Abstract
Mycobacterium tuberculosis NmtR is a Ni(II)/Co(II)-sensing metalloregulatory protein from the extensively studied ArsR/SmtB family. Two Ni(II) ions bind to the NmtR dimer to form octahedral coordination complexes with stepwise binding affinities of KNi1=1.2 (±0.1) × 1010 and KNi2=0.7 (±0.4) × 1010 M-1 (pH 7.0). A glutamine scanning mutagenesis approach reveals that Asp91, His93, His104 and His107, all contained within the C-terminal α5 helix, and His3 as part of the conserved α-NH2-Gly2-His3-Gly4 motif at the N-terminus make significant contributions to the magnitude of KNi. In contrast, substitution of residues from the C-terminal region, His109, Asp114 and His116, previously implicated in Ni(II) binding and metalloregulation in cells, gives rise to wild-type KNi and Ni(II)-dependent allosteric coupling free energies. Interestingly, deletion of residues 112-120 in the C-terminal region (Δ111 NmtR) reduces the Ni(II) binding stoichiometry to one per dimer and greatly reduces Ni(II) responsiveness. H3Q and Δ111 NmtRs also show clear perturbations in the rank order of metal responsiveness to Ni(II), Co(II) and Zn(II) that is distinct from wild-type NmtR. 15N relaxation experiments with apo-NmtR reveal that both N-terminal (residues 2-14) and C-terminal (residues 110-120) regions are unstructured in solution, and this property likely dictates the metal specificity profile characteristic of the Ni(II)-sensor NmtR relative to other ArsR family regulators.
The genome of the human pathogen Mycobacterium tuberculosis (Mtb)1 encodes an impressive diversity of known and putative metal ion transporters including 12 putative P-ATPases and several ABC transporters and cation diffusion facilitator (CDF) transporters.2 This diversity might have allowed a primordial soil-dwelling ancestor of Mtb to adapt to an environment of substrate complexity.1 At the present, this diversity might enable this obligate intracellular pathogen to efficiently respond to a range of host killing mechanisms,3 collectively known as nutritional immunity.4,5
A limited number of enzymes are known that require nickel as a cofactor.6,7Mtb1 encodes a urease (Rv1848), as well as a gene encoding a hypothetical Ni(II)-containing glyoxylase I (Rv0546c) that metabolizes the electrophile methylglyoxal.8 In M. bovis BCG, thought to be a progenitor strain of Mtb,9 the catalytic product of urease, ammonia, is believed to be important in host survival as it neutralizes the pH of the phagosome, inhibits lysosomal-phagosomal fusion, attenuates the exposure of the major histocompatibility complex class II in the host cell surface, and is used for nitrogen biosynthesis.10-12 Since Mtb does not compete against its host for nickel acquisition,13-16 in contrast for other essential metals in humans including Fe,17,18 this urease-dependent function may represent an important pathogenesis determinant in Mtb. As a result, Mtb is predicted to encode complete Ni-uptake and Ni-efflux systems. Although the importance of Ni homeostasis in Mtb pathogenesis remains poorly understood, it has been reported that total nickel levels as measured by x-ray fluorescence microscopy fall precipitously inside macrophages infected with Mtb.19,20
Nickel specific transcriptional regulation of genes encoding for uptake, efflux, detoxification, sequestration and regulatory proteins21 may well be required for adaptation and survival of Mtb in the human host. Some metalloregulators, transcriptional regulators that are activated or inhibited to bind DNA in response to a metal, encoded by the Mtb genome have been previously characterized (Table S1). There are two Ni(II) specific repressors in mycobacteria and both are members of the ArsR/SmtB family of metal sensors, of 10 total ArsR family regulators encoded by M. tuberculosis.21-23 These are NmtR and KmtR, which repress the transcription of ctpJ and cdf, which encode a P-type ATPase metal transporter and a putative cation diffusion facilitator (CDF) transporter, respectively, under low Ni(II). Under conditions of high Ni(II), NmtR and KmtR bind Ni(II) and dissociate from the DNA, allowing transcription of ctpJ and cdf, the gene products of which are predicted to mediate the export of Ni(II) and Co(II) from the cytoplasm.2,24 In other organisms, complete Ni(II) homeostasis systems have been characterized. In E. coli, rcnA encodes a Ni(II)/Co(II) efflux protein25 whose expression is controlled by RcnR,26,27 a member of a recently discovered CsoR/RcnR family of regulators26,28 while nikABCDE, is a high affinity ABC-type transporter that mediates acquisition of Ni(II), and is under the transcriptional control of NikR.29-31 In the actinomycete most closely related to Mtb, Streptomyces coelicolor Nur, a Fur family member, coordinates the expression of Ni(II) vs. Fe(II) superoxide dismutase (sodN and sodF) and of nikABCDE.32,33 The nickel-uptake regulator in Mtb has not yet been identified or functionally characterized.
Several members of the ArsR family of metalloregulators have now been extensively studied. Staphylococcus aureus Zn(II) sensor CzrA is currently the best characterized family member as to its functional, structural and thermodynamic mechanism of allosteric regulation.34-36 A current hypothesis is that transition metal selectivity in the cell is dictated primarily by coordination geometry and support of this model comes from comparative studies contrasting the metal center structures of two α5 site metalloregulators of the ArsR/SmtB family, NmtR, a Ni(II)/Co(II) sensor and CzrA, a Zn(II)/Co(II) sensor (see Fig. 1 for a schematic).21 CzrA binds Zn(II) in a tetrahedral geometry via ligands derived from the C-terminal α5 helix, and thus is designated an α5 site. NmtR binds Ni(II) and Co(II) with octahedral coordination which is proposed to include the analogous core of four CzrA zinc ligands supplemented by an additional two ligands from the C-terminus (designated α5C) to complete the six-coordinate complex (Fig. 1).37 These metal coordination geometries correspond to the most common coordination geometries found for Ni(II) and Zn(II) in proteins.38 The binding of the non-cognate metal to NmtR or CzrA results in formation of a non-native coordination geometry that gives rise to greatly reduced or eliminated metalloregulation of DNA binding.37 A residue substitution mutagenesis screen of CzrA revealed that two metal ligands were necessary to maintain the native coordination geometry and Zn(II)-mediated regulation (D84, H97; Fig. 1A), while two other ligands were necessary only for maintaining high metal affinity with no or small effects on regulation (H86, H100, Fig. 1A), in what was termed a “division of labor” organization.39 As a result, it has been hypothesized that the metal coordination geometry is a primary determinant of metal selectivity for metalloregulators40,41 and this hypothesis is supported by accumulating evidence from multiple families of metalloregulators, including CsoR and NikR.42
Figure 1.

Schematic rendering of symmetry-related metal sensing sites in ArsR family protein dimers. (A) Canonical interprotomer tetrahedral Zn(II) sensing sites structurally characterized in Staphylococcus aureus CzrA.36 Only the C-terminal α5 helices of each protomer are shown and ligand donor residues are indicated. (B) Previously proposed octahedral37 Ni(II)/Co(II) sensing site of M. tuberculosis NmtR denoted α5C.22 The C-terminal α5 helices and tail are shown as are proposed ligand donor residues. (C) Alternative Ni(II) sensing site in NmtR consistent with the experimental findings outlined here, with the region N-terminal to the α5 helices (residues 2–91) simply represented as a dashed line. It is unknown if coordination of Ni(II) by His3 is intra- or interprotomer; interprotomer coordination is shown. The identity of the sixth ligand (empty tan oval) is not identified here, but a strong candidate is the α-amino group of Gly2 (see text for details).
A previous in vivo study of NmtR identified functionally important metal ligands in the α5 helix, D91, H93, H104, H107 exactly coincident with the Zn(II) regulatory site of CzrA, as well as C-terminal residues H109 and H11622 (see Fig. 1B). In the present study, we further tested this model by measuring the affinity of the nickel center and metal-mediated negative allosteric regulation of DNA binding of a panel of conservative glutamine substitution mutants, i.e., single H-to-Q and D-to-Q substitutions. These studies provide new insights into the specific contributions of the N- and C-terminal regions in NmtR relative to CzrA in maintaining Ni(II) selectivity and Ni(II) mediated regulation in Mtb. In short, we provide evidence for an N-terminal Ni(II)-binding interaction involving His3 like that described for the Ni(II)/Co(II) sensor E. coli RcnR (Fig. 1C).26 An as yet uncharacterized putative NmtR in S. coelicolor (locus tag SCO6459) also conserves the N-terminal Gly2-His3-Gly4 motif found in Mtb NmtR. In contrast, previously implicated Ni(II)-ligands H109 and H116 are not conserved (Fig. 2) and we show here that conservative substitution of each shows that neither plays an identifiable role in negative allosteric regulation of DNA binding of Ni(II) binding in vitro. The implications of these findings are discussed.
Figure 2.

Multiple sequence alignment of SmtB/ArsR family members that were obtained with NmtR as a query. The results were filtered by selecting those sequences that possess a C-terminal extension past residue 114 of NmtR. Conserved residues are blue boxed. Arrows indicate residues that were substitute by glutamines at the present work, numeration as in NmtR. Mtb (Mtb), locus tag Rv3744, Nfar (Nocardia farcinica), locus tag nfa31050; Msm (Mycobacterium smegmatis), locus tag MSMEG_5405; Kse (Kitasataspora setae), locus tag KSE_71210; Sco (Streptomyces coelicolor), locus tag SCO6459. All belong to phylum Actinobacteria.
Experimental Procedures
Protein purification
Protein purification was essentially carried out as previously.22,37 This purification scheme involves PEI precipitation of lysis supernatant, followed by two successive ammonium sulfate cuts (35 and 70%) and dissolution in 25 mM MES buffer pH 6.0, 0.1 M NaCl, 5 mM EDTA in preparation for a SP (sulfopropyl)-sepharose column. Peak fractions were pooled and subjected to Q-sepharose chromatography in 25 mM Tris pH 8.0, 0.05 M NaCl, with final polishing by a G75 preparative grade gel filtration run in 10 mM Hepes, pH 7.0, 0.2 M NaCl buffer. The G75 elution volume of all the NmtR mutants is consistent with a dimer assembly state in all cases. All mutant NmtR were shown by ESI-MS to give the expected molecular weight if one assumes that in all cases the N-terminal Met1 is processed (Table S2) to yield an N-terminal α-NH2-Gly2-His3-Gly4 sequence.
Anisotropy experiments
These binding experiments and the purification of DNA was done as previously described.43 The protein sample was diluted to a concentration of 10 μM and 2-4 μL of this stock solution was titrated into 1800 μL 10 nM dsDNA in 10 mM Hepes, pH 7.0, 0.4 M NaCl (unless otherwise noted) and allowed to equilibrate for 3-6 min. These binding reactions contained either 500 μM EDTA or 10 μM or 100 μM Ni(II), Co(II) or Zn(II) as indicated. The DNA sequence corresponds to the wild type nmt operator containing the imperfect repeat known to be the binding site (underlined)22 and is fluorescein (FL) labeled at the 3′ end (5′-GAAATAAATGAACATATGATCATATATTCT-3′-FL). For non-specific DNA, 200 ng/ml of deoxyribonucleic acid (sodium salt) from salmon testes (SIGMA) was added to the buffer, diluted by stirring in a heating block at 60 °C for 6 hrs and autoclaved for preservation. Anisotropy measurements have a standad error of ≤0.001 with the slit set to 1.0 for experiments at 0.2 M NaCl and set to 2.0 for experiments at 0.4 M NaCl. The excitation monochrometer set to λ480, instrument in L-format, longpass filter λ515 in the emission and ‘G’ factor set to 1. All the data was fit to the model described in the text using DynaFit.44 The fit accounts for a dimerization constant of Kdimer,apo=1.9 × 105 M-1 or Kdimer, Ni=4.1 × 105 M-1.37
Tyrosine fluorescence experiments
Experiments were carried out as previously described.22 In 1800 μL of 10 mM Hepes buffer, pH 7.0, 25 °C and 0.2 M NaCl, wild-type NmtR and mutant variants were diluted to 5-10 μM and fluorescence measured every ith addition of 1-4 μL of 0.5-2 mM NiCl2, waiting 3 minutes after each addition to allow for equilibration. The excitation monochrometer was set at λ260 and emission collected through monochrometer set at λ300, and 2.0 slit, with a mesh used at the emission channel when the high protein concentration is used. The average of 100 iterations for each addition was recorded. The chelators nitrilotriacetic acid (NTA) or N,N′-ethyleneglycol tetraacetic acid (EGTA) were added to a concentration of 18-21 μM from a concentrated stock solution calibrated by metal addition monitored by isothermal titration calorimetry. Metal stocks were quantified by atomic absorption as described previously.22,37
Mag-fura 2 competition experiments
NmtR variants and competitor mag-fura 2 (mf2) were diluted in 1800 μL of buffer (25 mM Hepes pH 7.0, 0.2 M NaCl and 25 °C) to the concentration indicated in the text. Aliquots of 2 μL from a 0.5–1 μM Ni(II) stock were added to the mixture, equilibrated for 3 minutes and the excitation spectra (λex 377-383, slit 0.5) recorded (λex 497, slit 1.0) using the monochrometer. The average of 30 iterations for every ith addition was recorded using an ISS PC1 instrument.
Co(II) electronic absorption spectroscopy
These experiments were carried out as previously described for wild-type NmtR.37 A sample of NmtR or NmtR variants was diluted in buffer (10 mM MES, pH 6.0, 0.2 M NaCl, at room temperature) and titrated with 2-10 μL of 100 μM–2 mM Co(II) stock solution, and the UV-visible spectra recorded following every ith addition. The apo-protein spectra was subtracted and the resulting difference spectra corrected for dilution.
Calculation of NTA, EGTA and mf2 Ni(II) affinity constants
EGTA-Ni(II) and NTA-Ni(II) affinity constants was calculated by the Schwarzenbach's α-coefficients method (eqs 1-3)
| (1) |
| (2) |
| (3) |
and for the NTA-Ni(II) equilibria the NTA-Ω factor was also used:
| (4) |
| (5) |
where Kobs is equal to the observed affinity while K′ is the actual or corrected affinity of NmtR for Ni(II).
NMR spectroscopy
NMR spectra were recorded at 37°C on Varian 600 spectrometer equipped with a cryogenic probe and were referenced to DSS. Protein samples were prepared in NMR buffer (10 mM Hepes, pH 7.0, 100 mM NaCl) at concentration of 0.5 mM l3C/15N-labeled protein NMR data processing and analysis were performed using NMRPipe45 and NMRView46. Sequence-specific resonance assignments of the NmtR homodimer were obtained with uniformly 13C/15N-labeled NmtR using standard triple resonance NMR spectroscopy: HNCA,47 HN(CO)CA,48 HNCACB,49 and CBCA(CO)NH.47
Results
Apo-NmtR is characterized by flexible N-terminal and C-terminal “tails”
A multiple sequence alignment of Mtb NmtR with other closely related ArsR/SmtB family sensors with the highest pairwise similarity to that of NmtR is shown in Fig. 2. Although none of the other putative regulators have been characterized, this alignment reveals the Gly2-His3-Gly4 sequence as well as the previously characterized α5-site metal ligands Asp91, His93, His104 and His107 are invariant; in contrast, previously implicated regulatory Ni(II) ligands His109 and His116 are not. Asp114, not previously implicating in Ni(II) sensing in vivo is in contrast conserved. Here, we evaluate the importance of each of these residues in Ni(II) binding and Ni(II) regulation in vitro employing a Gln-scanning mutagenesis approach.
Complete backbone assignments were obtained for residues 4-120 in apo-NmtR, with residues 2, 3, 91, 92, and 93 broadened beyond detection in this conformational state. The secondary structure of apo-NmtR was obtained with an analysis of the backbone chemical shifts using TALOS+,50 and these data are shown superimposed on the primary structure in Fig. 3. As expected from previous solution and crystallographic structures of related ArsR/SmtB family sensors, NmtR has five helical segments (α1-α5) and two β-strands (β1-β2) encompassing residues 17-108 arranged in an α1-α2-α3-α4 (αR)-β1-β2-α5 as expected for a winged helix protein. Residues 2-14 and 110-120 are highly mobile in solution, as revealed by chemical shifts at or near random coil values, and a value of the 1H-{15N} heteronuclear NOE which is weakly positive or negative (Fig. 3). It is interesting to note that the extreme N-terminal residues 2-4 are strongly exchanged broadened in the apo state (with His3 unassigned), in contrast to C-terminal residues.
Figure 3.
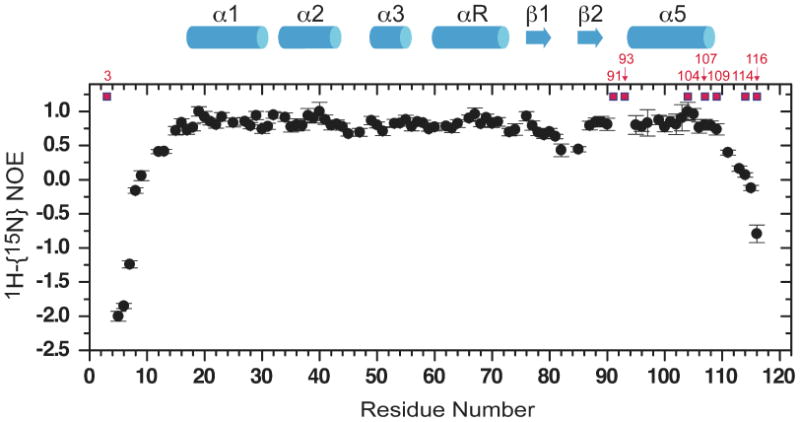
1H-{15N} heteronuclear NOE (hNOE) of apo-NmtR. The hNOEs of C-terminal two residues (residues 119 and 120) are strongly negative (≈–12) and are not shown on this plot for clarity. Secondary structural regions are α1: 17-30, α2: 33-43, α3: 49-55, α4 (αR): 60-72, β1: 76-80, β2: 85-88; α5: (94)-108 (since residues 91-93 are exchanged broadened, the actual start of the α5 helix is unknown). The residues targeted for substitution in this study are highlighted in red.
Wild-type NmtR metal binding affinity
The metal ligands were functionally identified using intrinsic fluorescence as a reporter of metal binding. On a sequence alignment based on structural homologs, three tyrosine residues (Y87, Y90, Y103) are predicted to flank the metal binding residues in NmtR (Fig. 2).21 Consistent with previous studies,37 NmtR binds Ni(II) with an concomitant 30-35% increase in the intrinsic tyrosine fluorescence, with a stoichiometry of two Ni(II) per dimer or one per protomer (Fig. 4A). Since this binding curve is essentially stoichiometric, only a lower limit of the Ni(II) binding affinity of ≥108 M-1 could be obtained. Thus, a competitor chelator was included in the binding experiments in order to extract quantitative metal stability constants. The chelators nitrilotriacetic acid (NTA)35 and N, N′-ethylene glycol tetraacetic acid (EGTA) were used for this purpose. The apparent stability of EGTA for Ni(II) corrected for the experimental pH used here (pH 7.0) is KEGTA-Ni=2.14×109 M-1 using the Schwarzenbach's α-coefficient method51 and pH-independent parameters taken from published NIST values.52 The affinity of Ni(II) for NTA was calculated (see Experimental Procedures) using the Ω-coefficient method (Ω=1.56 × 104, pH 7.0, 20 μM NTA) or the Schwarzenbach's α-coefficient (α L= 4 × 102, KNTA-Ni = 6.01 × 108 M-1, β2 = 7.7 × 1013 M-2 at pH 7.0), with both methods giving similar results.
Figure 4.

Ni(II) binding to wild-type (panels A-C) and H3Q (panels D-F) NmtRs at 5.0 μM protein monomer (2.5 μM dimer) in the absence (panels A and D) and presence of a NTA (panels B and E), EGTA (panel C) or mf2 (panel F). Ni(II) binding was monitored by a change in the intrinsic fluorescence intensity (I/Io) which was assumed to be linear function of Ni(II) occupancy. In the absence of chelator, the red line represents non-linear least-square fits to a 1:1 (Ni(II):monomer) binding model. In the presence of chelator, the red line represents non-linear least-square fits to a two-step 2:1 Ni(II):dimer binding model defined by KNi1 and KNi2, with the parameters summarized in Table 1. (A) NmtR wild-type, KNi≥8.9 × 108 M-1 (lower limit); (B) 5.0 μM NmtR wild-type and 21 μM NTA, KNi1=1.3 (±0.2) ×1010 M-1, KNi2=0.3 (±0.04) ×1010 M-1; (C) 5.0 μM NmtR wild-type and 10.0 μM EGTA, KNi1=1.1 (±0.03) ×1010 M-1, Kni2=1.1 (±0.03) ×1010 M-1 (D) H3Q NmtR, KNi∼ 1.50 × 108 M-1 (lower limit); (E) 5.0 μM H3Q NmtR and 20 μM NTA, KNi1=7. 3 (±0.8) ×108 M-1, KNi2=1.0 (±0.4) ×108 M-1; (F) 5.0 μM H3Q NmtR and 8.0 μM mf2, KNi1=1.0 (± 0.5) ×108 M-1, 1.0 (± 0.1) × 107 M-1. The dashed and dot-dashed discontinuous lines represent simulations if KNi,i values were 10-fold larger and 10-fold smaller that the fitted values, respectively.
The Ni(II) binding isotherms measured in the presence of NTA and EGTA for wild-type NmtR are very similar and each is indicative of an affinity for Ni(II) that is slightly higher than that of either competitor ligand (Fig. 4B-C). Based on the binding stoichiometry of 1:1 (metal:monomer) (Fig. 4A) and a dimeric assembly state (as determined by gel filtration in the μM range; see Experimental Procedures), we chose a Ni(II) binding model of two stepwise metal binding events (KNi1 and KNi2) to a nondissociable dimer to analyze these data. The results show that wild-type NmtR binds Ni(II) with an affinity of KNi1 ≈KNi2≈1010 M-1 (Fig. 4B-C; Table 1). Interestingly, KNi is ≈104 larger than KCo previously measured using the same approach.22
Table 1. Ni(II) binding affinities of NmtR wild-type and mutant variants.
| NmtR | KNi1(×1010 M-1) | KNi2(×1010 M-1) |
|---|---|---|
| Wild-type a | 1.15 (±0.10) | 0.70 (±0.40) |
| H3Q b | 0.042 (±0.03) | 0.001 (±0.0001) |
| D91Qc | 0.0120 (±0.0005) | 0.001 (±0.0001) |
| H93Qc | 0.0031 (±0.0001) | 0.001 (±0.0001) |
| H104Qc | 0.0004(±0.00003) | 0.001 (±0.0001) |
| H107Qc | 0.0034 (±0.0002) | 0.001 (±0.0001) |
| H109Qa | 0.75 (±0.25) | 1.00 (±0.01) |
| D114Qa | 0.76 (±0.04) | 0.60 (±0.13) |
| H116Qa | 0.77 (±0.18) | 0.55 (±0.05) |
| Δ111 NmtRa | 2.4 (±1.6) | - d |
Conditions: 10 mM Hepes pH 7.0, 0.2 M NaCl, 25 °C.
Average of KNi measured in presence of NTA or EGTA as competitor.
Average of KNi measured in presence of NTA or mf2 as competitor.
mf2 competition experiments.
not detected, i.e., stoichiometry of one per dimer
NmtR is allosterically regulated by Ni(II)>Co(II)>Zn(II)
NmtR binds to an intergenomic operator/promoter region in the absence of Ni(II), and represses the expression of a reporter gene in vivo.22 Transcription is derepressed upon addition of Ni(II) or Co(II) to the growth medium, but not by the addition of Zn(II).22 Here, we were interested in quantifying the metal affinity and allosteric response of NmtR to different metals and the extent to which single substitution mutants influence this response. Ni(II) affinities are compiled in Table 1 with DNA-binding properties summarized in Table 2. We employed an anisotropy-based experiment to measure allosteric regulation using a 30 bp fluorescein-labeled duplex DNA that harbors the quasi-perfect inverted repeat sequence to which NmtR binds.22
Table 2. NmtR wild-type DNA binding affinities for different metal complexes.
| NmtR WT | Δ rmaxa | KDNA (×109 M-1) | ΔGcb (kcal mol-1) |
|---|---|---|---|
| 1=0.2 | |||
| apoc | 0.027 | 28.6 (±0.4) | - |
| Ni(II)d | 0.027 | 1.5 (±0.1) | 1.7 (±0.04) |
| 1=0.4 and nonspecific competitore | |||
| apoc | 0.016 | 9.2 (±1.0) | - |
| Ni(II)d | 0.016 | 0.06 (±0.01) | 2.9 (±0.1) |
| 1=0.4 | |||
| apoc | 0.020 | 8.7 (±1.0) | - |
| Ni(II) f | 0.020 | 0.10 (±0.01) | 2.7 (±0.1) |
| Co(II) f | 0.020 | 0.21 (±0.03) | 2.2 (±0.1) |
| Zn(II) f | 0.027 | 1.5 (±0.2) | 1.1 (±0.1) |
Conditions: 10 mM Hepes, pH 7.0, 25 °C.
Δ rmax = maximum change in the fitted anisotropy (rcomplex – rfree).
ΔGc = -RT ln(KDNA-Ni/KDNA-apo).
For determination of apo KDNA 500 μM EDTA was added to the binding reaction.
For determination of Ni(II) KDNA 100 μM Ni(II) was added to the binding reaction.
Salmon sperm DNA.
For determination of metal KDNA 10 μM Ni(II), Co(II) or Zn(II) was added to the binding reaction.
To our surprise, Ni(II) was found to reduce the binding affinity by only ≈20-fold with respect to apo-NmtR vs. the expected 200-300 fold observed previously at higher monovalent salt concentration (0.4 M NaCl) (Table 2).37 This finding suggested that Ni(II)-NmtR forms a DNA complex characterized by significant non-specific binding interactions at 0.2 M NaCl. To test this, we measured the DNA binding affinity of NmtR for its operator at 0.4 M NaCl in the presence and absence of nonspecific DNA, in an effort to decrease the nonspecific electrostatic component of the interaction. We fit the anisotropy data to a dimer linkage model with the dimerization equilibrium constant fixed to the value previously measured37. In addition, the change in anisotropy that results from titrating apo-NmtR in to the DNA probe, (Δrmas) is 0.016 consistent with previous stoichiometric determinations of binding one dimer per DNA of similar length.34,37,53 These data (Fig. 5A; Table 2) reveal that apo-NmtR binds to DNA with a KDNA of 9.0 (±0.3) × 109 M-1 at 0.4 M NaCl independent of the presence of the competitor DNA; thus this is the specific binding affinity of apo-NmtR for its operator sequence.
Figure 5.

Allosteric negative regulation of DNA binding by various metals for (A) wild-type and (B) H3Q NmtRs. Representative data points of binding isotherms depicted by open and closed symbols for metal or apo- form respectively. The red line represents non-linear least-square fits to a model where a dissociable dimer binds to a single DNA. The total anisotropy change is normalized to the apo-NmtR value (Δrmax) so for each instance the anisotropy change observed (Δrobs) is a fraction of that for apo-NmtR. Tables 2 and 3 compile the KDNA values calculated from this analysis, along with values of Δrmax. Key to notation (as indicated in the figure): Ni(II); open circles, Co(II); diamonds, Zn(II); triangles. The data shown for metallated wild-type and H3Q NmtR were acquired in the presence of 10 μM Ni(II), Co(II) or Zn(II).
Addition of 10 μM Ni(II), sufficent to saturate all Ni(II) binding sites, shifts the DNA binding equilibrium to higher NmtR concentrations, decreasing the affinity by ≈ 100-fold to 8.0 (±2.0) ×107 M-1. The magnitude of this response is defined by the allosteric coupling free energy ΔGc,54 a quantitative measure of the degree to which metal regulates DNA binding. For wild-type NmtR ΔGc = 2.8 (±0.1) kcal mol-1, with the positive sign indicative of negative heterotropic linkage. The specific DNA binding affinity of NmtR is decreased by addition of 10 μM other metals, but the magnitude of the regulation is quantitatively different. Co(II), an inducer of the nmtA transcription in vivo,22 is characterized by a ΔGC = 2.2 (±0.1) kcal mol-1 which is followed by Zn(II), which reduces the affinity of NmtR for the DNA operator by just ≈6-fold. 10 μM Zn(II) is sufficient to fully saturate NmtR under these conditions given KZn≥108 M-1.22 10 μM Co(II) is approximately 10-fold larger than 1/KCo and is thus indicative of ≥90% saturation under these conditions.22
His3 contributes to the stability of the metal complex and its substitution alters the rank order of allosteric metal ions
Some members of the ArsR/SmtB family employ metal ligands from an N-terminal “extension” to coordinate metal to the α3 helix (α3N subfamily)21 and crystallographic studies suggest that this “tail” is not likely to be structurally organized.55 In he N-terminal domain of NmtR, the conserved residue His3 is an excellent candidate for a Ni(II) ligand (Fig. 2). This histidine is flanked to each side by Gly, forming a specific sequence (Gly-His-Gly) that can coordinate Ni(II) with one or two deprotonated main chain amides, the α-amino group and/or the side chain histidine Nδ1 or Nε2 atom.56-60 A bacterial metalloregulator thought to utilize such an arrangement is the Ni(II) efflux regulator E. coli RcnR.26
The N-terminal residue Met1 is processed during the expression of NmtR as determined by ESI-MS (Table S2) and as a result little steric clash is expected from the backbone Gly2-His3-Gly4 sequence, in the event of metal coordination. The functional importance of His3 was investigated using a Gln substitution mutant. NmtR H3Q binds Ni(II) with a stoichiometry of 1:1 (Ni(II):protomer) like wild-type NmtR; with a concomitant increase of 30% fluorescence intensity and a lower limit of KNi of ≈108 M-1 observed from direct titration in the absence of chelator (Fig. 4D). However, in the chelator competition experiments, the Ni(II) binding isotherm depicts a shape that is indicative of dramatically decreased affinity with respect to wild-type NmtR (Fig. 4E-F). The stepwise binding affinities of H3Q NmtR are reduced by ∼20 and ∼700 fold for KNi1 and KNi2 relative to wild-type NmtR (Table 1). Nevertheless, the DNA binding experiments show that apo H3Q NmtR binds DNA similarly to wild-type NmtR, with a change in anisotropy that is comparable to the wild-type NmtR (Fig. 5B, Table 3). H3Q binding to DNA is still strongly regulated by 10 μM Ni(II), and with ΔGcNi= 2.8 kcal mol-1 the same as that of wild-type protein NmtR. However, the striking finding is that, in contrast to wild-type NmtR, this mutant is significantly more sensitive to Zn(II)-mediated regulation relative to the cognate metal Co(II). These data reveal that His3 directly influences binding and Ni(II)/Co(II) selectivity of this allosteric switch; additional experiments are required to determine if other features of the N-terminus, e.g., the α-NH2 group or a main chain amide nitrogen, contribute coordination bonds to the Ni(II)26 (see Discussion).
Table 3. DNA binding affinities of H3Q NmtR with different metal complexes.
| H3Q NmtR | Δ rmaxa | KDNA (×109 M-1) | ΔGcb (kcal mol-1) | wild-type ΔGcc (kcal mol-1) |
|---|---|---|---|---|
| apod | 0.019 | 8.40 (±1.00) | - | - |
| 10 μM metale | ||||
| Ni(II) | 0.019 | 0.07 (±0.01) | 2.8 (±0.1) | 2.7 (±0.1) |
| Co(II) | 0.019 | 1.33 (±0.20) | 1.1 (±0.1) | 2.2 (±0.1) |
| Zn(II) | 0.019 | 0.35 (±0.02) | 2.0 (±0.1) | 1.1 (±0.1) |
Conditions: 10mM Hepes, pH 7.0, 0.4 M NaCl, 25 °C.
For this NmtR variant, Δrmax = 0.019 (rcomplex – rfree).
ΔGc = –RT ln(KDNA-Ni/KDNA-apo).
Wild-type NmtR values show for comparison.
For determination of apo KDNA 500 μM EDTA was added to the binding reaction.
For determination of KDNA, 100 μM metal was added to the binding reaction.
Conserved residues in the α5 helix are essential for Ni(II) binding affinity and allosteric regulation of DNA binding
Mutations in the α5 helix of metal sensors CzrA and SmtB are characterized by loss of function and/or a decrease of Zn(II) binding affinity.39 NmtR is predicted to coordinate Ni(II) with the four ligands conserved in the α5 helical region relative to the CzrA sequence (D91, H93, H104 and H107) (see Fig. 1), and on the basis of Asp-to-Ala and His-to-Arg substitution mutations, all were previously shown to be essential for regulation in an in vivo experiment.22 Here we substitute each of these residues with a non-liganding and conservative Gln to test the requirement of each for stabilization of the metal complex and allosteric regulation. An increase in intrinsic Tyr fluorescence in response to Ni(II) binding was not detected for any of these α5-site NmtR variants, suggestive of non-native structure found in each case. Therefore, the fluorescent indicator mag-fura-2 (mf2) was used instead to monitor binding of Ni(II) in competition experiments analogous to an approach used previously.26,61 Calibration of the affinity of mf2 for Ni(II) using NTA as a competitor gives an affinity Kmf2-Ni = 2.0 (±0.1) ×107 M-1 (Fig. S2) that is in reasonable agreement with the previously tabulated affinity KNi-Mf2 =7.7 × 106 M-1 measured at the same salt concentration.26,61
The results of these competition experiments with α5-helix NmtR variants (Fig. S2) are indicative of detectable competition with mf2, albeit with a pronounced reduction of KNi1 and KNi2, in each α5-site variant (Table 1). H104Q NmtR is the most compromised in nickel affinity, with KNi1 decreased by ≥10-fold relative to other missense mutants. As above, the Ni(II) dependent allosteric regulation of DNA binding was quantified by measuring DNA binding affinity in the absence and presence of 0.1 mM Ni(II), a concentration sufficient to saturate all Ni(II) regulatory sites in each case (Fig. S3, Table 4). Surprisingly, the presence of excess metal increases the value of Δrmax by two to three-fold, with the possible exception of H104Q NmtR which binds with an affinity and Δrmax similar to that of wild-type apo NmtR. These findings are perhaps indicative of a different assembly state on the DNA relative to metal-bound wild-type NmtR. Thus, not only are these mutants refractive to Ni(II)-dependent inhibition of DNA binding, but Ni(II) seems to slightly increase the DNA binding affinity, effectively reversing the sign on ΔGc if analyzed in exactly the same way (Fig. S3; Table 4).
Table 4. DNA binding affinities of α5 missense mutant NmtRs in presence of Ni(II).
| NmtR | Δrmaxa | KDNA (×109 M-1) | ΔGcb,c (kcal mol-1) | |
|---|---|---|---|---|
| D91Q | apod | 0.026 | 5.0 (±0.4) | - |
| Ni(II)e | 0.046 | 7.5 (±0.1) | –0.2 (±0.1) | |
| H93Q | apod | 0.019 | 4.2 (±0.7) | - |
| Ni(II)e | 0.050 | 16 (±3.0) | –0.8 (±0.1) | |
| H104Q | apod | 0.003 | 0.85 (±0.22) | - |
| Ni(II)e | 0.010 | 0.77 (±0.10) | –0.1 (±0.2) | |
| H107Q | apod | 0.024 | 50.0 (±1.0) | - |
| Ni(II)e | 0.043 | 24.0 (±0.5) | 0.4 (±0.01) |
Conditions: 10 mM Hepes, pH 7.0, 0.4 M NaCl, 25.0 °C.
Δrmax, anisotropy change fixed as maximal response in the fit (rcomplex – rfree) to value for wild-type NmtR.
ΔGc = –RT ln(KDNA-Ni/KDNA-apo).
For comparison ΔGc Ni(II)NmtR wild-type = 2.7 (±0.2) kcal mol-1.
For determination of apo KDNA 500 μM EDTA was added to the binding reaction.
For determination of Ni(II) KDNA, 100 μM Ni(II) was added to the binding reaction.
This observation was further explored in H104Q NmtR. The presence of other metals also appears to stabilize DNA complex formation, with Ni(II) and Co(II) the most efficient in increasing the anisotropy of the DNA in a way that is dependent on metal concentration (Fig. S4, Table S3). Interestingly, this binding has residual elements of DNA specificity since an excess of nonspecific DNA reduces the affinity of apo-H104Q NmtR only by ≈10 fold when an unrelated DNA probe is used in place of the nmt operator (Table S3).
C-terminal NmtR Gln substitution mutants are functionally silent
On the basis of arginine substitution mutants, His109 and His116 were previously reported to be essential for metal dependent regulation in vivo (Fig. 1B).22 Here we investigate Ni(II) binding affinity and metal-dependent regulation of DNA binding of conservative Gln-substitution mutants of H109, D114, H116 as well as a truncated C-terminal NmtR that deletes residues 112-120 and is designated Δ111 NmtR. For Δ111 NmtR, intrinsic fluorescence experiments conducted in absence of chelator reveal a metal stoichiometry of just one metal per dimer for Ni(II) and Zn(II), with the second binding event undetected by this method. The change in fluorescence intensity is about half that of wild-type NmtR, also consistent with occupancy at 1 of 2 sites in the dimer by Ni(II) (Fig. 6A). Interestingly, Co(II) binding as monitored by UV electronic absorption suggests a stoichiometry of ≈1.4 equivalents per monomer (Fig. 6B). The shape of these binding curves suggests that Ni(II) and Co(II) bind stoichiometrically to the remaining site(s) under these conditions, with the magnitude of KNi very similar to that of wild-type NmtR (Table 1). These results suggest a perturbation of the structural symmetry of the molecule upon deletion of most of the tail despite the fact that this region is flexible in solution (Fig. 3).
Figure 6.

Stoichiometry and affinity of Δ111 NmtR for different metals. (A) Ni(II) (circles) and Zn(II) (triangles) binding to Δ111NmtR were monitored by the intrinsic fluorescence intensity change (I/Io). The red line represents non-linear least-square fit to 0.5:1 (metal:monomer). KΔ111_NmtR-Zn(II) ≥1.4 × 108 M-1, KΔ111_NmtR-Ni(II) ≥9.1 × 107 M-1; both are lower limits given the stoichiometric nature of these binding isotherms. (B) Co(II) binding was monitored by a change in the visible absorption at 550 nm. The red line represents non-linear least-square fits to 1.5:1 (Co(II):monomer). KΔ111_NmtR-Co(II) ≥8.4 × 107 M-1.
Apo-Δ111 NmtR binds the DNA operator with an affinity comparable to that of wild-type NmtR (Fig. 7; Table 5). However, this mutant is nearly refractive to Ni(II)-mediated regulation, with the coupling free energy decreased to a level comparable to that of non-cognate metal Zn(II). Interestingly, Co(II) is the most potent regulator of Δ111 NmtR and is indistinguishable from that of Co(II)-regulation of wild-type NmtR (Table 5). One possible explanation of this finding is that the stoichiometry of metal binding may well be a primary determinant of regulation in this mutant rather than the precise identity of the residues in the tail. To test this, we prepared single Gln substitution mutants H109Q, D114Q and H116Q. To our surprise, each of the single C-terminal mutant variants binds stoichiometric Ni(II) (Fig. S5) and the chelator competition experiments reveal a Ni(II)-binding affinity that is indistinguishable from wild-type NmtR (Fig. S6 and Table 1). The Ni(II)-dependent negative allosteric regulation of DNA by these variants are also wild-type-like at 100 μM added Ni(II) (Fig. 8 and Table 6).
Figure 7.

Δ111 NmtR is nearly refractive to Ni(II)-dependent regulation but not to Co(II) regulation. Fluorescence anisotropy experiments were used to monitor DNA binding of the truncated variant. Open and closed symbols represent metal or apo- form, respectively. The red line represents non-linear least-square fits to a model where a dissociable dimer binds to a single DNA with the parameters complied in Table 5. Apoprotein, filled circles; Ni(II), open circles; Zn(II), triangles; Co(II), diamonds. The experiments were performed in 100 μM metal.
Table 5. DNA binding affinities of Δ111 NmtR complexed with various metals ions.
| Δ111 NmtR | Δ rmaxa | Δ robsb | KDNA (×109 M-1) | ΔGcc (kcal mol-1) | Wild-type ΔGcd (kcal mol-1) |
|---|---|---|---|---|---|
| apoe | 0.014 | 0.013 | 5.6 (±1.0) | - | |
| 100 μM metal | |||||
| Ni(II) | 0.012 | 1.00 (±0.02) | 1.0 (±0.1) | 2.7 (±0.1) | |
| Co(II) | 0.005 | 0.12 (±0.01) | 2.4 (±0.1) | 2.2 (±0.1) | |
| Zn(II) | 0.024 | 0.020 | 1.20 (±0.20) | 0.9 (±0.1) | 1.1 (±0.1) |
Conditions: 10 mM Hepes, 0.4 M NaCl, 25.0 °C.
Δrmax = measured anisotropy maximal response (rcomplex – rfree).
Δrobs = observed anisotropy change at highest protein concentration used in the analysis.
ΔGc = –RT ln(KDNA-Ni/KDNA-apo).
Wild-type NmtR values shown for comparison.
For determination of apo KDNA 500 μM EDTA was added to the binding reaction.
For determination of KDNA 100 μM metal was added to the binding reaction.
Figure 8.
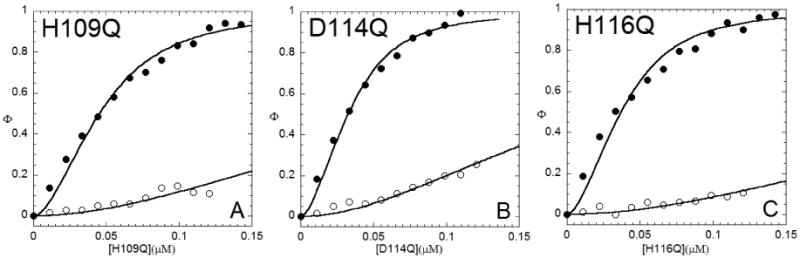
Ni(II) dependent allosteric regulation of DNA binding by C-terminal variant (A) H109Q NmtR, (B) D114Q NmtR, and (C) H116Q NmtR. Shown are binding isotherms monitored by a change in the fluorescence anisotropy of the DNA. Titrations carried out in the in presence (open symbols) or absence (closed symbols) of 100 μM Ni(II) are shown. The red line represent non-linear least-square fits to a model where a dissociable dimer binds to a DNA operator. The data are normalized to the maximum anisotropy change (Δrmax) that corresponds to apoprotein in each case. Table 6 complies the KDNA values calculated from this analysis, along with values of Δrmax.
Table 6. DNA binding affinities of C-terminal NmtR variants.
| NmtR | Δrmaxa | Δrobsb | KDNA (×109 M-1) | ΔGcc,d (kcal mol-1) |
|---|---|---|---|---|
| H109Q | ||||
| apoe | 0.014 | 0.013 | 4.3 (±0.6) | - |
| Ni(II)f | 0.014 b | 0.001 | 0.04 (±0.01) | 2.8 (±0.2) |
| D114Q | ||||
| apoe | 0.020 | 0.019 | 10.0 (±0.2) | - |
| Ni(II)f | 0.020 | 0.005 | 0.08 (±0.004) | 3.0 (±0.03) |
| H116Q | ||||
| apoe | 0.017 | 0.017 | 7.3 (±0.1) | - |
| Ni(II)f | 0.017 | 0.002 | 0.02 (±0.002) | 3.5 (±0.1) |
Conditions: 10 mM Hepes, pH 7.0, 0.4 M NaCl, 25.0 °C.
Δrmax = measured anisotropy maximal response (rcomplex – rfree).
Δrobs = observed anisotropy change at highest protein concentration used in the analysis.
ΔGc = –RT ln(KDNA-Ni/KDNA-apo).
For comparison ΔGc Ni(II)NmtR wild-type = 2.7 (±0.2) kcal mol-1
For determination of apo KDNA 500 μM EDTA was added to the binding reaction.
For determination of Ni(II) KDNA 100 μM Ni(II) was added to the binding reaction.
UV-visible absorption spectroscopy of Co(II)-substituted NmtR variants
The UV-visible absorption spectrum of Co(II)-substituted wild-type NmtR is consistent with a five- or six-coordinate Co(II)-NmtR complex37,62 (Fig. S7A). Although all mutant NmtRs bind Co(II) with a stoichiometry of 1.0-1.5 per protomer, the spectrum obtained for each saturated complex differs from wild-type NmtR to varying degrees. H3Q NmtR (Fig. S7B) has a decreased optical absorptivity and is characterized by a spectrum that most closely resembles that of filling of the second site in the wild-type dimer. H109Q NmtR (Fig. S7D) is also strongly perturbed, with D114Q and H116Q NmtRs (Fig. S7E-F) most similar to that of wild-type NmtR. The spectrum of Co(II)-substituted Δ111 NmtR is most strongly perturbed of any mutant, characterized by two maxima at 475 and 570 nm, and a significantly increased molar absorptivity of ε570 = 140 M-1 cm-1 (Fig. S7C); this is suggestive of a reduction in the coordination number in this mutant. It is interesting to note that this Δ111 NmtR is characterized by the strongest Co(II)-dependent regulation of DNA binding of any NmtR (Table 5).
Discussion
The structural basis of metal selectivity by a family of metal sensor proteins, individuals of which respond to distinct metal inducers, is strongly dictated by the detailed structure of the first metal coordination shell and is thus of intrinsic interest.21,42 In this work, we employ a glutamine-scanning mutagenesis approach with the Ni(II) sensor NmtR to investigate cognate Ni(II) vs. Co(II) regulation and how the sensing site in NmtR has evolved to functionally discriminate against a highly competitive metal like Zn(II).42,63 Biochemical and structural studies of α5 prototype Zn(II) sensing members from the ArsR/SmtB family, e.g. SmtB and CzrA,36,62 reveal that two Zn(II) bind to a pair of 2-fold symmetric tetrahedral sites that straddle the C-terminal α5 helices of the dimerization domain (Fig. 1). Previous studies established a model of metal selectivity based primarily on coordination geometry since binding of the noncognate metal Ni(II) to CzrA to form a non-native octahedral coordination geometry abolishes allosteric negative regulation of protein-DNA interactions. Likewise, Zn(II) is poorer allosteric regulator of NmtR and was shown to adopt a tetrahedral coordination geometry.37 In support of this model, evaluation of single substitution mutants of the zinc sensor CzrA revealed 1:1 correspondence between the ability of a mutant sensor to adopt a native-like tetrahedral coordination geometry and a near-wild-type allosteric coupling free energy irrespective of metal affinity, KMe.37 In the cellular milieu of course, KMe is predicted to have a substantial impact on metal sensing, since this value may well “tune” a sensor to detect changes in metal availability over a physiological range.42
In this work, we corroborate and significantly extend previous observations22 and show that the affinity of NmtR for Ni(II) in its octahedral center37 is on the order of 1010 M-1, a value approximately between that of two other characterized Ni(II) metalloregulators. E. coli NikR binds Ni(II) in a square planar geometry, coordinated by 3 His and 1 Cys with a KNi= 1012 M-1 64 while its functional counterpart, RcnR is reported to bind Ni(II) with an octahedral geometry with a correspondingly lower affinity ≈108 M-1.26 In RcnR, the Ni(II) is modeled to be bound by a Cys and one His at the periphery of what is anticipated to be a four-helix bundle, the side chain of His3, the α-NH2 group of residue 2, and a main chain amide and a solvent molecule.26 Co(II), a metal inducer of RcnR in vivo, also adopts an octahedral coordination geometry with Cys as a ligand donor, but this residue is only essential for Co(II) regulation in vivo, not Ni(II).26 This suggests that although both metals are sensed by RcnR, the functional determinants of the coordination structure may not be identical in each case.
KmtR is the second Ni(II) sensor from the ArsR/SmtB family that is expressed in Mtb cytoplasm.24 KmtR binds Ni(II) with an as yet uncharacterized coordination geometry, although mutagenesis experiments suggest that the Ni(II) is also six-coordinate with a mixture of carboxylate and imidazole ligands.24 In an experiment designed to estimate the relative affinities of KmtR and NmtR for Ni(II), it was shown that the former has a ≥100-fold higher affinity for Ni(II).24 Thus, based on the binding affinity reported here for NmtR, KmtR should bind Ni(II) with an affinity of ≥1012 M-1. The presence of two regulators in the same cytoplasm makes it possible to sense a metal at distinct concentration ranges (sensitivities) that allows for a graded metal response.24 For example, Mtb encodes two structurally homologous Cu(I) sensors that regulate distinct sets of genes in response to Cu(I) stress at apparently different extracellular Cu(I) regimes.65,66 It is expected that in vivo de-repression will be observed only after Ni(II) concentrations have reached their activation threshold, which for NmtR may be two-three orders of magnitude higher concentration than for KmtR.
The allosteric negative heterotropic linkage that characterizes Ni(II) NmtR (ΔGc≈2.8 kcal mol-1) is of a magnitude similar to that of other ArsR/SmtB members including the α3N Cd(II) sensor CadC,67 and α3N Zn(II) site of Cu(I)/Zn(II) sensor BxmR,68 which recruit one or more ligands from the N-terminal region to coordinate their respective metals. However, this allosteric energy coupling is small relative to the paradigm α5-site zinc sensor CzrA, ΔGc ≈6 kcal mol-1.35,39 The origin of this difference is not yet known but strongly suggests that the structural mechanism may well differ between CzrA and NmtR as anticipated based on their distinct metal specificity profile. All of the anticipated metal ligands in the α5 helix are necessary, but not sufficient, to establish a native-like metal site structure and metal response profile, and each is required to drive allosteric negative regulation of DNA operator binding. These observations establish a functional difference with the α5 Zn(II) sensing CzrA, where conservation of just two of the four metal ligands is sufficient for maintenance of tetrahedral complex and the Zn(II)-mediated allosteric switching mechanism.39 A novel observation is that the metallated NmtR α5 variants seem to stabilize the formation of higher order protein-DNA complexes, which may be indicative of metal mediated stabilization of an allosterically impaired dimeric interface.69,70
Our characterization of H3Q NmtR provides strong support for the proposal that His3 in the N-terminus is a direct ligand to the Ni(II) ion since KNi is greatly decreased in this mutant (Fig. 1C). However, provided sufficient Ni(II) is present, H3Q NmtR is characterized by a wild-type ΔGc. The striking finding is that the primary role of His3 may be to prevent regulation of DNA binding by the non-cognate metal Zn(II), since mutagenesis of His3 allows NmtR to recover substantial Zn(II)-responsiveness in vitro. H3Q NmtR also exhibits an attenuated Co(II) absorption spectrum and poor Co(II)-dependent allosteric regulation, consistent with the idea that the Co(II) complex may also be strongly functionally dependent on His3 for wild-type activity than Ni(II). There is precedent for distinct coordination complexes found by pairs of cognate metals, e.g. Co/Ni in E. coli RcnR.26 On the other hand, Co(II) binds much more weakly than Ni(II) (KCo≈106 M-1) to NmtR22 and loss of His3 may simply destabilize the complex to a degree such that not all binding sites are saturated at 10 μM Co(II) (Table 3). All we can conclude is that XAS reveals a Ni(II) center in NmtR containing a mixture of histidines and other as yet unidentified N/O ligands.37 If His3 is indeed a ligand to the Ni(II) ion, this would be a unique structural future among the ArsR/SmtB members containing a core α5 metalloregulatory site (Fig. 1). Since this N-terminal extension adopts a dynamic, random coil conformation in the absence of Ni(II), it is clearly of sufficient length to reach the C-terminal α5 helix of the opposite protomer.53 The structure of Zn(II)-bound CzrA suggests that less than 9 Å will separate the residues analogous to Pro14 and His107 in NmtR.34
If indeed this is the case, a plausible mechanism of allosteric regulation may involve a reorganization of the N-terminal tail which may be in direct contact with the DNA in the bound apoprotein complex, e.g, Arg8 and Arg10. In fact, in the solution structure of CzrA in complex with DNA, residues in the N-terminus of the α1 helix contact the intervening minor groove of consecutive major grooves which harbor 5′-TGAA recognition elements.34 Although little structural information is available for members of the α3N family in complex with DNA, other metalloregulators including Helicobacter pylori NikR71,72 and H. pylori Fur73,74 possess a flexible N-terminal “arm” that modulates specific DNA interactions.
Our results fail to provide direct support for the involvement of His109 and His116 in the C-terminal tail as directly involved in metal binding and allosteric regulation, in contrast to the conclusions reached on the basis of the cellular nickel responsiveness of H109R and H116R NmtRs;22 these studies also showed that D114A NmtR exhibited wild-type Ni(II) regulation in vivo. This discrepancy may be attributed to a number of possible explanations and structural studies will be required to discriminate among them; in any case, they point out the potential shortcomings of assigning function to a residue on the basis of a single substitution. The insertion of a bulky and positively charge arginine residue near the metal binding site could compete for binding of the metal ion (steric inhibition) and/or create an electrostatic repulsive interaction (charge repulsion). Interesting, such a precedent exists in the case of an arginine substitution mutant of a non-Ni(II) ligand in E. coli RcnR that was found to abrogate Ni(II) sensing in vivo.26 The second possibility is the recruitment of a non-native ligand from multiple ligands donors available, similar to what is believed to happen in other metal sensors SmtB and RcnR.26,75
The C-terminal tail is however required to maintain a Ni(II) binding stoichiometry of 2:1 as well as a native coordination geometry; indeed, the Δ111 NmtR truncation mutant is characterized by a decreased metal mediated allosteric regulation (ΔGc = 1 kcal mol-1) relative to wild-type NmtR. A Zn(II) sensor CzrA (fCzrA) was previously engineered by the fusion of the N- and C-terminal domains by a flexible peptide inter-dimeric linker, with the objective of measuring the effects of the perturbations in the dimeric interface on allosteric regulation.53 In a fCzrA variant where the shortest linker was used and the strongest perturbation in the dimer interface is expected, binding of only one Zn(II) per dimer was detected, analogous to the Δ111NmtR. However this fCzrA variant is competent to Zn(II) regulation, in contrast to the results show for Ni(II) Δ111 NmtR. However, this truncation mutant binds Co(II) with four or five ligands and a stoichiometry of up to three metals per dimer which efficiently negatively regulates DNA binding (ΔGc = 2.4 kcal mol-1) to an extent similar to that observed for the Co(II)-complex in wild-type NmtR.
These data taken collectively suggest the C-terminal tail functions indirectly in Ni(II)/Co(II) sensing by NmtR, and effectively controls the stoichiometry and coordination geometry of the Ni(II) and Co(II) complexes to varying degrees, but is unlikely to donate a primary coordination bond to the regulatory Ni(II). This conclusion is consistent with the sequence alignment. Indeed the tail appears essential for Ni(II) regulation, but less so for Co(II)-regulation. The regulatory Ni(II) complex in NmtR is characterized by features of a N-terminal Ni(II)-binding motif that may be structurally similar to the “nickel hook” motif in Ni(II) superoxide dismutase (SodN)58 and inferred from the mutagenesis studies of E. coli RcnR.26 High resolution structural studies are required to provide further support for what may be a unique metalloregulatory model in ArsR-SmtB family sensors.21,23
Supplementary Material
Acknowledgments
This work has been submitted by H. R.-C. to Texas A&M University in partial fulfillment of the requirements for the Ph.D. in Biochemistry.
Abbreviations used are
- ArsR
arsenic repressor
- CsoR
copper-sensitive operon repressor
- CzrA
chromosomally-encoded zinc-regulated repressor
- Mtb
Mycobacterium tuberculosis
- EGTA
N,N′-ethyleneglycol tetraacetic acid
- Fur
iron uptake regulator
- mf2
mag-fura-2
- NikR
nickel-responsive regulator of the nik operon
- NmtR
nickel-dependent Mycobacterium tuberculosis repressor
- NTA
nitrilotriacetic acid
- Nur
nickel uptake regulator
- PEI
polyethylenimine
- RcnR
resistance to cobalt and nickel repressor
- SmtB
Synechococcus metallothionein
Footnotes
This work was supported by a grant from the NIH (GM042569) to D. P. G.
Supporting Information. Supplementary Tables S1-S3 and Supplementary Figures S1-S7 are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 2.Agranoff D, Krishna S. Metal ion transport and regulation in mycobacterium tuberculosis. Front Biosci. 2004;9:2996–3006. doi: 10.2741/1454. [DOI] [PubMed] [Google Scholar]
- 3.Brosch R, Gordon SV, Eiglmeier K, Garnier T, Tekaia F, Yeramian E, Cole ST. In: Genomics, Biology, and Evolution of the Mycobacterium tuberculosis Complex, in Molecular Genetics of Mycobacteria. Hatfull GF, Jacobs WR Jr, editors. ASM Press; Washingron, D.C.: 2000. pp. 19–36. [Google Scholar]
- 4.Weinberg ED. Iron availability and infection. Biochim Biophys Acta. 2009;1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulrooney SB, Hausinger RP. Nickel uptake and utilization by microorganisms. FEMS Microbiol Rev. 2003;27:239–261. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 7.Doukov TI, Iverson TM, Seravalli J, Ragsdale SW, Drennan CL. A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Science. 2002;298:567–572. doi: 10.1126/science.1075843. [DOI] [PubMed] [Google Scholar]
- 8.Sukdeo N, Clugston SL, Daub E, Honek JF. Distinct classes of glyoxalase I: metal specificity of the Yersinia pestis, Pseudomonas aeruginosa and Neisseria meningitidis enzymes. Biochem J. 2004;384:111–117. doi: 10.1042/BJ20041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith NH, Gordon SV, de la Rua-Domenech R, Clifton-Hadley RS, Hewinson RG. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nature Rev Microbiol. 2006;4:670–681. doi: 10.1038/nrmicro1472. [DOI] [PubMed] [Google Scholar]
- 10.Clemens DL, Lee BY, Horwitz MA. Purification, characterization, and genetic analysis of Mycobacterium tuberculosis urease, a potentially critical determinant of host-pathogen interaction. J Bacteriol. 1995;177:5644–5652. doi: 10.1128/jb.177.19.5644-5652.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sendide K, Deghmane AE, Reyrat JM, Talal A, Hmama Z. Mycobacterium bovis BCG Urease Attenuates Major Histocompatibility Complex Class II Trafficking to the Macrophage Cell Surface. Infect Immun. 2004;72:4200–4209. doi: 10.1128/IAI.72.7.4200-4209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baena A, Porcelli SA. Evasion and subversion of antigen presentation by Mycobacterium tuberculosis. Tissue Antigens. 2009;74:189–204. doi: 10.1111/j.1399-0039.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoi K, Uthus EO, Nielsen FH. Nickel deficiency diminishes sperm quantity and movement in rats. Biol Trace Elem Res. 2003;93:141–154. doi: 10.1385/BTER:93:1-3:141. [DOI] [PubMed] [Google Scholar]
- 14.Denkhaus E, Salnikow K. Nickel essentiality, toxicity, and carcinogenicity. Crit Rev Oncol Hematol. 2002;42:35–56. doi: 10.1016/s1040-8428(01)00214-1. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen FH. Nutritional requirements for boron, silicon, vanadium, nickel, and arsenic: current knowledge and speculation. FASEB J. 1991;5:2661–2667. [PubMed] [Google Scholar]
- 16.Nielsen FH. Ultratrace elements of possible importance for human health: an update. Prog Clin Biol Res. 1993;380:355–376. [PubMed] [Google Scholar]
- 17.Agranoff DD, Krishna S. Metal ion homeostasis and intracellular parasitism. Mol Microbiol. 1998;28:403–412. doi: 10.1046/j.1365-2958.1998.00790.x. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez GM, Smith I. Mechanisms of iron regulation in mycobacteria: role in physiology and virulence. Mol Microbiol. 2003;47:1485–1494. doi: 10.1046/j.1365-2958.2003.03384.x. [DOI] [PubMed] [Google Scholar]
- 19.Wagner D, Maser J, Moric I, Boechat N, Vogt S, Gicquel B, Lai B, Reyrat JM, Bermudez L. Changes of the phagosomal elemental concentrations by Mycobacterium tuberculosis Mramp. Microbiology. 2005;151:323–332. doi: 10.1099/mic.0.27213-0. [DOI] [PubMed] [Google Scholar]
- 20.Wagner D, Maser J, Lai B, Cai Z, Barry CE, III, Honer zu Bentrup K, Russell DG, Bermudez LE. Elemental Analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing Phagosomes Indicates Pathogen-Induced Microenvironments within the Host Cell's Endosomal System. J Immunol. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- 21.Ma Z, Jacobsen FE, Giedroc DP. Coordination Chemistry of Bacterial Metal Transport and Sensing. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavet JS, Meng W, Pennella MA, Appelhoff RJ, Giedroc DP, Robinson NJ. A nickel-cobalt-sensing ArsR-SmtB family repressor. Contributions of cytosol and effector binding sites to metal selectivity. J Biol Chem. 2002;277:38441–38448. doi: 10.1074/jbc.M207677200. [DOI] [PubMed] [Google Scholar]
- 23.Osman D, Cavet JS. Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat Prod Rep. 2010;27:668–680. doi: 10.1039/b906682a. [DOI] [PubMed] [Google Scholar]
- 24.Campbell DR, Chapman KE, Waldron KJ, Tottey S, Kendall S, Cavallaro G, Andreini C, Hinds J, Stoker NG, Robinson NJ, Cavet JS. Mycobacterial Cells Have Dual Nickel-Cobalt Sensors. J Biol Chem. 2007;282:32298–32310. doi: 10.1074/jbc.M703451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigue A, Effantin G, Mandrand-Berthelot MA. Identification of rcnA (yohM), a Nickel and Cobalt Resistance Gene in Escherichia coli. J Bacteriol. 2005;187:2912–2916. doi: 10.1128/JB.187.8.2912-2916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwig JS, Leitch S, Herbst RW, Maroney MJ, Chivers PT. Ni(II) and Co(II) Sensing by Escherichia coli RcnR. J Am Chem Soc. 2008;130:7592–7606. doi: 10.1021/ja710067d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwig JS, Rowe JL, Chivers PT. Nickel homeostasis in Escherichia coli -the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol Microbiol. 2006;62:252–262. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu T, Ramesh A, Ma Z, Ward SK, Zhang LM, George GN, Talaat AM, Sacchettini JC, Giedroc DP. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 29.De Pina K, Desjardin V, Mandrand-Berthelot MA, Giordano G, Wu LF. Isolation and Characterization of the nikR Gene Encoding a Nickel-Responsive Regulator in Escherichia coli. J Bacteriol. 1999;181:670–674. doi: 10.1128/jb.181.2.670-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chivers PT, Sauer RT. Regulation of high affinity nickel uptake in bacteria. Ni2+-Dependent interaction of NikR with wild-type and mutant operator sites. J Biol Chem. 2000;275:19735–19741. doi: 10.1074/jbc.M002232200. [DOI] [PubMed] [Google Scholar]
- 31.Schreiter ER, Wang SC, Zamble DB, Drennan CL. NikR-operator complex structure and the mechanism of repressor activation by metal ions. Proc Natl Acad Sci U S A. 2006;103:13676–13681. doi: 10.1073/pnas.0606247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn BE, Cha J, Lee EJ, Han AR, Thompson CJ, Roe JH. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol Microbiol. 2006;59:1848–1858. doi: 10.1111/j.1365-2958.2006.05065.x. [DOI] [PubMed] [Google Scholar]
- 33.An YJ, Ahn BE, Han AR, Kim HM, Chung KM, Shin JH, Cho YB, Roe JH, Cha SS. Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition. Nucl Acids Res. 2009;37:3442–3451. doi: 10.1093/nar/gkp198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arunkumar AI, Campanello GC, Giedroc DP. Solution structure of a paradigm ArsR family zinc sensor in the DNA-bound state. Proc Natl Acad Sci U S A. 2009;106:18177–18182. doi: 10.1073/pnas.0905558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grossoehme NE, Giedroc DP. Energetics of allosteric negative coupling in the zinc sensor S. aureus CzrA. J Am Chem Soc. 2009;131:17860–17870. doi: 10.1021/ja906131b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, Sacchettini JC, Giedroc DP. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol. 2003;333:683–695. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Pennella MA, Shokes JE, Cosper NJ, Scott RA, Giedroc DP. Structural elements of metal selectivity in metal sensor proteins. Proc Natl Acad Sci U S A. 2003;100:3713–3718. doi: 10.1073/pnas.0636943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuppuraj G, Dudev M, Lim C. Factors Governing Metal-Ligand Distances and Coordination Geometries of Metal Complexes. J Phys Chem B B. 2009;113:2952–2960. doi: 10.1021/jp807972e. [DOI] [PubMed] [Google Scholar]
- 39.Pennella MA, Arunkumar AI, Giedroc DP. Individual metal ligands play distinct functional roles in the zinc sensor Staphylococcus aureus CzrA. J Mol Biol. 2006;356:1124–1136. doi: 10.1016/j.jmb.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 40.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Micro. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 41.Chen PR, He C. Selective recognition of metal ions by metalloregulatory proteins. Curr Opin Chem Biol. 2008;12:214–221. doi: 10.1016/j.cbpa.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 42.Reyes-Caballero H, Campanello GC, Giedroc DP. Metalloregulatory Proteins: Metal selectivity and allosteric switching. Biophys Chem. 2011;156:103–114. doi: 10.1016/j.bpc.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reyes-Caballero H, Guerra AJ, Jacobsen FE, Kazmierczak KM, Cowart D, Koppolu UMK, Scott RA, Winkler ME, Giedroc DP. The Metalloregulatory Zinc Site in Streptococcus pneumoniae AdcR, a Zinc-activated MarR Family Repressor. J Mol Biol. 2010;403:197–216. doi: 10.1016/j.jmb.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 45.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;5:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 47.Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J Magn Reson. 1992;96:432–440. [Google Scholar]
- 48.Bax A, Ikura M. An efficient 3D NMR technique for correlating the proton and 15N backbone amide resonances with the alpha-carbon of the preceding residue in uniformly 15N/13C enriched proteins. J Biomol NMR. 1991;1:99–104. doi: 10.1007/BF01874573. [DOI] [PubMed] [Google Scholar]
- 49.Wittekind M, Mueller L. HNCACB, a high-sensititity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonaces in proteins. J Magn Reson. 1993;101:201–205. [Google Scholar]
- 50.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perrin DD, Dempsey B. Buffers for pH and Metal Ion Control. 2nd. Chapman and Hall; New York: 1979. [Google Scholar]
- 52.Martell AE, Smith RM, editors. NIST Standard Reference Database 46 Version 8 0. NIST; Gaithersburg, MD: 2003. [Google Scholar]
- 53.Lee S, Arunkumar AI, Chen X, Giedroc DP. Structural Insights into Homo- and Heterotropic Allosteric Coupling in the Zinc Sensor S. aureus CzrA from Covalently Fused Dimers. J Am Chem Soc. 2006;128:1937–1947. doi: 10.1021/ja0546828. [DOI] [PubMed] [Google Scholar]
- 54.Giedroc DP, Arunkumar AI. Metal sensor proteins: nature's metalloregulated allosteric switches. Dalton Trans. 2007;7:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- 55.Ye J, Kandegedara A, Martin P, Rosen BP. Crystal Structure of the Staphylococcus aureus pI258 CadC Cd(II)/Pb(II)/Zn(II)-Responsive Repressor. J Bacteriol. 2005;187:4214–4221. doi: 10.1128/JB.187.12.4214-4221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farkas E, Sovago I, Gergely A. Studies on transition-metal peptide complexes. 8 Parent and mixed-ligands complexes of histidine-containing dipeptides. Journal of the Chemical Society-Dalton Trans. 1983:1545–1551. [Google Scholar]
- 57.Sovago I, Osz K. Metal ion selectivity of oligopeptides. Dalton Trans. 2006:3841–3854. doi: 10.1039/b607515k. [DOI] [PubMed] [Google Scholar]
- 58.Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA, Getzoff ED. Nickel Superoxide Dismutase Structure and Mechanism. Biochemistry. 2004;43:8038–8047. doi: 10.1021/bi0496081. [DOI] [PubMed] [Google Scholar]
- 59.Herbst RW, Guce A, Bryngelson PA, Higgins KA, Ryan KC, Cabelli DE, Garman SC, Maroney MJ. Role of conserved tyrosine residues in NiSOD catalysis: a case of convergent evolution. Biochemistry. 2009;48:3354–3369. doi: 10.1021/bi802029t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryan KC, Johnson OE, Cabelli DE, Brunold TC, Maroney MJ. Nickel superoxide dismutase: structural and functional roles of Cys2 and Cys6. J Biol Inorg Chem. 2010;15:795–807. doi: 10.1007/s00775-010-0645-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Golynskiy MV, Gunderson WA, Hendrich MP, Cohen SM. Metal binding studies and EPR spectroscopy of the manganese transport regulator MntR. Biochemistry. 2006;45:15359–15372. doi: 10.1021/bi0607406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.VanZile ML, Chen X, Giedroc DP. Structural characterization of distinct α3N and α5 metal sites in the cyanobacterial zinc sensor SmtB. Biochemistry. 2002;41:9765–9775. doi: 10.1021/bi0201771. [DOI] [PubMed] [Google Scholar]
- 63.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 64.Wang SC, Dias AV, Zamble DB. The “metallo-specific” response of proteins: a perspective based on the Escherichia coli transcriptional regulator NikR. Dalton Trans. 2009:2459–2466. doi: 10.1039/b818167p. [DOI] [PubMed] [Google Scholar]
- 65.Ward SK, Hoye EA, Talaat AM. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J Bacteriol. 2008;190:2939–2946. doi: 10.1128/JB.01847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Festa RA, Jones MB, Butler-Wu S, Sinsimer D, Gerads R, Bishai WR, Peterson SN, Darwin KH. A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol Microbiol. 2011;79:133–148. doi: 10.1111/j.1365-2958.2010.07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Busenlehner LS, Weng TC, Penner-Hahn JE, Giedroc DP. Elucidation of primary (α3N) and vestigial (α5) heavy metal-binding sites in Staphylococcus aureus pI258 CadC: Evolutionary implications for metal ion selectivity of ArsR/SmtB metal sensor proteins. Journal of Molecular Biology. 2002;319:685–701. doi: 10.1016/S0022-2836(02)00299-1. [DOI] [PubMed] [Google Scholar]
- 68.Liu T, Chen XH, Ma Z, Shokes J, Hemmingsen L, Scott RA, Giedroc DP. A CuI-sensing ArsR family metal sensor protein with a relaxed metal selectivity profile. Biochemistry. 2008;47:10564–10575. doi: 10.1021/bi801313y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


