three single nucleotide polymorphisms (SNPs) were reported to contribute to the interethnic African–Caucasian difference in Lp(a) levels.23 One Lp(a)-increasing SNP (G-21A, which increases promoter activity) was more common in African Americans, whereas two Lp(a)-lowering SNPs [T3888P and G+1/inKIV-8A, which inhibits Lp(a) assembly] were more common in Caucasians. Although the apo(a) size spectrum does not differ, ESRD patients have increased Lp(a) levels irrespective of ethnicity, underscoring the importance to extend these findings to non-ESRD subjects.
Table 1.
Recent Studies that Have Investigated Apo(a) Size and Non-Size Polymorphisms
| Authors [Ref] | Year | Major polymorphisms | LPA gene location/function | Population | Association with Lp(a) |
|---|---|---|---|---|---|
| Rubin et al. [19] | 2006 | a C/T variation | promoter region | CA and AA | T allele was less and small PNR allele was more common in AA than CA. A stepwise decrease in Lp(a) level with increasing PNR number >8 was observed in CA, but not in AA. |
| a pentanucleotide repeat (TTTTAn) | 1 kb upstream | ||||
| Chretien et al. [23] | 2006 | G-21A | increases promoter activity | AA and CA | Lp(a)-increasing SNP G-21A was common in AA, whereas Lp(a)-lowering SNPs T3888P and G+ 1/inKIV-8A were common in CA. All 3 SNPs contributed to higher Lp(a) levels in AA. |
| T3888P and G+ 1/inKIV-8A | inhibits Lp(a) assembly | ||||
| Luke et al. [24] | 2007 | I4399M/rs3798220 | protease-like domain | CA | Risk allele-carriers had 5-fold higher median Lp(a) level and significantly smaller apo(a) isoform (17 K4 vs. 22 K4) vs. non-carriers. |
| Clarke et al. [20] | 2009 | rs10455872 | maps to intron 25 | CA | 16 SNPs had significant effects on Lp(a) level. The two SNPs had the strongest associations with an elevated Lp(a) level and a reduced copy number of K4 repeat and explained 36% of the variance in Lp(a) level. |
| rs3798220 | protease-like domain | ||||
| Ober et al. [30] | 2009 | rs6919346 | maps to intron 37 | Hutterites and CA | In Hutterites, both SNPs were significantly associated with an elevated Lp(a) level, independent of the apo(a) size and had a combined effect size of 4% on Lp(a) level. In CA, rs6919346 was associated with Lp(a) level. |
| rs1853021 (+93C/T) | 5′ untranslated region | ||||
| Lanktree et al. [27] | 2010 | rs10455872 | maps to intron 25 | South Asians, Chinese, CA | Prevalent only in CA and was associated with both Lp(a) level and K4 repeat number. |
| rs6415084 | the same haplotype block as the K4 type 2 variation | Prevalent in all 3 ethnicities and was associated with both Lp(a) level and K4 repeat number. SNPs and apo(a) size polymorphism together explained 36% of variation in Lp(a) levels in CA, 27% in Chinese and 21% in South Asians. | |||
| Ronald et al. [28] | 2011 | rs3798220 | protease-like domain | CA | 9 SNPs were predictive of Lp(a) level and accounted for 30% of Lp(a) variance. The two SNPs were associated with Lp(a) level after accounting for K4 repeat number and explained 22% of Lp(a) variance. SNPs and apo(a) size polymorphism together explained 60% Lp(a) variance. |
| rs10455872 | maps to intron 25 | ||||
| Deo et al. [33] | 2011 | rs9457951 | intronic | AA | A number of common SNPs were associated with Lp(a) level accounting for up to 7% of the variation, as well as >70% of the African-Caucasian interethnic difference in Lp(a) level. SNP rs9457951 expressed the strongest association and alone explained 5% of Lp(a) level variance. |
| rs6930542 | intronic | ||||
| rs10455872 | maps to intron 25 | ||||
| rs6922216 | intronic | ||||
| rs1801693 | exonic, K4 type 9 | ||||
| T3888P | inhibits Lp(a) | ||||
| G+ 1/inKIV-8A | assembly |
AA, African Americans; CA, caucasians; K, kringle; SNP, single-nucleotide polymorphism; apo(a), apolipoprotein(a); Lp(a), lipoprotein(a); PNR, pentanucleotide repeat.
Several recent studies have addressed a role of a cluster of SNPs at the LPA locus in predicting Lp(a) levels. Among these SNPs, rs3798220, located in the protease-like domain of apo(a) and rs10455872, which maps to intron 25, have repeatedly been associated with an increased Lp(a) level and a reduced copy number of K4 repeats. In a study by Luke et al. conducted among Caucasians, carriers of the I4399M, rs3798220 risk allele had five-fold higher median Lp(a) level and significantly smaller apo(a) isoform (17 K4 vs. 22 K4) compared to noncarriers.24 Also, the risk allele carriers had an adjusted odds ratio (OR) for severe coronary artery disease (CAD) of 3.14, independent of the apo(a) gene size. A subsequent study from the same group of investigators demonstrated higher plasma oxidized phospholipid (OxPL)/apoB levels, which primarily represent OxPL in Lp(a), in Caucasian carriers of 14339M SNP compared to noncarriers.25 The relationship was strongly dependent on apo(a) isoform size, because carriers of the 14339M SNP manifesting small apo(a) isoforms had the highest OxPL/apoB levels.
Integrated Role of apo(a) Gene Size and Non-Size Polymorphisms and Lp(a)
In a more recent study by Clarke et al., where a custom genotyping chip containing 48,742 SNPs in 2,100 candidate genes was tested in 6,497 CAD and control subjects, the genomic region 6q26–q27 spanning the LPA locus was significantly associated with CAD.20 The finding was replicated in three independent cohorts (PROCARDIS, SHEEP, and SCARF) involving an additional 9,440 subjects. Further investigation revealed that of 16 SNPs at the LPA locus with significant effects on Lp(a) levels rs10455872 and rs3798220 had the strongest associations, with an increased Lp(a) level and a lower number of K4 repeats. Together they explained 36% of the overall variance in plasma Lp(a) levels and were associated with increased CAD risk. A total of 7 SNPs, including the former two SNPs, associated with Lp(a) levels in stepwise regression analysis, and collectively explained 40% of the variance in Lp(a) levels.
The findings of this study have several important implications. First, in agreement with previous findings, the results demonstrate that the largest variability in Lp(a) levels is seen for smaller apo(a) sizes.14,26 Thus, variability in genetic loci mapped to small-size apo(a) can be expected to be a major predictor of Lp(a) levels. Second, the results confirmed earlier observations on the association of SNP rs3798220 with the Lp(a) level.24 Third, the results also confirmed the well-demonstrated previous association between small-size apo(a) and CVD. However, after adjustment for the Lp(a) levels, the association between LPA genotypes and CAD was abolished.
Furthermore, a comprehensive analysis of genomic variation in the LPA locus conducted in a multiethnic population comprising of South Asians, Chinese, and Caucasians reported that the SNP rs10455872 reported by Clarke et al. was prevalent only among Caucasians.27 This SNP was associated with both plasma Lp(a) level and K4 repeat number, consistent with the previous finding.20 In addition, SNP rs6415084 within the same haplotype block as the K4 type 2 variation was significantly associated with both plasma Lp(a) level and K4 type 2 repeat number in all three ethnicities. SNPs and apo(a) size polymorphism together explained a greater proportion of variation in the Lp(a) level in Caucasians (36%) than in Chinese (27%) or South Asians (21%).
More recently, Ronald et al.,28 in a carotid artery disease (CAAD) cohort identified a set of 9 SNPs that accounted for 30% of the variation in Lp(a) level, 5 of which overlapped with the set of 7 SNPs described by Clarke et al.20 Furthermore, 6 of these SNPs, 4 of which had been reported previously by others, were predictive of Lp(a) level conditional on the number of K4 repeats.24,27 Again, after adjustment for K4 repeat number, SNPs rs3798220 and rs10455872 were strongly associated with Lp(a) levels, and together explained 22% of Lp(a) variance, in general agreement with previous observations.24,27 It has been proposed that the nonsynonymous SNP rs3798220 may affect protein stability,24 whereas rs10455872 may be in LD with regulatory variants.29 There has been a considerable heterogeneity in estimating the portion of variance in Lp(a) level explained by SNPs alone or in conjunction with copy number K4 repeat. Thus, two SNPs (rs10455872 and rs3798220) were reported to explain 36% of variance in Lp(a) level in the study by Clarke et al., whereas the corresponding contribution of these two SNPs was only 22% in the study by Ronald et al.20,28 Furthermore, the combination of SNPs and K4 repeat polymorphism explained 36% of the variance in Lp(a) levels in the study by Lanktree et al.,27 whereas the corresponding contribution was higher (above 60%) in the study by Ronald et al.28 An early study by Boerwinkle et al. indicated that 90% of the variance in Lp(a) level was attributable to variation at the LPA locus, of which K4 repeat polymorphism accounted for 69% of the variance.1 The variability in these reports illustrates the difficulty in accurately assessing the complex relationship between Lp(a), apo(a) gene size (copy number of K4 repeat), and other genetic variants at the LPA locus, and underscores the importance of identifying other common as well as rare genetic markers in the region.
In Hutterites, two additional SNPs at the LPA locus [rs6919346 in intron 37 and rs1853021 (+93C/T) in the 5′ untranslated region] were significantly associated with an elevated Lp(a) level independent of the apo(a) size polymorphism.30 The combined effect size of these two polymorphisms on Lp(a) level was approximately 4% in this population. A replication study in unrelated Caucasian males overlapping with the CAAD cohort from the study by Ronald et al.28 confirmed a significant association between Lp(a) level and SNP rs6919346, independent of the apo(a) gene size. The association between LPA +93C/T SNP and Lp(a) levels has been reported in African Americans,31 but in the opposite direction to that seen in Hutterites.32
Recently, Deo et al. investigated genetic variants in LPA that might contribute to the interethnic difference in Lp(a) levels in 4,464 African Americans from the Jackson Heart Study.33 The authors used a panel of ancestry informative markers that allowed an accurate estimation of the African ancestry proportion. A number of common SNPs, strongly associated with Lp(a) level, accounted for up to 7% of the variation in Lp(a) level, as well as >70% of the African-Caucasian interethnic difference in Lp(a) level. SNP rs9457951 expressed the strongest association and alone explained 5% of Lp(a) level variance. In addition, the finding was replicated in 1,726 African Americans from the Dallas Heart Study. In contrast to previous findings in Caucasians,20,27 no single common SNP has been found to explain a large portion of variation in Lp(a) levels in African Americans. These findings might reflect a possibility of limited LD between K4 repeat number and common SNPs on the African ancestral background and are in agreement with earlier observations in Chinese and South Asians.27 In the Dallas Heart Study, the K4 repeat number explained >40% of Lp(a) variance.33 Despite the strong association with Lp(a) levels, there was no association of any LPA SNP with incidence of CHD in 3,225 African Americans from the Atherosclerosis Risk in Communities Study. This negative finding might be, in part, due to the minor contribution of individual SNP on Lp(a) level in African Americans as compared to that seen for Caucasians.20 Altogether, further studies are warranted to reach a firm conclusion on the role of common LPA variants in CHD development among African Americans.
Non-LPA Gene Polymorphism and Lp(a)
In addition to genetic variants in the apo(a) gene, recent studies have suggested a more complex genetic regulation of Lp(a) levels involving multiple other genes on chromosome 6. A genome-wide association study by Ober et al. identified eight other genes (SYNE1, TIAM2, ARID1B, SYTL3, IGF2R, PLG, PARK2, and PACRG), in addition to the LPA gene, on chromosome 6q26–q27 that had significant effects on Lp(a) levels in Hutterites.30 SNPs in these genes each accounted for up to 8.7% of the total variance in Lp(a) levels. Variations at least in six genes were significantly associated with Lp(a) levels independently of each other and of the apo(a) size polymorphism in this population. A replication study in Caucasian males reported that only a single SNP (rs14224) in the PLG gene was associated with Lp(a) levels, and that the association was in LD with the copy number of K4 repeats.30 Further studies are needed to explore the impact of genetic variability beyond the apo(a) gene on Lp(a) levels across different ethnic populations.
Apo(a) Phenotype–Genotype Relationship: Allele-Specific Lp(a) Level
In most studies to date, plasma Lp(a) levels represent a sum of the total amount of circulating Lp(a) particles, irrespective of the apo(a) size isoform distribution. Due to extensive apo(a) size heterogeneity, in any given individual, the plasma Lp(a) level represents the sum of two different Lp(a) populations with likely different size apo(a) particles. The relative contribution of each apo(a) size isoform to the overall Lp(a) level may vary substantially, and this pattern variability may significantly impact susceptibility to cardiovascular risk. In this sense, if size determines risk, the situation is somewhat analogous to LDL and high-density lipoprotein (HDL) particles, where atherogenicity and antiatherogenic potential, respectively, vary between different populations. However, although these differences for HDL and LDL largely affect the lipid composition, the size variability in apo(a) is due to protein heterogeneity. Use of size allele-specific Lp(a) levels determining the amount of Lp(a) associated with each apo(a) size allele therefore provides opportunities for better insight into the risk factor role of Lp(a), because they take into account both phenotypic and genotypic characteristics of apo(a).
The most common tool in assessing allele-specific Lp(a) levels has been to estimate protein isoforms by sodium dodecyl sulfate (SDS)–agarose gel electrophoresis followed by immunoblotting. This methodology is feasible and defines the phenotypic pattern, but it has some limitations. Only a single detectable isoform is present in many subjects, necessitating use of complex methodologies to infer allele sizes. In many cases, the size information cannot be inferred, forcing exclusion of a substantial number of study subjects. In a report on apo(a) polymorphisms contributing to interethnic differences, in spite of using a computerized algorithm to infer missing allele sizes, nearly a quarter of both African-American and Caucasians subjects had to be excluded.23
At present, two different genotyping procedures are available for determining apo(a) size polymorphism: (1) Pulsed field gel electrophoresis (PFGE) and (2) real-time polymerase chain reaction (RT-PCR). Although the PFGE procedure is cumbersome, labor intensive, and necessitates the availability of intact cells embedded in agarose plugs,15,34 it offers several advantages, including an accurate discrimination of subtle differences in apo(a) K4 repeats, determination of the number of K4 repeats on each individual allele, and identification of unexpressed “null” alleles that are not detected on the protein level (pseudo-homozygosity phenomenon). Recently, Kamstrup et al.35 reported on the use of a RT-PCR analysis genotyping Lp(a) as the sum of K4 type 2 repeats from both alleles. Use of this genotypic approach alone might be potentially limiting, because studies have demonstrated variable expressions of different K4 type 2 repeats.36 However, despite this limitation, the K4 type 2 genotype result based on RT-PCR explained 21% and 27% of the total interindividual variation in Lp(a) levels in two independent samples of the general population.35
Simultaneous determination of both apo(a) phenotype and genotype (by PFGE) in an individual can potentially provide more precise information in assessing the risk factor role of Lp(a). This approach led to recognition that nonexpressed alleles were present in approximately 11% of Indian Asian subjects.37 While taking into account both apo(a) size and Lp(a) levels, it was reported that elevated Lp(a) levels for smaller apo(a) sizes, but not the apo(a) size alone, were significantly associated with CAD in African-American and Caucasian men.13 Notably, some studies that have failed to detect an association between Lp(a) and CAD in African Americans were based on either Lp(a) levels or apo(a) isoform sizes alone,38,39 underscoring the importance of taking into account both the Lp(a) level and apo(a) size, e.g., allele-specific Lp(a) levels in assessing a relationship with CAD.
A recent systematic review of 40 studies involving over 58,000 participants confirmed the relationship between small-size apo(a) and CHD.40 The study reported that individuals with smaller apo(a) isoforms have an approximately two-fold higher risk of CHD and ischemic stroke than those with larger isoforms. Considering the fact that apo(a) size polymorphism contributes between 40% and 70% of the variation in Lp(a) level, with a fewer number of K2 type 2 repeats being associated with higher Lp(a) level in Caucasians,8,20,21 it is likely that at least part of the association observed between apo(a) size and CHD risk was mediated by the Lp(a) concentration. Only three of these studies reported relative risks (RR) adjusted for Lp(a) concentrations with a decrease in the combined RR from 2.26 to 1.48.40 Furthermore, the RR for CHD differed substantially between studies, allowing analysis of individual versus combined apo(a) isoform sizes. The combined RR for CHD in subjects with smaller compared to larger apo(a) isoforms was 2.08 for the former and 1.19 for the latter.
The above-mentioned assay considerations regarding the use of RT-PCR may account for the considerably lower RRs for CHD seen in the studies, where apo(a) genotype was expressed as the sum of apo(a) K4 type 2 repeats. In addition, six studies focusing on ischemic stroke generated a combined RR of 2.14 among subjects with smaller versus larger apo(a) isoform size. A recent report from a multiethnic cohort (The Northern Manhattan Stroke Study) comprising Hispanics (52%), African Americans (31%), and Caucasians (17%) confirmed the risk factor role of Lp(a) in predicting ischemic stroke independent of other established risk factors across ethnicity/gender.41 Notably, the study found the strongest association between Lp(a) and ischemic stroke among men and African Americans.
Allele-Specific Lp(a) Level and Inflammation
Inflammation contributes to all phases of atherosclerosis, from fatty streak initiation, growth, and complication of the atherosclerotic plaque to CVD events.42 Thus, virtually every step in atherosclerosis is believed to involve cytokines, bioactive molecules, and proinflammatory cells. The inflammatory response is mediated by systemic acute-phase reactants such as C-reactive protein (CRP), fibrinogen, and serum amyloid A (SAA),43–45 as well as vascular inflammatory biomarkers such as pentraxin 3 (PTX-3) and lipoprotein-associated phospholipase A2 (Lp-PLA2).46,47 The apo(a) gene contains response elements to inflammatory factors such as interleukin-6 (IL-6), and Lp(a) stimulates release of proinflammatory cytokines from vascular endothelial and smooth muscle cells, as well as from monocytes and macrophages.48,49 Importantly, OxPLs which possess a strong proinflammatory potential, are preferentially carried on Lp(a) particles.50 Of note, the correlation between OxPL and Lp(a) level was stronger in individuals with smaller apo(a) isoforms than in individuals with larger apo(a) isoforms.51 OxPLs can alter intracellular redox status and activate proinflammatory genes, leading to inflammatory cascades in the vessel wall.52,53 The findings suggest a synergy between inflammation and Lp(a), and the magnitude of this relationship may differ across various ethnic populations.
The impact of inflammation as detected by increased levels of systemic or vascular inflammatory biomarkers on allele-specific Lp(a) levels across African-American–Caucasian ethnicity has been explored in several studies. Increased levels of CRP and fibrinogen were significantly associated with higher allele-specific Lp(a) levels for smaller (<26 K4 repeats) apo(a) sizes in African Americans.54 Furthermore, in this cohort, higher Lp-PLA2 activity levels were significantly associated with elevated allele-specific Lp(a) levels for smaller (<26 K4) apo(a) sizes in both African Americans and Caucasians.55 A significant association between elevated SAA, an HDL-associated systemic inflammatory biomarker, and higher allele-specific Lp(a) levels for smaller apo(a) size was found in African Americans.56 Collectively, the findings suggest a potential for an additive effect between molecular properties of Lp(a), in particular small-size apo(a), and inflammation that might enhance Lp(a) proatherogenic properties. At present, less is known regarding to what extent systemic or vascular inflammation might affect Lp(a) levels in the general population, and further studies are needed to explore such associations across ethnicity. Overall, beyond underscoring an impact of inflammation on Lp(a) levels, the findings reinforce the concept that inflammation-associated events may contribute to the Lp(a) interethnic difference.
Summary
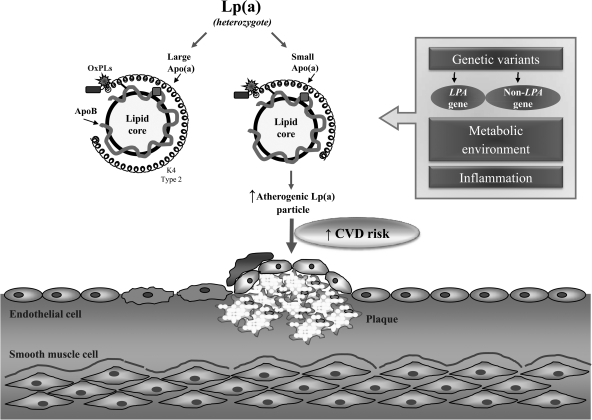
Almost five decades after its initial discovery,57 Lp(a) has been robustly identified as an established risk factor for CVD,58 and interest has grown regarding its potential causative role. Recent methodological and analytical advances have made it possible to identify important common genetic variants in LPA gene, as well as in some other genes, that are significantly associated with an elevated Lp(a) level and a reduced copy number of K4 repeats (Fig. 1). The findings confirm and extend the long-term observation that smaller apo(a) sizes are associated with increased CVD risk. At present, there is a remaining knowledge gap regarding underlying mechanisms that explain these results. The findings have also significantly informed our knowledge in explaining the well-known interethnic differences in Lp(a) levels. Taken together, recent reports and previous findings underscore the importance of genotypic and phenotypic characteristics of Lp(a). It is likely that a key to unlocking the mysteries of Lp(a) will depend on a thorough understanding of both these components. One step in this direction involves use of the allele-specific Lp(a) level in assessing CVD risk.
FIG. 1.
A proposed mechanism underlying plasma lipoprotein(a) [Lp(a)] level and its impact on atherogenic risk. Lp(a) is under a strong genetic regulation of the LPA locus. In addition to an apolipoprotein(a) [apo(a)] gene size polymorphism attributable to a variable number of K2 type 2 repeats, other common genetic variants in the LPA gene, as well as in some other genes, play an important role in regulating Lp(a). Also, metabolic and environmental factors together with proinflammatory conditions modulate risk factor properties of Lp(a), in particular, Lp(a) associated with small-size apo(a). Furthermore, proinflammatory oxidized phospholipids are preferentially attached to Lp(a). These alterations in Lp(a) may enhance its proatherogenic properties and lead to an increased risk for cardiovascular disease (CVD). OxPL, oxidized phospholipid; ApoB, apolipoprotein B.
Acknowledgment
Dr. Berglund was supported by grant number HL 62705 from the National Heart, Lung and Blood Institute, Davis, CA. This work was supported in part by the University of California Davis Clinical and Translational Science Center (RR 024146).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Boerwinkle E. Leffert CC. Lin J. Lackner C. Chiesa G. Hobbs HH. Apolipoprotein(a) gene accounts for greater than 90% of the variation in plasma lipoprotein(a) concentrations. J Clin Invest. 1992;90:52–60. doi: 10.1172/JCI115855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Utermann G. Weber W. Protein composition of Lp(a) lipoprotein from human plasma. FEBS Lett. 1983;154:357–361. doi: 10.1016/0014-5793(83)80182-3. [DOI] [PubMed] [Google Scholar]
- 3.Gaubatz JW. Heideman C. Gotto AM., Jr. Morrisett JD. Dahlen GH. Human plasma lipoprotein [a]. Structural properties. J Biol Chem. 1983;258:4582–4589. [PubMed] [Google Scholar]
- 4.Koschinsky ML. Marcovina SM. Lipoprotein(a): Structural implications for pathophysiology. Int J Clin Lab Res. 1997;27:14–23. doi: 10.1007/BF02827238. [DOI] [PubMed] [Google Scholar]
- 5.McLean JW. Tomlinson JE. Kuang WJ. Eaton DL. Chen EY. Fless GM. Scanu AM. Lawn RM. cDNA sequence of human apolipoprotein(a) is homologous to plasminogen. Nature. 1987;330:132–137. doi: 10.1038/330132a0. [DOI] [PubMed] [Google Scholar]
- 6.Utermann G. The mysteries of lipoprotein(a) Science. 1989;246:904–10. doi: 10.1126/science.2530631. [DOI] [PubMed] [Google Scholar]
- 7.Gavish D. Azrolan N. Breslow JL. Plasma Ip(a) concentration is inversely correlated with the ratio of Kringle IV/Kringle V encoding domains in the apo(a) gene. J Clin Invest. 1989;84:2021–2027. doi: 10.1172/JCI114395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraft HG. Kochl S. Menzel HJ. Sandholzer C. Utermann G. The apolipoprotein (a) gene: A transcribed hypervariable locus controlling plasma lipoprotein (a) concentration. Hum Genet. 1992;90:220–230. doi: 10.1007/BF00220066. [DOI] [PubMed] [Google Scholar]
- 9.Kamstrup PR. Lipoprotein(a) and ischemic heart disease—a causal association? A review. Atherosclerosis. 2010;211:15–23. doi: 10.1016/j.atherosclerosis.2009.12.036. [DOI] [PubMed] [Google Scholar]
- 10.Bennet A. Di Angelantonio E. Erqou S. Eiriksdottir G. Sigurdsson G. Woodward M. Rumley A. Lowe GD. Danesh J. Gudnason V. Lipoprotein(a) levels and risk of future coronary heart disease: Large-scale prospective data. Arch Intern Med. 2008;168:598–608. doi: 10.1001/archinte.168.6.598. [DOI] [PubMed] [Google Scholar]
- 11.Gudnason V. Lipoprotein(a): A causal independent risk factor for coronary heart disease? Curr Opin Cardiol. 2009;24:490–495. doi: 10.1097/HCO.0b013e32832f0a5b. [DOI] [PubMed] [Google Scholar]
- 12.Rubin J. Paultre F. Tuck CH. Holleran S. Reed RG. Pearson TA. Thomas CM. Ramakrishnan R. Berglund L. Apolipoprotein [a] genotype influences isoform dominance pattern differently in African Americans and Caucasians. J Lipid Res. 2002;43:234–244. [PubMed] [Google Scholar]
- 13.Paultre F. Pearson TA. Weil HF. Tuck CH. Myerson M. Rubin J. Francis CK. Marx HF. Philbin EF. Reed RG. Berglund L. High levels of Lp(a) with a small apo(a) isoform are associated with coronary artery disease in African American and white men. Arterioscler Thromb Vasc Biol. 2000;20:2619–2624. doi: 10.1161/01.atv.20.12.2619. [DOI] [PubMed] [Google Scholar]
- 14.Marcovina SM. Albers JJ. Wijsman E. Zhang Z. Chapman NH. Kennedy H. Differences in Lp[a] concentrations and apo[a] polymorphs between black and white Americans. J Lipid Res. 1996;37:2569–2585. [PubMed] [Google Scholar]
- 15.Griffioen PH. Zwang L. van Schaik RH. Engel H. Lindemans J. Cobbaert CM. Optimization of apolipoprotein(a) genotyping with pulsed field gel electrophoresis. Clin Chem. 1999;45:771–776. [PubMed] [Google Scholar]
- 16.Lackner C. Cohen JC. Hobbs HH. Molecular definition of the extreme size polymorphism in apolipoprotein(a) Hum Mol Genet. 1993;2:933–940. doi: 10.1093/hmg/2.7.933. [DOI] [PubMed] [Google Scholar]
- 17.White AL. Lanford RE. Cell surface assembly of lipoprotein(a) in primary cultures of baboon hepatocytes. J Biol Chem. 1994;269:28716–28723. [PubMed] [Google Scholar]
- 18.Bonen DK. Hausman AM. Hadjiagapiou C. Skarosi SF. Davidson NO. Expression of a recombinant apolipoprotein(a) in HepG2 cells. Evidence for intracellular assembly of lipoprotein(a) J Biol Chem. 1997;272:5659–5667. doi: 10.1074/jbc.272.9.5659. [DOI] [PubMed] [Google Scholar]
- 19.Rubin J. Kim HJ. Pearson TA. Holleran S. Ramakrishnan R. Berglund L. Apo[a] size and PNR explain African American-Caucasian differences in allele-specific apo[a] levels for small but not large apo[a] J Lipid Res. 2006;47:982–989. doi: 10.1194/jlr.M500359-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Clarke R. Peden JF. Hopewell JC. Kyriakou T. Goel A. Heath SC. Parish S. Barlera S. Franzosi MG. Rust S. Bennett D. Silveira A. Malarstig A. Green FR. Lathrop M. Gigante B. Leander K. de Faire U. Seedorf U. Hamsten A. Collins R. Watkins H. Farrall M. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 21.Kraft HG. Lingenhel A. Pang RW. Delport R. Trommsdorff M. Vermaak H. Janus ED. Utermann G. Frequency distributions of apolipoprotein(a) kringle IV repeat alleles and their effects on lipoprotein(a) levels in Caucasian, Asian, and African populations: The distribution of null alleles is non-random. Eur J Hum Genet. 1996;4:74–87. doi: 10.1159/000472175. [DOI] [PubMed] [Google Scholar]
- 22.Rubin J. Kim HJ. Pearson TA. Holleran S. Berglund L. Ramakrishnan R. The apolipoprotein(a) gene: Linkage disequilibria at three loci differs in African Americans and Caucasians. Atherosclerosis. 2008;201:138–147. doi: 10.1016/j.atherosclerosis.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chretien JP. Coresh J. Berthier-Schaad Y. Kao WH. Fink NE. Klag MJ. Marcovina SM. Giaculli F. Smith MW. Three single-nucleotide polymorphisms in LPA account for most of the increase in lipoprotein(a) level elevation in African Americans compared with European Americans. J Med Genet. 2006;43:917–923. doi: 10.1136/jmg.2006.042119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luke MM. Kane JP. Liu DM. Rowland CM. Shiffman D. Cassano J. Catanese JJ. Pullinger CR. Leong DU. Arellano AR. Tong CH. Movsesyan I. Naya-Vigne J. Noordhof C. Feric NT. Malloy MJ. Topol EJ. Koschinsky ML. Devlin JJ. Ellis SG. A polymorphism in the protease-like domain of apolipoprotein(a) is associated with severe coronary artery disease. Arterioscler Thromb Vasc Biol. 2007;27:2030–2036. doi: 10.1161/ATVBAHA.107.141291. [DOI] [PubMed] [Google Scholar]
- 25.Arai K. Luke MM. Koschinsky ML. Miller ER. Pullinger CR. Witztum JL. Kane JP. Tsimikas S. The I4399M variant of apolipoprotein(a) is associated with increased oxidized phospholipids on apolipoprotein B-100 particles. Atherosclerosis. 2010;209:498–503. doi: 10.1016/j.atherosclerosis.2009.09.077. [DOI] [PubMed] [Google Scholar]
- 26.Paultre F. Tuck CH. Boden-Albala B. Kargman DE. Todd E. Jones J. Paik MC. Sacco RL. Berglund L. Relation of Apo(a) size to carotid atherosclerosis in an elderly multiethnic population. Arterioscler Thromb Vasc Biol. 2002;22:141–146. doi: 10.1161/hq0102.101097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanktree MB. Anand SS. Yusuf S. Hegele RA. Comprehensive analysis of genomic variation in the LPA locus and its relationship to plasma lipoprotein(a) in South Asians, Chinese, and European Caucasians. Circ Cardiovasc Genet. 2010;3:39–46. doi: 10.1161/CIRCGENETICS.109.907642. [DOI] [PubMed] [Google Scholar]
- 28.Ronald J. Rajagopalan R. Cerrato F. Nord AS. Hatsukami T. Kohler T. Marcovina S. Heagerty P. Jarvik GP. Genetic variation in LPAL2, LPA, and PLG predicts plasma lipoprotein(a) level and carotid artery disease risk. Stroke. 2011;42:2–9. doi: 10.1161/STROKEAHA.110.591230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schadt EE. Molony C. Chudin E. Hao K. Yang X. Lum PY. Kasarskis A. Zhang B. Wang S. Suver C. Zhu J. Millstein J. Sieberts S. Lamb J. GuhaThakurta D. Derry J. Storey JD. Avila-Campillo I. Kruger MJ. Johnson JM. Rohl CA. van Nas A. Mehrabian M. Drake TA. Lusis AJ. Smith RC. Guengerich FP. Strom SC. Schuetz E. Rushmore TH. Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ober C. Nord AS. Thompson EE. Pan L. Tan Z. Cusanovich D. Sun Y. Nicolae R. Edelstein C. Schneider DH. Billstrand C. Pfaffinger D. Phillips N. Anderson RL. Philips B. Rajagopalan R. Hatsukami TS. Rieder MJ. Heagerty PJ. Nickerson DA. Abney M. Marcovina S. Jarvik GP. Scanu AM. Nicolae DL. Genome-wide association study of plasma lipoprotein(a) levels identifies multiple genes on chromosome 6q. J Lipid Res. 2009;50:798–806. doi: 10.1194/jlr.M800515-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraft HG. Windegger M. Menzel HJ. Utermann G. Significant impact of the +93 C/T polymorphism in the apolipoprotein(a) gene on Lp(a) concentrations in Africans but not in Caucasians: confounding effect of linkage disequilibrium. Hum Mol Genet. 1998;7:257–264. doi: 10.1093/hmg/7.2.257. [DOI] [PubMed] [Google Scholar]
- 32.Newman DL. Hoffjan S. Bourgain C. Abney M. Nicolae RI. Profits ET. Grow MA. Walker K. Steiner L. Parry R. Reynolds R. McPeek MS. Cheng S. Ober C. Are common disease susceptibility alleles the same in outbred and founder populations? Eur J Hum Genet. 2004;12:584–590. doi: 10.1038/sj.ejhg.5201191. [DOI] [PubMed] [Google Scholar]
- 33.Deo RC. Wilson JG. Xing C. Lawson K. Kao WH. Reich D. Tandon A. Akylbekova E. Patterson N. Mosley TH. Boerwinkle E. Taylor HA. Single-nucleotide polymorphisms in LPA explain most of the ancestry-specific variation in Lp(a) levels in African Americans. PLoS One. 2011;6:e14581. doi: 10.1371/journal.pone.0014581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lackner C. Boerwinkle E. Leffert CC. Rahmig T. Hobbs HH. Molecular basis of apolipoprotein (a) isoform size heterogeneity as revealed by pulsed-field gel electrophoresis. J Clin Invest. 1991;87:2153–2161. doi: 10.1172/JCI115248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamstrup PR. Tybjaerg-Hansen A. Steffensen R. Nordestgaard BG. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA. 2009;301:2331–2339. doi: 10.1001/jama.2009.801. [DOI] [PubMed] [Google Scholar]
- 36.Kraft HG. Lingenhel A. Kochl S. Hoppichler F. Kronenberg F. Abe A. Muhlberger V. Schonitzer D. Utermann G. Apolipoprotein(a) kringle IV repeat number predicts risk for coronary heart disease. Arterioscler Thromb Vasc Biol. 1996;16:713–719. doi: 10.1161/01.atv.16.6.713. [DOI] [PubMed] [Google Scholar]
- 37.Geethanjali FS. Luthra K. Lingenhel A. Kanagasaba-Pathy AS. Jacob J. Srivastava LM. Vasisht S. Kraft HG. Utermann G. Analysis of the apo(a) size polymorphism in Asian Indian populations: Association with Lp(a) concentration and coronary heart disease. Atherosclerosis. 2003;169:121–130. doi: 10.1016/s0021-9150(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 38.Moliterno DJ. Jokinen EV. Miserez AR. Lange RA. Willard JE. Boerwinkle E. Hillis LD. Hobbs HH. No association between plasma lipoprotein(a) concentrations and the presence or absence of coronary atherosclerosis in African-Americans. Arterioscler Thromb Vasc Biol. 1995;15:850–855. doi: 10.1161/01.atv.15.7.850. [DOI] [PubMed] [Google Scholar]
- 39.Sorrentino MJ. Vielhauer C. Eisenbart JD. Fless GM. Scanu AM. Feldman T. Plasma lipoprotein (a) protein concentration and coronary artery disease in black patients compared with white patients. Am J Med. 1992;93:658–662. doi: 10.1016/0002-9343(92)90199-l. [DOI] [PubMed] [Google Scholar]
- 40.Erqou S. Thompson A. Di Angelantonio E. Saleheen D. Kaptoge S. Marcovina S. Danesh J. Apolipoprotein(a) isoforms and the risk of vascular disease: Systematic review of 40 studies involving 58,000 participants. J Am Coll Cardiol. 55:2160–2167. doi: 10.1016/j.jacc.2009.10.080. [DOI] [PubMed] [Google Scholar]
- 41.Boden-Albala B. Kargman DE. Lin IF. Paik MC. Sacco RL. Berglund L. Increased stroke risk and lipoprotein(a) in a multiethnic community: The Northern Manhattan Stroke Study. Cerebrovasc Dis. 30:237–243. doi: 10.1159/000319065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross R. Atherosclerosis—an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 43.Danesh J. Wheeler JG. Hirschfield GM. Eda S. Eiriksdottir G. Rumley A. Lowe GD. Pepys MB. Gudnason V. C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med. 2004;350:1387–1397. doi: 10.1056/NEJMoa032804. [DOI] [PubMed] [Google Scholar]
- 44.Danesh J. Lewington S. Thompson SG. Lowe GD. Collins R. Kostis JB. Wilson AC. Folsom AR. Wu K. Benderly M. Goldbourt U. Willeit J. Kiechl S. Yarnell JW. Sweetnam PM. Elwood PC. Cushman M. Psaty BM. Tracy RP. Tybjaerg-Hansen A. Haverkate F. de Maat MP. Fowkes FG. Lee AJ. Smith FB. Salomaa V. Harald K. Rasi R. Vahtera E. Jousilahti P. Pekkanen J. D'Agostino R. Kannel WB. Wilson PW. Tofler G. Arocha-Pinango CL. Rodriguez-Larralde A. Nagy E. Mijares M. Espinosa R. Rodriquez-Roa E. Ryder E. Diez-Ewald MP. Campos G. Fernandez V. Torres E. Marchioli R. Valagussa F. Rosengren A. Wilhelmsen L. Lappas G. Eriksson H. Cremer P. Nagel D. Curb JD. Rodriguez B. Yano K. Salonen JT. Nyyssonen K. Tuomainen TP. Hedblad B. Lind P. Loewel H. Koenig W. Meade TW. Cooper JA. De Stavola B. Knottenbelt C. Miller GJ. Cooper JA. Bauer KA. Rosenberg RD. Sato S. Kitamura A. Naito Y. Palosuo T. Ducimetiere P. Amouyel P. Arveiler D. Evans AE. Ferrieres J. Juhan-Vague I. Bingham A. Schulte H. Assmann G. Cantin B. Lamarche B. Despres JP. Dagenais GR. Tunstall-Pedoe H. Woodward M. Ben-Shlomo Y. Davey Smith G. Palmieri V. Yeh JL. Rudnicka A. Ridker P. Rodeghiero F. Tosetto A. Shepherd J. Plasma fibrinogen level and the risk of major cardiovascular diseases and nonvascular mortality: An individual participant meta-analysis. JAMA. 2005;294:1799–1809. doi: 10.1001/jama.294.14.1799. [DOI] [PubMed] [Google Scholar]
- 45.Uhlar CM. Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. Eur J Biochem. 1999;265:501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- 46.Rolph MS. Zimmer S. Bottazzi B. Garlanda C. Mantovani A. Hansson GK. Production of the long pentraxin PTX3 in advanced atherosclerotic plaques. Arterioscler Thromb Vasc Biol. 2002;22:e10–e14. doi: 10.1161/01.atv.0000015595.95497.2f. [DOI] [PubMed] [Google Scholar]
- 47.Lavi S. McConnell JP. Rihal CS. Prasad A. Mathew V. Lerman LO. Lerman A. Local production of lipoprotein-associated phospholipase A2 and lysophosphatidylcholine in the coronary circulation: association with early coronary atherosclerosis and endothelial dysfunction in humans. Circulation. 2007;115:2715–2721. doi: 10.1161/CIRCULATIONAHA.106.671420. [DOI] [PubMed] [Google Scholar]
- 48.Klezovitch O. Edelstein C. Scanu AM. Stimulation of interleukin-8 production in human THP-1 macrophages by apolipoprotein(a). Evidence for a critical involvement of elements in its C-terminal domain. J Biol Chem. 2001;276:46864–46869. doi: 10.1074/jbc.M107943200. [DOI] [PubMed] [Google Scholar]
- 49.Ramharack R. Barkalow D. Spahr MA. Dominant negative effect of TGF-beta1 and TNF-alpha on basal and IL-6-induced lipoprotein(a) and apolipoprotein(a) mRNA expression in primary monkey hepatocyte cultures. Arterioscler Thromb Vasc Biol. 1998;18:984–990. doi: 10.1161/01.atv.18.6.984. [DOI] [PubMed] [Google Scholar]
- 50.Bergmark C. Dewan A. Orsoni A. Merki E. Miller ER. Shin MJ. Binder CJ. Horkko S. Krauss RM. Chapman MJ. Witztum JL. Tsimikas S. A novel function of lipoprotein [a] as a preferential carrier of oxidized phospholipids in human plasma. J Lipid Res. 2008;49:2230–2239. doi: 10.1194/jlr.M800174-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.Tsimikas S. Witztum JL. The role of oxidized phospholipids in mediating lipoprotein(a) atherogenicity. Curr Opin Lipidol. 2008;19:369–377. doi: 10.1097/MOL.0b013e328308b622. [DOI] [PubMed] [Google Scholar]
- 52.Tsimikas S. Brilakis ES. Miller ER. McConnell JP. Lennon RJ. Kornman KS. Witztum JL. Berger PB. Oxidized phospholipids, Lp(a) lipoprotein, and coronary artery disease. N Engl J Med. 2005;353:46–57. doi: 10.1056/NEJMoa043175. [DOI] [PubMed] [Google Scholar]
- 53.Gargalovic PS. Imura M. Zhang B. Gharavi NM. Clark MJ. Pagnon J. Yang WP. He A. Truong A. Patel S. Nelson SF. Horvath S. Berliner JA. Kirchgessner TG. Lusis AJ. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc Natl Acad Sci USA. 2006;103:12741–12746. doi: 10.1073/pnas.0605457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anuurad E. Rubin J. Chiem A. Tracy RP. Pearson TA. Berglund L. High levels of inflammatory biomarkers are associated with increased allele-specific apolipoprotein(a) levels in African-Americans. J Clin Endocrinol Metab. 2008;93:1482–1488. doi: 10.1210/jc.2007-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Enkhmaa B. Anuurad E. Zhang W. Pearson TA. Berglund L. Association of Lp-PLA(2) activity with allele-specific Lp(a) levels in a bi-ethnic population. Atherosclerosis. 2010;211:526–530. doi: 10.1016/j.atherosclerosis.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enkhmaa B. Anuurad E. Ozturk Z. Zhang W. Pearson TA. Berglund L. Differential associations of serum amyloid A and pentraxin-3 with allele-specific lipoprotein(a) levels in African Americans and Caucasians. Trans Res. 2011 doi: 10.1016/j.trsl.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berg K. A New Serum Type System in Man–the Lp System. Acta Pathol Microbiol Scand. 1963;59:369–382. doi: 10.1111/j.1699-0463.1963.tb01808.x. [DOI] [PubMed] [Google Scholar]
- 58.Nordestgaard BG. Chapman MJ. Ray K. Boren J. Andreotti F. Watts GF. Ginsberg H. Amarenco P. Catapano A. Descamps OS. Fisher E. Kovanen PT. Kuivenhoven JA. Lesnik P. Masana L. Reiner Z. Taskinen MR. Tokgozoglu L. Tybjaerg-Hansen A. Lipoprotein(a) as a cardiovascular risk factor: current status. Eur Heart J. 2010;31:2844–2853. doi: 10.1093/eurheartj/ehq386. [DOI] [PMC free article] [PubMed] [Google Scholar]