Abstract
A 900-KB inversion exists within a large region of conserved linkage disequilibrium (LD) on chromosome 17. CRHR1 is located within the inversion region and associated with inhaled corticosteroid response in asthma. We hypothesized that CRHR1 variants are in LD with the inversion, supporting a potential role for natural selection in the genetic response to corticosteroids. We genotyped 6 single nucleotide polymorphisms (SNPs) spanning chr17:40,410,565–42,372,240, including 4 SNPs defining inversion status. Similar allele frequencies and strong LD were noted between the inversion and a CRHR1 SNP previously associated with lung function response to inhaled corticosteroids. Each inversion-defining SNP was strongly associated with inhaled corticosteroid response in adult asthma (p-values 0.002–0.005). The CRHR1 response to inhaled corticosteroids may thus be explained by natural selection resulting from inversion status or by long-range LD with another gene. Additional pharmacogenetic investigations into to regions of chromosomal diversity, including copy number variation and inversions, are warranted.
Keywords: CRHR1, tau haplotype, MAPT, inversion, asthma, corticosteroid, pharmacogenetics
Introduction
Areas of the chromosome displaying diversity such as copy number variation, duplications, and inversions continue to garner interest as potential functional regions. To date, associations of such areas with drug response have been limited to the CYP2D6 duplication[1]. Inversions are regions in which the chromosomal order is inverted compared to normal sequence; these have been associated with natural selection both within and across species[2, 3]. Recently, a 900-kb inversion on chromosome 17q21 was described[3]. Uniquely, this inversion lies in the middle of a large region (~2 megabases) of extended linkage disequilibrium (LD). This region has been used to characterize the H1 and H2 extended haplotypes associated with the microtubule-associated protein tau (MAPT) gene [3–5] and is the largest region of LD described in humans to date[3, 5]. The H2 haplotype is uniquely associated with inversion status within this region, forming a so-called “inversion polymorphism”[3] found in 20–25% of mixed European populations.
The corticotropin-releasing hormone receptor one (CRHR1) gene is also located within the chromosome 17 inversion region. We previously described an association of CRHR1 with inhaled corticosteroid response in asthma[6]. However, we have yet to identify a functional variant explaining this association. Since the inversion polymorphism frequency is similar to that of the CRHR1 variants (a haplotype and a single nucleotide polymorphism or SNP, rs1876828) associated with the pharmacogenetic responses, we hypothesized that CRHR1 would be in LD with the inversion, supporting a role for natural selection in the therapeutic response to corticosteroids. We evaluated this hypothesis in two asthma clinical trial cohorts, representing the haplotypic and SNP associations from our prior study.
Methods
Populations and Outcome Definition
CAMP was a multicenter, randomized, double-blinded clinical trial testing the safety and efficacy of inhaled budesonide vs. nedocromil vs. placebo over a mean of 4.3 years. Trial design, methodology, and primary clinical outcome have been previously published[7, 8]. Of 1,041 children with mild-moderate asthma, 311 were randomized to budesonide (an inhaled corticosteroid). Each patient’s parent or guardian signed a consent statement, with each child providing assent. IRB approval was obtained for all participating CAMP centers and the data coordinating center.
Two completed trials conducted by the ACRN, salmeterol or corticosteroids (SOCS) [9] and salmeterol ± inhaled corticosteroids (SLIC) [10], had a common initial 6-week run-in utilizing 4 inhalations twice daily of triamcinolone (another inhaled corticosteroid) prior to separate randomization to one of the trials. All patients met American Thoracic Society criteria for treatment with inhaled corticosteroids. Of subjects randomized, 336 had DNA available, forming the basis of our adult sample. All ACRN subjects analyzed provided both clinical trial and genetics studies consent.
Our primary outcome was the change in forced expiratory volume at one second (FEV1) over 6–8 weeks in response to inhaled corticosteroids:
The time intervals during which change was measured were chosen to replicate our previous experiments [6].
Genotyping
CRHR1 SNPs were genotyped via a SEQUENOM MassARRAY MALDI-TOF mass spectrometer (Sequenom, San Diego, CA), as previously described[6]. Six other chromosome 17 SNPs were genotyped by the TaqMAN 5′→3′ exonuclease assay (Applied Biosystems, Foster City, California)[11]. Of the SNPs, two (rs894685 and rs758391) were located within the region of extended LD, but outside of the inversion region[4]; four (rs1396862, rs1800547, rs9468, and rs1528072) represented both H2 haplotype and inversion status[3, 4]. Protocol details, SNP flanking sequence, and primer data are available from the authors. Duplicate genotyping performed on ~10% of the samples demonstrated <1% discordance. Genotype completion rates were ≥95% for all loci; each locus was in Hardy-Weinberg equilibrium.
Analysis
We compared the minor allele frequencies and LD patterns of the CRHR1 SNPs and haplotypes with those of the other chromosome 17 SNPs. We dichotomized haplotype assignments to “GAT”/“non-GAT” (based on imputed alleles from rs1876828, rs242939, and rs242941, respectively, per our previous report)[6]. We used an additive linear regression model, adjusting for age, gender, and baseline FEV1, to investigate change in FEV1 with chromosome 17 SNPs, limiting our analyses to Caucasian probands taking inhaled corticosteroids. Analysis of an unlinked panel of 50 markers genotyped within CAMP and ACRN found no overt evidence for population stratification (data not shown). Statistical analyses used SAS, version 8 (Cary, NC); LD was assessed using Haploview, version 3.32 (http://www.broad.mit.edu/mpg/haploview/index.php).
Results
Compared with CAMP, ACRN consisted of older asthmatics with more severe disease (Table 1). Both trials had significant variability in FEV1 response to inhaled corticosteroids, as evidenced by the large standard deviations.
Table 1.
| CAMP | ACRN | |
|---|---|---|
| N on Inhaled Corticosteroids | 201 | 224 |
| Inhaled Corticosteroid Used | Budesonide | Triamcinolone |
| Age | 8.8 ± 2.1 | 34.2 ± 11.7 |
| Sex – n (%) | ||
| - Male | 111 (55.2) | 87 (38.8) |
| - Female | 90 (44.8) | 137 (61.2) |
| Mean Baseline FEV1‡ | 94.4 ± 14.3% | 77.8 ± 16.0 % |
| Mean Change in FEV1§ | 8.7 ± 14.7% | 6.7 ± 19.8% |
Plus-minus values are means ± standard deviations
Due to concerns over possible population stratification and small numbers of subjects in other racial groups, only genotypic information from Caucasians were analyzed; their characteristics are shown
As a percent of predicted
Change in FEV1 while on inhaled corticosteriods evaluated at 8 weeks in CAMP and 6 weeks in ACRN
SNP allele frequencies and imputed GAT haplotype frequencies are shown in Table 2. One of the CRHR1 SNPs, rs1876828, demonstrated minor allele frequencies similar to the inversion SNPs. The other CRHR1 haplotype-tag SNPs and “GAT” haplotype and the two SNPs outside the inversion region varied from the inversion SNPs in frequency.
Table 2.
Allele Frequencies*
| Position† | SNP‡ | Gene | Variant | MAF - CAMP | MAF - ACRN |
|---|---|---|---|---|---|
| chr17:40410565 | RS894685 | C1QL1 | [A/G] | 0.38 | 0.30 |
| chr17:41248300 | RS242941 | CRHR1 | [G/T] | 0.30 | 0.32 |
| chr17:41251360 | RS242939 | CRHR1 | [A/G] | 0.03 | 0.07 |
| chr17:41258778 | RS1396862 | CRHR1 | [C/T] | 0.23 | 0.23 |
| chr17:41267306 | RS1876828 | CRHR1 | [G/A] | 0.22 | 0.24 |
| chr17:41407682 | RS1800547 | MAPT | [A/G] | 0.22 | 0.23 |
| chr17:41457408 | RS9468 | MAPT | [T/C] | 0.22 | 0.24 |
| chr17:41592502 | RS1528072 | LOC284058 | [C/A] | 0.22 | 0.24 |
| chr17:42372240 | RS758391 | GOSR2 | [G/A] | 0.40 | 0.43 |
CRHR1 “GAT” haplotype (based on ordered imputed alleles from rs1876828, rs242939, and rs242941) frequency 0.25 and 0.26 in CAMP and ACRN, respectively.
Position based upon version HG18 of human genome browser (http://genome.ucsc.edu)
The CRHR1 haplotype tag SNPs are italicized. SNPs known to be defining for the H2 haplotype and/or the chromosome 17 inversion region are bolded.
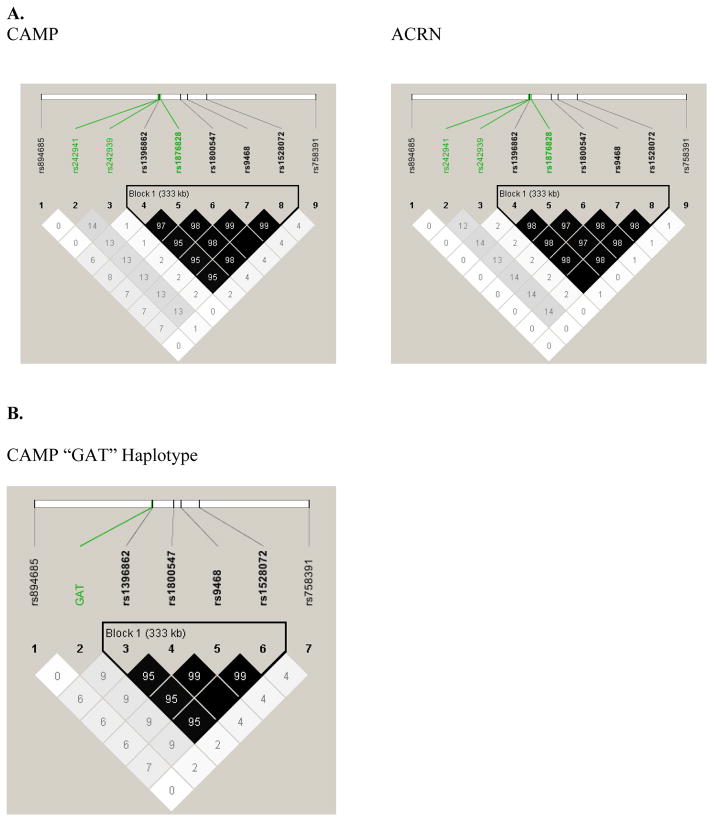
Linkage disequilibrium was evaluated on all SNPs using r2 (Figures 2a and 2b). In both populations, nearly complete LD was found for all four inversion SNPs and the CRHR1 haplotype-tag SNP (rs1876828) that had similar allele frequency to the inversion SNPs. However, little LD existed between the “GAT” haplotype or the other CRHR1 haplotype-tag SNPs and the inversion region. Differences in LD between rs1876828 and the “GAT” haplotype (of which rs1876828 is a component SNP) are likely related to other CRHR1 haplotypes involving the rs1876828 “G” allele.
The association of chromosome 17 SNPs with FEV1 response to inhaled corticosteroids is shown in Table 3. In the ACRN population, after covariate adjustment, homozygosity for the mutant allele of each inversion SNP, along with CRHR1 rs1876828, was significantly associated with an approximately three-fold increase in response (p-values 0.002–0.005). None of the non-CRHR1 CAMP SNPs demonstrated a significant association; the mean homozygous mutant inversion SNP response was approximately 1½ times that of those homozygous wild type.
Table 3.
Association of Extended LD Region SNPs with Change in FEV1 on Inhaled Corticosteroids
| Change in FEV1 on Therapy* - CAMP | Change in FEV1 on Therapy* - ACRN | |||||
|---|---|---|---|---|---|---|
| SNP† | Wildtype | Heterozygote | Mutant | Wildtype | Heterozygote | Mutant |
| RS894685 | 5.21 ± 3.93 | 11.38 ± 2.78 | 19.53 ± 7.41 | 7.97 ± 1.86 | 6.35 ± 1.98 | 7.36 ± 4.55 |
| RS242941‡ | 7.56 ± 1.49 | 8.48 ± 1.56 | 18.21 ± 3.45 | 8.52 ± 2.02 | 5.56 ± 1.82 | 12.21 ± 4.93 |
| RS242939 | 9.45 ± 1.05 | 4.68 ± 4.22 | - | 7.04 ± 1.42 | 9.52 ± 3.70 | 10.20 ± 18.36 |
| RS1396862§ | 9.37 ± 1.36 | 7.69 ± 1.63 | 12.86 ± 5.58 | 7.03 ± 1.68 | 4.69 ± 2.16 | 21.11 ± 4.69 |
| RS1876828§ | 9.31 ± 1.37 | 8.25 ± 1.71 | 13.83 ± 5.33 | 6.21 ± 1.65 | 5.54 ± 2.18 | 19.52 ± 4.31 |
| RS1800547§ | 9.58 ± 1.31 | 8.22 ± 1.65 | 13.56 ± 5.24 | 7.12 ± 1.64 | 4.90 ± 2.19 | 22.60 ± 4.52 |
| RS9468§ | 9.29 ± 1.30 | 8.59 ± 1.66 | 13.53 ± 5.23 | 6.91 ± 1.67 | 4.69 ± 2.15 | 20.45 ± 4.52 |
| RS1528072§ | 9.54 ± 1.31 | 8.20 ± 1.65 | 13.53 ± 5.23 | 6.96 ± 1.68 | 4.68 ± 2.16 | 20.50 ± 4.52 |
| RS758391 | 14.58 ± 3.11 | 4.99 ± 4.02 | 7.52 ± 4.13 | 9.38 ± 2.26 | 5.43 ± 1.83 | 9.02 ± 3.06 |
Least squares mean change, adjusted for age, gender, height, and baseline FEV1 (+/− S.E.M.)
CRHR1 haplotype tag SNPs are italicized. SNPs known to be defining for the H2 haplotype and/or the chromosome 17 inversion region are underlined.
p < 0.05 in CAMP
p ≤ 0.005 in ACRN
Discussion
We postulated that inversion status and/or long-range LD might affect pharmacogenetic response to inhaled corticosteroids in asthma, since the chromosome 17 inversion polymorphism region encompasses the CRHR1 gene. In both our populations, SNP variants within CRHR1 were indeed noted to be in tight LD with H2 inversion haplotype–defining SNPs, supporting an “inversion polymorphism” in the region. Moreover, in our adult population, each inversion SNP was associated with a marked increase in lung function response to inhaled corticosteroids. In our pediatric cohort, inversion SNPs were also associated with increases in average response to inhaled steroids, though not with statistical significance. The CRHR1 “GAT” haplotype previously associated with the pharmacogenetic response was not in significant LD with the inversion region. To our knowledge, this is the first description of an association between a chromosomal inversion and a pharmacogenetic response.
Chromosomal inversions have been associated with speciation and natural selection[2, 3]. As part of a large inversion region, the association of CRHR1 variation with response to exogenous corticosteroids may originate in developmental biology. CRHR1 is the primary receptor for corticotropin-releasing factor (CRF), a primary mediator of several hormones (including corticosteroids) critical to fetal development and timing of birth[12]. Indeed, placentally-derived CRF regulates the fetal response to stress leading to premature birth [13]. Therefore, inversion status may allow for an optimal response to stress, including increases in antenatal corticosteroid levels, leading to positive selection. Thus, a CRHR1 variant in LD with inversion status associated with enhanced corticosteroid response might be an extension of protective responses innate to fetal development.
While inversion status may influence the pharmacogenetic association, a more plausible explanation might be long-range LD of the region with CRHR1 variants. While we have yet to ascertain the functional CRHR1 variant despite resequencing (variants are posted on the PharmGKB website: www.pharmgkb.org), our adult study results suggest that the functional variant may be in LD with the H2 haplotype. In our pediatric cohort, while the CRHR1 “GAT” haplotype was not correlated with inversion status, it may be in LD with a portion of the H1 haplotype. The H1 haplotype, unlike H2, can be phylogenetically divided into multiple sub-haplotypes[3, 5]. In turn, H1 sub-haplotypes also demonstrate extensive, long-range LD, including the region containing CRHR1 [5].
Our study has several potential limitations. We did not determine the full extent of LD for either H1 or H2 haplotypic status. Instead, we focused on a limited number of SNPs representing H2 haplotype and/or inversion status. However, noting near-complete LD for inversion-defining SNPs from the literature in both of our populations in a region spanning >300 kilobases, our two populations likely have that same extensive distribution of LD. While we report similar direction of the inversion SNP results in both our cohorts, the pediatric findings were not significant and, thus, do not constitute a true replication. Although this could be due to the slightly smaller sample size, children and adults do have inherent differences in response to asthma therapy [14, 15].
In conclusion, SNPs defining chromosome 17q21.31 inversion status are associated with increased response to inhaled corticosteroids in asthma. Though inversion status may simply be a marker for the effects of extensive LD, continued investigation into associations of regions of chromosomal diversity with response to therapy are soon likely to yield novel insights in the field of pharmacogenetics.
Figure 1. LD Patterns.
Linkage disequilibrium measured as r2 between the chromosome 17 extended haplotype/inversion SNPs and CRHR1 haplotype tag SNPs (A) and “GAT” haplotype (B). There is near complete LD between the inversion SNPs, including one of the CRHR1 haplotype tag SNPs (rs1876828). The “GAT” haplotype, while of similar allele frequency to the inversion SNPs, was not in tight LD with the inversion polymorphism.
Acknowledgments
Support: This work was supported by NIH U01 HL65899, P01 HL083069, and K23 HG3983. We thank all families for their enthusiastic participation in the CAMP Genetics Ancillary Study, supported by the National Heart, Lung, and Blood Institute, NO1-HR-16049. We also acknowledge the CAMP investigators and research team, supported by NHLBI, for collection of CAMP Genetic Ancillary Study data. Additional support for this research came from grants N01 HR16044, HR16045, HR16046, HR16047, HR16048, HR16049, HR16050, HR16051, and HR16052 from the National Heart, Lung and Blood Institute. We also acknowledge the Asthma Clinical Research Network (ACRN) investigators and research teams supported by U01 HL51510, U01 HL51834, U01 HL51831, U01 HL51845, U01 HL 51843, M01 RR00079, M01 RR03186, from the NHLBI. All work on data from the CAMP Genetics Ancillary Study was conducted at the Channing Laboratory of the Brigham and Women’s Hospital under appropriate CAMP policies and human subjects protections.
References
- 1.Kirchheiner J, Henckel HB, Franke L, Meineke I, Tzvetkov M, Uebelhack R, et al. Impact of the CYP2D6 ultra-rapid metabolizer genotype on doxepin pharmacokinetics and serotonin in platelets. Pharmacogenet Genomics. 2005;15:579–87. doi: 10.1097/01.fpc.0000167331.30905.9e. [DOI] [PubMed] [Google Scholar]
- 2.Navarro A, Barton NH. Chromosomal speciation and molecular divergence--accelerated evolution in rearranged chromosomes. Science. 2003;300:321–4. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- 3.Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–37. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 4.Pittman AM, Myers AJ, Duckworth J, Bryden L, Hanson M, Abou-Sleiman P, et al. The structure of the tau haplotype in controls and in progressive supranuclear palsy. Hum Mol Genet. 2004;13:1267–74. doi: 10.1093/hmg/ddh138. [DOI] [PubMed] [Google Scholar]
- 5.Oliveira SA, Scott WK, Zhang F, Stajich JM, Fujiwara K, Hauser M, et al. Linkage disequilibrium and haplotype tagging polymorphisms in the Tau H1 haplotype. Neurogenetics. 2004;5:147–55. doi: 10.1007/s10048-004-0180-5. [DOI] [PubMed] [Google Scholar]
- 6.Tantisira KG, Lake S, Silverman ES, Palmer LJ, Lazarus R, Silverman EK, et al. Corticosteroid Pharmacogenetics: Association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004 doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- 7.The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- 8.Long-term effects of budesonide or nedocromil in children with asthma. The Childhood Asthma Management Program Research Group. N Engl J Med. 2000;343:1054–63. doi: 10.1056/NEJM200010123431501. [DOI] [PubMed] [Google Scholar]
- 9.Lazarus SC, Boushey HA, Fahy JV, Chinchilli VM, Lemanske RF, Jr, Sorkness CA, et al. Long-acting beta2-agonist monotherapy vs continued therapy with inhaled corticosteroids in patients with persistent asthma: a randomized controlled trial. Jama. 2001;285:2583–93. doi: 10.1001/jama.285.20.2583. [DOI] [PubMed] [Google Scholar]
- 10.Lemanske RF, Jr, Sorkness CA, Mauger EA, Lazarus SC, Boushey HA, Fahy JV, et al. Inhaled corticosteroid reduction and elimination in patients with persistent asthma receiving salmeterol: a randomized controlled trial. Jama. 2001;285:2594–603. doi: 10.1001/jama.285.20.2594. [DOI] [PubMed] [Google Scholar]
- 11.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′----3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A. 1991;88:7276–80. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crespi EJ, Denver RJ. Ancient origins of human developmental plasticity. Am J Hum Biol. 2005;17:44–54. doi: 10.1002/ajhb.20098. [DOI] [PubMed] [Google Scholar]
- 13.Pike IL. Maternal stress and fetal responses: evolutionary perspectives on preterm delivery. Am J Hum Biol. 2005;17:55–65. doi: 10.1002/ajhb.20093. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins HA, Cherniack R, Szefler SJ, Covar R, Gelfand EW, Spahn JD. A comparison of the clinical characteristics of children and adults with severe asthma. Chest. 2003;124:1318–24. doi: 10.1378/chest.124.4.1318. [DOI] [PubMed] [Google Scholar]
- 15.Verberne AA. Options for bronchodilation in children: can we rely on adult data? Allergy. 1999;54 (Suppl 49):51–4. doi: 10.1111/j.1398-9995.1999.tb04388.x. [DOI] [PubMed] [Google Scholar]