Abstract
Chromosome condensation is established and maintained by the condensin complex. The mechanisms governing loading of condensin onto specific chromosomal sites remain unknown. To elucidate the molecular pathways that determine condensin binding to the nucleolar organizer, a key condensin binding site, we analyzed the properties of condensin-bound sites within the rDNA repeat in budding yeast and demonstrated that the bulk of mitotic condensin binding to rDNA is reduced or eliminated when Pol I transcription is elevated. Conversely, when Pol I transcription is repressed either by rapamycin treatment or by promoter shut-off, condensin binding to rDNA is increased. This novel potential role for constitutive and/or periodic repression of Pol I transcription in rDNA condensin loading is an important factor in determining the segregation proficiency of NOR-containing chromosomes.
Keywords: condensin, nucleolus, transcription, chromosome segregation, chromosome condensation, RFB, FOB1
INTRODUCTION
The condensin complex drives chromosome condensation in Eukaryota and Prokaryota, thus facilitating sister chromatid compartmentalization and faithful chromosome segregation during cell division. While nematodes and yeast have a single condensin complex, vertebrates and plants have two condensin complexes: condensin I and condensin II.1 Eukaryotic condensin complexes have similar structures and are composed of five conserved subunits. These subunits are Smc2p and Smc4p, which form a heterodimer, and three auxiliary subunits, Brn1p, Ycs4p and Ycs5p in budding yeast,2 that bind to the globular (ATPase) domains of Smc2p and Smc4p.3,4 Dysfunction of any condensin subunit results in impaired mitotic and meiotic chromosome segregation.2,5–10 While condensin can positively supercoil DNA in vitro in the presence of ATP,11 the actual mechanism of chromosome condensation in vivo is not understood.
Eukaryotic condensin complexes are distributed over the length of mitotic chromosomes with a specific pattern.6,7,12–14 In higher eukaryotes, condensin is enriched in the nucleoli only in interphase, with notable distinct dynamics for condensin I and II, while in S. cerevisiae, the nucleolar organizer (NOR) has been identified as a main target of mitotic condensin activity.2 Condensin binding dramatically increases in the nucleolar area in mitosis2 and particularly in anaphase.15 This enrichment is a reflection of an essential role condensin plays in the mitotic segregation of the rDNA-containing chromosome XII2,15–18 and of any other chromosome in which rDNA repeats have been inserted.2 However, in contrast to higher eukaryotes and fission yeast, where condensin defects lead to missegregation of all chromosomes, the functional role of budding yeast condensin binding to non-NOR chromosomal sites14 remains uncharacterized.
The single rDNA repeat in budding yeast includes all genes for ribosomal RNAs, with 5S gene transcribed by Pol III and the rest by Pol I as a large 35S precursor. In addition to the transcribed portions and their regulatory sequences rDNA also includes functional sites crucial for nucleolus formation and stable maintenance: replication fork block (RFB), origin of replication and amplification control elements.19 S. cerevisiae presents a unique opportunity to dissect the functional interface between the features of rDNA chromatin and condensin activity. First, it is possible to rearrange/homogenize the tandemly repeated NOR locus, so that all rRNA genes are episomal, and thus amenable to mutation analysis in cis.20–22 Second, it is possible to manipulate the copy number23,24 and transcription25,26 of tandem chromosomal rDNA repeats. Finally, multiple conserved rDNA elements19 required for nucleolar maintenance have been identified in budding yeast, so they can be tested for a potential role in the recognition of the nucleolar chromatin sites by condensin. Existing studies on condensin function in rDNA do not address the role that NOR architecture plays in the nucleolar function of condensin. In this report we identify the mechanism behind the significant role condensin plays in rDNA segregation and propose a role for transcriptional heterogeneity of individual rDNA repeats27,28 in NOR segregation.
MATERIALS AND METHODS
Yeast culture and genetic methods
Yeast culture conditions were as described.29,30 Yeast strains are shown in Table 1. For G1 arrest, cells were treated with 5 μM alpha factor (US Biological). After 3 h incubation, up to 95% cells were arrested. For mitotic arrest, cells were synchronized in G1, alpha factor was washed off and cells were released into medium with 15 μg/ml nocodazole (Sigma) for 2.5 hr. After a 2-hr incubation, 99% cells achieved mitotic arrest, as determined by cell morphology and FACS. For time course experiments, cdc15-ts cells were arrested in late-mitotic phase after incubation for 3 h at the nonpermissive temperature (37°C), and then released by shifting to 23°C. Samples were taken every 20 min for FACS, ChIP and transcript analysis.
Table 1.
S. cerevisiae strains
| Strain | Genotype |
|---|---|
| W303 | MATa ade2 leu2 can1 his3 trp1 ura3 |
| 532-SLJ127 | MATa ade2 bar1 can1 cdc15-2 his3 leu2 trp1 ura3 SMC2:6His:3HA::LEU2 |
| 733-W303 | MATa ade2 can1 his3 leu2 trp1 ura3 smc2-8::LEU2 |
| 532-TAK300 | MATa ade2 can1 his3 leu2 trp1 ura3 20-RDN1 SMC2:6His:3HA::LEU2 |
| 532-W303 | MATa ade2 can1 his3 leu2 trp1 ura3 SMC2:6His:3HA::LEU2 |
| 532-W303-1 | MATa ade2 can1 his3 leu2 trp1 ura3 fob1Δ|::TRP1 SMC2:6His:3HA::LEU2 |
| 640-3-AS426 | MATα cdc15-1 his leu2 ura3 SMC4:GFP::URA3 |
| 1cAS452 | MATa cdc15-1 leu2 trp1 ura3 BRN1:6His:3HA::URA3 |
| 532-NOY891/pNOY353 | MATa ade2 ura3 leu2 his3can1rdn1-Δ|::HIS3 SMC2:12His:3HA::LEU2//TRP1 pGAL7:RDN |
| 640-NOY396s | MATα ade2 his 3 trp1 ura3 leu2 fob1Δ|::TRP1 |
| 640-NOY891s/pRDNwt | MATa ade2 can1 his3 leu2 ura3 rdn1-Δ|::HIS3 SIK1:mRFP::kanMX SMC4:GFP::URA3/TRP1 leu2-d rDNA |
| 532-NOY891/pRDNwt | MATa ade2 can1 his3 leu2 ura3 rdn1-Δ|::HIS3 SMC2:6His:3HA::LEU2/TRP1 leu2-d rDNA |
| YP377 | MATa ade2 his3 leu2 trp1 ura3 rdn1-Δ|/URA3 rDNA32 |
| 733-YP377 | MATa ade2 his3 leu2 trp1 ura3 rdn1-Δ| smc2-8::LEU2/URA3 rDNA |
| TAK300E | MATa ade2 can1 his3 leu2 trp1 ura3 RDN1 |
For rapamycin (200 nM) and cycloheximide (25 mg/ml) treatment, cdc15-ts cells were arrested for 2.5 hr at 37°C, drugs were added to cultures for 30 min, and cells were then released to 23°C in the presence of drugs. Samples were harvested every 30 min for microscopy (Smc4p-GFP and Sik1p-mRFP) or ChIP/qPCR (Smc2p-HA). All cultures contained DMSO (1μg/ml) to mimic the rapamycin solvent.
For glucose shut-off of 35S rRNA transcription from pGAL7: RDN, the ErDNA strain 532-NOY891/pNOY353 was cultured in YPGal medium and arrested in mitosis with nocodazole. Cells were spun down, washed twice with fresh YPGal medium and released into the fresh YPD medium. A time course ChIP analysis was performed with Smc2p-HA after cells were transferred to YPD.
Flow cytometry analysis (FACS) was done as following: yeast cells (1 ml, OD600 of 0.3–0.4) were pelleted and resuspended in 300 μl of 50 mM Tris pH 7.5 and 700 μl of EtOH, repelleted and resuspended in 100 μl RNAse (10mg/mL in 50 mM Tris pH 7.5) for 2 hr at 37°C. The cell suspension was treated with 5 μl of proteinase K (15 mg/ml) for 60 min at 50°C. The suspension was mixed with 900 μl of 15 μg/ml propidium iodide, sonicated and analyzed with a BD FACSCalibur System (BD Biosciences).
The smc2–8 allele marked with LEU2 was introduced into strains via SMC2 gene replacement as in.14 FOB1 disruption was done by transformation with fob1-Δ::TRP1or fob1-Δ::HIS3PCR products made using specific FOB1-targeting primers. All integration/disruptions were verified by diagnostic PCR.
rDNA plasmids were as following: pRDNwt was from,20 pNOY353 was from,22 pBW1065 was constructed by inserting the rDNA repeat (PstI-BamHI fragment) from pNOY37322 into pRS416 cut with NsiI and BamHI. YP377 strain and derivatives contain unnamed 2-micron-based URA3 rDNA plasmid as in.32 Minichromosome loss rates were quantified as in,14,31 except cells were shifted to 37°C for 3 h before plating.
Microscopy
Still image microscopy was performed with a wide-field AxioVert (Zeiss) microscope with epifluorescence equipped with a cooled CCD camera. Twenty 0.2 μm-step Z-axis frames were captured for each field. Prior to microscopy, cells were briefly (5 min) fixed with 4% paraformaldehyde, washed with PBS and kept on ice. Live cells time-lapse microscopy was done with Olympus inverted IX2-SP microscope equipped with the Perkin Elmer Ultra View ERS Rapid Confocal Imager (“spinning disc”). For prolonged imaging cells were placed on gelatine slab as described.33 Twenty eight 0.2 μm-step Z-axis frames were captured for each timepoint (1 min). Volocity software package was used for image deconvolution (with fine setting) and 3D rendering. The SMC4:GFP fusion/replacement (pLF640 plasmid integration) was as described.2 The SIK1:mRFP fusion/replacement was constructed as in.15 The pMET:SCC3:GFP plasmid was constructed using the p413Met vector.
Molecular methods
ChIP and quantitative real time PCR (qPCR) analyses were performed as described.14 For Pol I transcript analysis, 10 ml of culture were harvested at each time point, pelleted and resuspended in 400 μl of AE buffer (50 mM Na acetate, pH 5.3, 10 mM EDTA). Cells were transferred to a 1.5 ml microcentrifuge tube, 40 μl of 10% SDS was added with mixing. An equal volume of phenol was added with mixing, and tubes were incubated for 4 minutes at 65°C. Tubes were then rapidly chilled in dry ice/ethanol bath until phenol crystals appeared, and were centrifuged for 2 minutes at 18000g at room temperature. The aqueous phase was transferred to a new tube, and an equal volume of phenol/chloroform/isoamyl alcohol (24:24:1) was added, mixed by vortexing, and spun for 5 minutes at 18000 g at room temperature. The aqueous phase was transferred to a new tube, mixed with 1/10 volume of 3 M Na Acetate (pH 5.3), 2.5 volumes of ethanol, and then ethanol-precipitated. The RNA pellet was washed with 80% ethanol, dried, and resuspended in 20 μl diethylpyrocarbonate-treated H2O.
Quantitative RT-PCR reactions were performed using a Stratagene MX3000P Real-time PCR System. RT-PCR reactions (50 μl) contained 1μl template RNA (0.2–0.5 μg/μl), 25 μl 2x SYBR QRT-PCR master mix (Stratagene), and 50nM primers. One-step RT-PCR parameters were 1 cycle of 50°C for 30 min, 1 cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min. Primer concentrations and PCR cycle parameters was optimized to eliminate formation of primer dimers. Fluorescence values, Ct values and all quantifications were obtained using the MX3000P software. TUB2 transcript levels were used as internal control for normalization.
Psoralen crosslinking was as in,34 with modifications. Yeast cells were collected, washed and resuspended in TE buffer. 1.5 ml of the cell suspension was UV-irradiated (366nm) in open dishes (3.3 cm diameter) on ice in the presence of 4,5',8-trimethylpsoralen (Sigma) for 125 min, with additional psoralen addition four times every 25 min. Crosslinked cells were washed and resuspended in 1M sobitol, 40 mM potassium phosphate pH 7.5, 20 mM β-mercaptoethanol with 0.1 mg/ml zymolase, then incubated for 30 min at 37°C. Spheroplasts were washed, lysed in TE buffer with 0.5% SDS and treated with proteinase K (1 mg/ml) for 3 h at 50°C. Phenol/chlorophorm extracted DNA was treated with RNAse and digested with EcoRI for 10 h. with extra enzyme added after 5 h. After separation in agarose gel electrophoresis DNA was irradiated for 20 min with UV (254nm) to reverse the crosslinks and the 2.8 rDNA fragment was detected by Southern blotting.
RESULTS
Condensin occupancy of rDNA sites depends on Pol I transcription levels
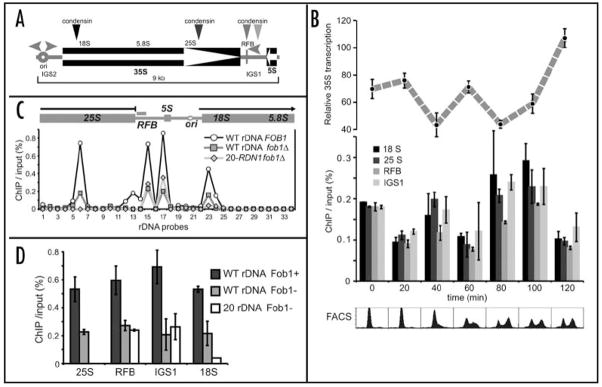
Two of condensin binding sites in rDNA2,15 are positioned within the Pol I transcribed rRNA gene and two other in the close proximity to the region of Pol I transcription termination, next to the RFB (Fig. 1A). While in mammalian cells Pol I transcription is active during S and G2 phases, low in G1 and shut off in mitosis,35 in budding yeast it continues throughout the cell cycle.36 To investigate how condensin binding can cohabit with strong transcription we determined condensin enrichment at all four hotspots by ChIP in a cdc15-synchronized cell population in parallel with 35S transcription analysis by real-time RT-PCR (RT-qPCR) (Fig. 1B). Condensin was mildly enriched in S-phase (40 min point) with an additional increase in mitosis (80–100 min). The four condensin peaks showed similar cell cycle profiles of condensin association, independent of their being in the transcribed or non-transcribed part of rDNA. However, condensin occupancy was inversely correlated with the level of Pol I transcription at a given timepoint (Fig. 1B). This suggests that rDNA transcription level is related to condensin binding to rDNA, particularly in mitosis, when condensin binding there reaches its maximum.2,15 As active transcription is not compatible with stable binding of many multisubunit complexes to the transcribed regions,37–39 condensin must either directly compete for binding sites with transcription machinery or bind to repeats that are not transcribed. The latter could be accomplished as about half of rDNA repeats in wild type cells are not transcribed.34,40,41 These transcriptionally silent repeats could thus be specialized for segregation of the transcriptionally active nucleolus in yeast.
Figure 1.
Condensin binding in the NOR is controlled by rDNA transcription and Fob1p. (A) Schematic of an rDNA repeat. Positions of transcripts (35S and 5S) and intergenic sequences (IGS) are shown in relation to the replication fork block (RFB) and origin of replication (ori). Horizontal arrowheads denote DNA replication, vertical arrowhead correspond to condensin binding hotspots. (B) Cell cycle analysis of Smc2-HA binding to rDNA in relation to Pol I transcription. Smc2p-HA ChIP data are from qPCR analysis of Smc2p-ChIP samples from cdc15-2 SMC2:HA cells (532-SLJ127 strain). Quantitative real-time RT-PCR was used to determine Pol I transcription levels. Samples were taken every 20 min after release from arrest at 37°C. Primers pair spanning the 18S RNA 5' processing site were used for RT-PCR analysis to avoid detecting mature ribosomal RNA. RT-PCR values were normalized to TUB2 transcript levels to account for cell growth during the time course (data not shown). Error bars are standard deviations of multiple determinations. Cell cycle progression was monitored by DNA flow cytometry (lower panel), nuclear division and budding indices (not shown). Entry in mitosis was detected 80 min after release from the arrest. (C) ChIP/qPCR analysis of Smc2p-HA binding to the rDNA repeat in fob1Δ 20-RDN1 (532-TAK300), fob1Δ with wild type rDNA (532-W303-1) and wild type strain (532-W303). Cells were arrested in G2/M by nocodazole. Primers are as in.58 (D) FOB1 dependence of Smc2p-HA binding at the four rDNA hotspots, analyzed by ChIP/qPCR. Hotspot peaks correspond to those shown in (A and B). Error bars are standard deviations from three independent experiments.
If Pol I transcription and condensin binding are inversely correlated, then condensin binding to the NOR should be reduced if Pol I transcription from every repeat were maximized and transcriptionally silent rDNA repeats were eliminated. Testing this prediction requires a strain where the number of rDNA repeats is reduced to a minimum, so that every repeat is transcribed.23 However, such cells with reduced rDNA copy number undergo rapid expansion of repeats by amplification, unless the FOB1gene is inactivated.42 We therefore analyzed the condensin binding to rDNA by ChIP in a fob1Δ strain, which contains 20 rDNA repeats (20-RDN1),23 in contrast to the approximately 150 rDNA repeats present in wild type.42 In the 20-RDN1 strain we observed a near-complete elimination of condensin from the transcribed area of rDNA and a marked reduction in condensin occupancy at nontranscribed sites, as compared to the isogenic wild type (Fig. 1C). To address a possible contribution of FOB1 deletion in this reduction we also assayed a fob1Δ strain with the normal-size rDNA locus (Fig. 1C). Surprisingly, fob1Δ itself reduced condensin occupancy, not only at the RFB site (where Fob1p binds DNA) but at all four hotspots. We conducted several additional experiments assaying condensin binding to four hotspots in Fob+, Fob1− and 20-repeat/Fob1− cells. These assays confirmed that fob1 deletion in the strain with a wild type rDNA repeat number caused a 2 to 3-fold reduction in condensin binding at all four loci, while the 20-repeat strain displayed a further reduction in condensin binding to the point of elimination, but only at transcribed sites (Fig. 1D). This differential reduction in condensin occupancy at rDNA in fob1DELTA cells and in the 20-repeat strain was reflected in the different degree of their sensitivity to condensin dysfunction. In a 20-RDN1 strain, a fob1Δ smc2-8 double mutant is inviable, while smc2-8 fob1Δ double mutants are viable with the wild-type rDNA array (data not shown). These results indicate that Fob1p and transcription of rDNA both have a role in regulating condensin occupancy in the nucleolus. Fob1p acts as a facilitator of condensin targeting to NOR in general, and elevated transcription levels reduce condensin binding to the transcribed regions. However these two regulatory pathways are not entirely independent, as fob1-Δ itself has a direct effect on Pol I transcription: rDNA transcription is elevated in the fob1-Δ cells (Supplement 1).
Repression of rDNA transcription increases condensin binding to the NOR
While the data in Figure 1 suggest an inverse correlation between condensin binding to rRNA-encoding regions and their transcription, these studies either involved analyses of a transcriptionally heterogeneous rDNA array (Fig. 1B) or were done in a fob1Δ background (Fig. 1C and D). In order to verify the dependence of condensin binding on downregulation of rDNA transcription without these interfering factors, we conducted experiments where Pol I transcription levels were artificially modulated, and the effect on condensin binding to rDNA was examined.
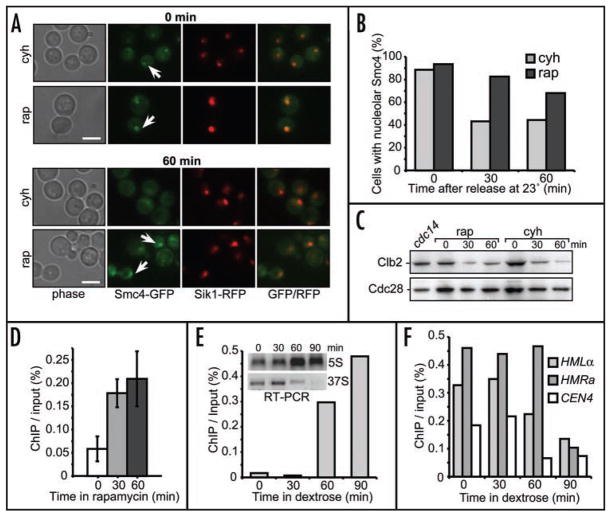
We first asked if repressing transcription of the 35S rRNA precursor would increase condensin binding to rDNA. We used rapamycin, an inhibitor of the TOR pathway,43,44 to repress rDNA transcription, and assayed the relative condensin enrichment at rDNA under these conditions. To avoid potential indirect interference of repressing ribosomal RNA gene transcription with the complex pathway of condensin loading onto rDNA at the onset of mitosis, we monitored the release of condensin from the nucleolus, which normally occurs at the exit from mitosis.2,15 In cells released from the cdc15-2 arrest in the absence of drug, condensin (Smc4p-GFP) appears to exit the nucleolus within minutes after release (data not shown). In rapamycin treated cells condensin is still concentrated in the nucleolus of most cells as late as one hour after release from the arrest (Fig. 2A and B). This retention persists despite the fact that most cells exit mitosis, as determined by the decrease in Clb2p levels (Fig. 2C). By contrast, in cdc15-2/SMC4:GFP cells treated with cycloheximide (used here as a control to offset a possible rapamycin effect on translation45) it takes 30 min for Smc4p-GFP diffusion from the nucleus to occur in 50% of cells (Fig. 2B). Even in cells counted as retaining Smc4p in the nucleus the signal is much more diffuse (Fig. 2A) than in rapamycin-treated cdc15-2/SMC4:GFP cells. Retention of condensin in the post-mitotic nucleolus in the rapamycin-treated cells was further confirmed by quantitative ChIP at the 25 S rRNA site (Fig. 2D). Moreover, this higher-resolution analysis reveals that condensin binding is actually increased in rapamycin-treated cells (Fig. 2D). These results indicate that rDNA transcription, which is inhibited by rapamycin but not by cycloheximide, is needed to remove condensin from rDNA.
Figure 2.
Reducing Pol I transcription causes condensin accumulation in the nucleolus. (A) Retention of nucleolar condensin in rapamycin-treated cells. The test strain (640-3-AS426) contains a cdc15 ts allele for synchronization, a SIK1-RFP fusion as a nucleolar marker,48 and the SMC4-GFP to allow condensin detection. For the time (0) panel the cells were imaged after a 2.5 hr arrest at 37°C, with the last 30 min in the presence of rapamycin or cycloheximide.45 Arrows point to the nucleolar Smc4p-GFP signal. The same cells were then imaged 1hr after shift from 37°C to 23° in the presence of rapamycin or cycloheximide. Arrows point to the nucleolar Smc4p-GFP signal still present in rapamycin-exposed cells. Both rapamycin and cycloheximide-treated cells failed to enter the next cell cycle (data not shown). (B) Quantitative analysis of nucleolar Smc4p-GFP signal for the experiment in (A). A minimum of 200 cells were scored. (C) Both rapamycin and cycloheximide-treated cells exit mitosis. Western blotting for protein extracts corresponding to timepoints in (A) was probed for Clb2p degradation with specific antibodies. Anti-PSTAIRE antibodies (Cdc28p) were used for protein loading control. (D) ChIP/qPCR analysis of condensin binding to the 18S rRNA gene site in rapamycin-treated cells. The cdc15ts BRN1-HA strain (1cAS452)15 was used to prepare ChIP under conditions as in (A). (E) Condensin enrichment at rDNA upon transcription shut-off. Chromatin extracted from cells expressing the 35S rRNA precursor under control of the GAL7 promoter (532-NOY891/pNOY353) was analyzed for Smc2p-HA binding to the 18S site (Fig. 1A) by ChIP/qPCR. Cells were arrested at mitosis in the presence of galactose and released into glucose-containing medium; samples were collected at 0, 30, 60 and 90 minutes after release. Insert—agarose gels showing products of RT-PCR for the 5S RNA (which is expressed from its own promoter) and the 35S rRNA precursor which is expressed from the GAL7 promoter. (F) Condensin depletion from chromosomal sites upon rDNA transcription shut-off. ChIP samples from (E) were analyzed for condensin binding at HM loci and peri-CEN4 site, hotspots showing dynamic changes in condensin enrichment.14
To complement the rapamycin experiments, we used cells where the 35S precursor is transcribed only from a galactose-controlled Pol II (pGAL7) promoter.22 In such cells, upon dextrose addition, which rapidly represses transcription from pGAL7, condensin occupancy at the 25S rRNA site increases significantly with time to almost 50-fold over the level seen in galactose-containing media (Fig. 2E). The increase in condensin binding is greater than that normally observed in the wild type mitosis, when condensin relocalizes from some non-rDNA sites to the nucleolus.14 This suggests that increased availability of binding sites in rDNA upon transcription shutoff (Fig. 2E) may drain some condensin from other sites. Indeed, three sites known to undergo cell-cycle changes in condensin occupancy14 are depleted for condensin binding under these conditions (Fig. 2F). These results show that transcription of rRNA genes should be an important factor in regulating condensin binding to rDNA chromatin as well as elsewhere in the genome.
Condensin function in NOR segregation is regulated by rDNA transcription
Assaying the relationship between rDNA transcription, condensin binding and the segregation proficiency of the nucleolus is technically difficult in the wild-type rDNA array, due to above-mentioned need to rely on fob1 mutants to maintain a low copy number of rDNA repeats. In addition, in fob1Δ cells, due to RFB elimination, replication timing of rDNA changes as origin activation remains at the wild type level.46 We therefore explored a system where rDNA repeats are homogenized by placing a single repeat on a multicopy plasmid (episomal rDNA, ErDNA) and deleting the native rDNA locus.20,22 In such a system, the link between Pol I transcription, condensin binding/function and the rDNA segregation fidelity can be readily tested. These strains display a more diffuse condensin localization and a less prominent condensin enrichment in the nucleolar area in mitosis (Fig. 3A). However, as the nucleolus itself becomes more diffuse21 it is difficult to determine whether condensin binding to rDNA actually changes by cytological criteria alone. qPCR analysis showed that in these cells the rDNA copy number decreases to 58% of wild type, while rRNA levels remain the same as in wild type (Fig. 3B). This indicates that in ErDNA cells, the amount of rRNA produced from a repeat is at least two-fold higher than in wild type (Fig. 3B). Psoralen crosslinking34 experiment also shows that higher fraction of rDNA repeats is open for transcription/crosslinking in ErDNA cells and hence gives rise to more DNA with reduced electrophoretic mobility (Fig. 3C). As the repeats are individualized (i.e., not in a tandem array) in ErDNA strains it is likely that every repeat has equal chances of being transcribed, unlike in wild type rDNA where Pol I transcription occurs only in unsilenced repeats that are open for transcription.40,47
Figure 3.
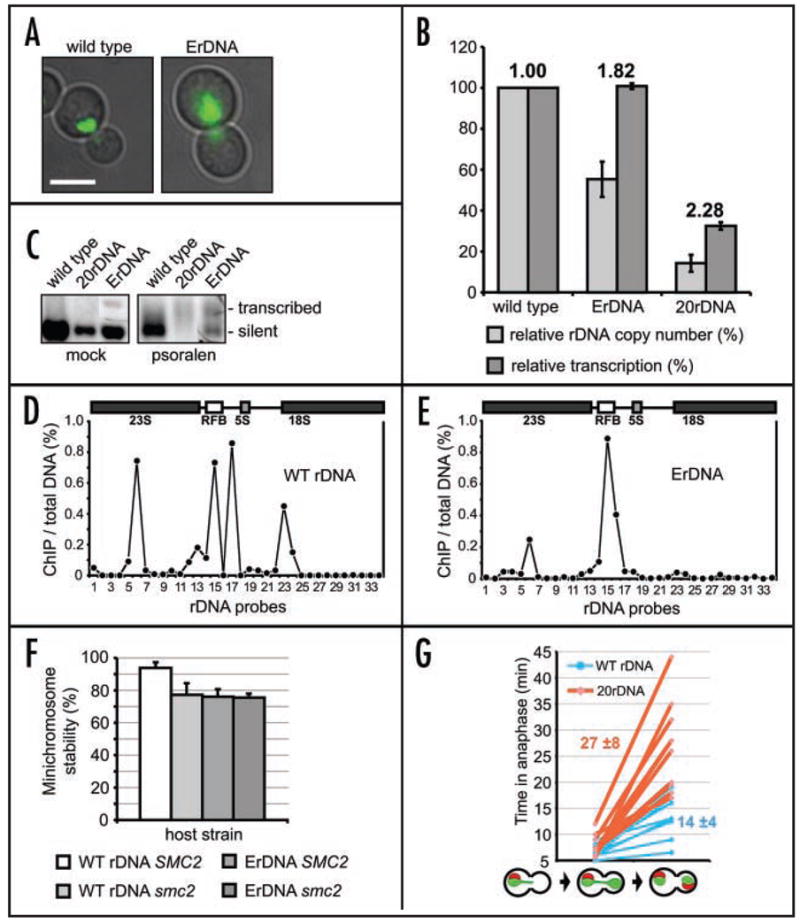
Competition between condensin binding and Pol I transcription results in reduced segregation fidelity. (A) Comparison of condensin localization in ErDNA cells and wild type. Smc4p-GFP localization (green fluorescence) is shown in representative mitotic cells with wild type rDNA (640-NOY396) and ErDNA cells (640-NOY891/pRDNwt). Cells were grown asynchronously and imaged as described above. The cell cycle stage was identified by nucleus position and bud size. Scale bar 5 μm. (B) Increased ratio between Pol I transcription and rDNA copy number in ErDNA cells. Relative rDNA copy number (normalized to wild type rDNA strain NOY396) and relative levels of Pol I transcription (also normalized to NOY396) are shown for ErDNA cells (YP377 strain). The 20-repeat rDNA strain (TAK300) analysis is shown as a reference. Indexes above the histograms represent relative increase of transcription per rDNA repeat, compared to wild type. (C) Increased Pol I transcription in ErDNA cells. Psoralen crosslinking and Southern blotting with the rDNA probe (2.8 kb EcoRI fragment) were used as in34 to detect the ratio between transcribed (heavily crosslinked slow mobility band) and silent (lightly crosslinked faster mobility band) rDNA repeats. To facilitate comparison between the strains the conditions were chosen so that crosslinking is minimal in wild type rDNA. Strains are as in (B). The 20 repeat rDNA data shown as a reference for fully transcribed rDNA. (D) (E) ChIP/qPCR analysis of condensin binding to rDNA. Cells with wild type rDNA (532-W303) in (D) and ErDNA (532-NOY891/pRDNwt) in (E) were arrested with nocodazole and processed for Smc2p-HA ChIP/qPCR analysis. Primers/probes are as in (Fig. 1C).58 (F) Mitotic stability of rDNA minichromosome as a function of condensin proficiency and rDNA organization in smc2-8 and ErDNA cells. The URA3 rDNA minichromosome (pBW1065) stability was analyzed as in14 in wild type (W303), smc2-8 (733-W303), ErDNA (YP377) and smc2-8 ErDNA (733-YP377) cells after a 3-hr shift to 37°C in selective media. Error bars are standard deviations from analysis of at least eight independent clones. (G) Delay of rDNA segregation in cells with minimal NOR. Time lapse video microscopy was used to monitor segregation of the nucleolus (Sik1p-mRFP marker) relatively to anaphase initiation (time 0) and full separation of non-rDNA chromatin (Scc3p-GFP marker). Corresponding cell types are shown below the graph. Ten cells of asynchronous population were analyzed for each strain: TAK300E (with wild type rDNA locus length) and TAK300 (20 rDNA repeats). Imaging was done with one-minute intervals, red and green channels were recorded simultaneously. Sample video files from this analysis are in Supplement 2. Average times of rDNA segregation after anaphase initiation (min) are shown in boldface.
The observed increase in relative Pol I transcription per rDNA repeat in ErDNA may result in changing condensin binding within the individual repeat. Indeed, qPCR/ChIP comparison of condensin binding in wild type rDNA and in ErDNA revealed a striking difference between binding profiles. Wild-type rDNA was bound by condensin in mitosis with a pattern consistent with previous reports2,14,15 (Fig. 3D). In contrast, in ErDNA, only the nontranscribed RFB site was bound by condensin in mitosis, while condensin binding to the transcribed portion of rDNA was considerably reduced (Fig. 3E). However, as the episomal NORs are still essential and are present in multiple copies, the system does not allow direct testing of the transcription effect on rDNA segregation, or to test previous conclusions that condensin is essential for mitotic segregation of tandem chromosomal rDNA repeats but is not essential for mitotic stability of 2-micron-based ErDNA.2 We therefore constructed a single-copy rDNA minichromosome with an alternative marker and introduced it in the ErDNA cells. In such a system the single extra copy of rDNA in minichromosome should be in the same transcriptional state as the other extra-chromosomal copies, but should also be nonessential for ribosomal biogenesis and present in a single copy, allowing accurate analysis of its mitotic stability. After a transient (3-hr) shift to nonpermissive temperature (to enable comparison with smc2-8 cells), the segregation proficiency of the rDNA minichromosome was reproducibly lower in an ErDNA strain than a wild type strain (Fig. 3F), suggesting that active transcription is detrimental for rDNA segregation in wild type cells. Placing the same minichromosome in the smc2-8 mutant or smc2-8 ErDNA cells resulted in equally decreased minichromosome stability, showing that elevated transcription compromises segregation of single rDNA repeat and that this effect is likely mediated by condensin. In agreement with previous results,14 no notable decrease in stability of non-rDNA minichromosomes was detected in the smc2-8 or ErDNA strains (data not shown).
As we showed that condensin function and transcription downregulation are likely required even for segregation of an individual rDNA repeat (Fig. 3F) it is conceivable that segregation of the nucleolus is negatively affected when condensin is forced to compete for rDNA binding with active Pol I transcription. Such a condition occurs in the 20-repeat strain.23 We used a cytological assay to evaluate the effect of maximization of rDNA transcription on the nucleolar segregation timing. First we promoted expansion of rDNA in a 20-repeat strain (TAK300) by transient transformation with a FOB1-containing plasmid. After removal of the plasmid the resulting strain TAK300E was used as an isogenic full-length rDNA control. Two fluorescent markers were introduced into both strain: Scc3p-GFP, under control of an uninduced MET15 promoter, as a general chromatin stain; and the nucleolar marker Sik1p-mRFP.48 Asynchronous live populations of these strains were imaged at 23°C by time-lapse microscopy (Supplement 2). Quantification of this experiment (Fig. 3G) demonstrates that both strains spend a comparable time (5–10 min) to progress from anaphase initiation (determined by beginning of GFP signal separation) to complete separation of the bulk of chromatin (Scc3p-GFP). However, segregation of the nucleolus is significantly delayed (by 13 min on average) in the 20-repeat strain (Fig. 3G). This results argues in favor of a model in which that transcriptionally silent rDNA repeats, present in wild-type but not in the ErDNA strain, facilitate nucleolar segregation (Fig. 4B), while (Fig. 3F) indicates that this effect is likely mediated by condensin binding and function there.
Figure 4.
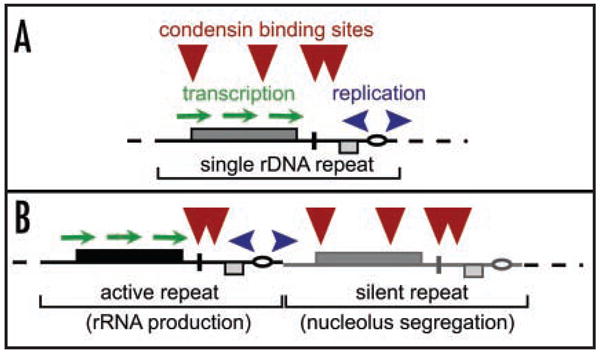
Putative mechanisms of combining condensing binding with active rDNA transcription. (A) Competition between condensin bunding and transcription. This scenarion is likely avoided in the wild-type rDNA, but is likely the only possible mechanism for rDNA segregation in the 20-repeat strain. (B) rDNA repeat specialization for rRNA production (active Pol I transcription) and nucleolar segregation (silent for Pol I transcription) in wild type budding yeast.
DISCUSSION
Condensin complexes have been shown to form a reproducible binding pattern on all chromosomes in higher eukaryotes.12,13 The recent analysis of condensin distribution in S. cerevisiae14 suggests that yeast condensin also has a unique distribution pattern across the genome, similarly to the specific distribution pattern in the rDNA locus described earlier.2,15 However, the molecular mechanisms governing placement of condensin at the specific sites are far from being understood. In case of rDNA condensin targeting, at least three prerequisites of condensin loading to rDNA in mitosis were identified using genetic analysis: proficient sister chromatid cohesion,49,50 proper balance of SUMO modification51 and mitotic activation of Cdc14p phosphatase.15,17 A condensin-binding site in the replication fork termination region was also described in rDNA.2,15 However, it is not presently known whether these pathways act directly on condensin, or if they ensure the general structural integrity of the nucleolus, thus providing a permissive environment for chromosome condensation at the NOR. It was therefore important to elucidate the mechanism of condensin loading onto rDNA in more detail.
Using quantitative ChIP analysis we established that condensin binding across the rDNA is affected by two factors: one is a reduction in RNA polymerase I transcription (constitutive or cell-cycle dependent), and another is Fob1p. In this work, we mainly focused on the role of rDNA transcription in regulating condensin binding. With respect to condensin targeting to rDNA transcription regulation and Fob1p likely act together to mediate significant mitotic enrichment of condensin binding at the four NOR sites. However, at the present time it is difficult to completely separate the effect of Fob1p from that of rDNA transcription, as a fob1 deletion increases rDNA transcription levels (Supplement 1). On the other hand, detailed analysis of condensin binding to rDNA, in Fob1+ and Fob1− strains (Fig. 1D), combined with the fact that that smc2-8 and fob1Δ are synthetically lethal only in a strain where all repeats are actively transcribed, indicate that Fob1p has a role in condensin targeting independent of Pol I transcription interference. However, it is unlikely that Fob1p recruits condensin by direct physical contact: unlike condensin Fob1p is not evolutionary conserved, condensin is not found in complexes with Fob1p,52–54 Fob1p only binds DNA at the RFB site55 and condensin is able to bind RFB sites even in the absence of Fob1p (Figs. 1C, 1D). Instead, the many roles Fob1p plays as a general facilitator of nucleolar organization23,42,46,56,57 may potentially explain why it is required for proper condensin targeting to the nucleolus.
We provide a comprehensive analysis of the role of transcription in controlling condensin binding to rDNA, especially to the transcribed portion of rRNA genes (Figs. 1 and 2). Enrichment for condensin at rDNA is increased when rDNA transcription subsides in mitosis (Fig. 1B), when rDNA transcription is blocked by rapamycin (Figs. 2A, B and D), or when GAL7 promoter-driven transcription of rDNA is repressed (Fig. 2E). These findings, combined with the observed loss of condensin when all tandem rDNA repeats are transcribed (Fig. 1D) are consistent with the suggestion that transcription negatively regulates condensin binding during the normal cell cycle. Moreover, our data support the suggestion that rDNA repeats silent for Pol I transcription possibly act as specialized sites for constitutive condensin binding (Fig. 4B) and play a crucial role in mitotic condensin function—segregating sister chromatids in the NOR region.2 Indeed, rDNA transcription activation in an ErDNA background causes a notable reduction in the mitotic stability of artificial minichromosomes with a single rDNA repeat (Fig. 3F). If this result can be extrapolated to the tandem rDNA arrays present in normal cells, it would suggest that the cell’s ability to repress rDNA transcription and/or to have some repeats constitutively silent may be an important mechanism assuring NOR segregation. Consistent with this suggestion, we found a significant delay in rDNA segregation in the 20 rDNA repeat strain, where all rDNA repeats are transcribed (Fig. 3G and Supplement 2), showing that Pol I transcription is detrimental for rDNA segregation. The latter could be an evolutionary impetus to develop a pathway for mitotic shutdown of Pol I transcription in Metazoa. The fact that condensin is released from rDNA when the number of silent repeats is reduced (Figs. 3A and E) gives additional indirect support to the hypothesis that that silent rDNA repeats are normally bound by condensin (Fig. 4). The possible shift of the cellular role of condensin due to its relocalization in ErDNA cells may allow more targeted study of condensin’s activity at non-rDNA chromosomal sites, independent of its effect on rDNA segregation.
We have shown that the level of Pol I transcription is a powerful control mechanism determining condensin localization in the nucleolus (Fig. 4), and possibly across all chromosomes. Thus, condensin localization to chromatin is dynamic: it may change in response to essentially an epigenetic factor (Pol I transcription patterns within the rDNA array). We suggest that the silencing of rDNA transcription plays an important role in the homeostatic control of condensin localization and, possibly, other nucleolus-enriched proteins.
Supplementary Material
Quantitative real-time RT-PCR analysis of rDNA transcript levels in Fob1+ and Fob1− cells. RNA was prepared from asynchronous populations of two isogenic strains 532-W303 (FOB1) and 532-W303-1 (fob1 I) and analyzed as in Figure 1C.
Examples of time-lapse video microscopy of anaphase in 20rDNA (TAK300) and wild type rDNA (TAK300E) cells. Total nuclear chromatin is visualized by green fluorescence: ectopically expressed Scc3p-GFP fusion (under control of partially repressed MET15 promoter). Red fluorescence – nucleolar marker: Sik1-mRFP fusion expressed from integrated single copy SIK1:mRFP gene. (A) TAK300E (wild type rDNA length). (B) TAK300 (20 rDNA repeats).
Acknowledgments
We thank M. Nomura, T. Kobayashi, S. Roeder and S. Liebman for strains and plasmids; Micheal Lichten and A. Hinnebusch for critical reading of the manuscript; K. Bloom and A. Arnaoutov for time-lapse microscopy advice; R. Mehta and J. Panebianco for technical help. This research was supported by the Intramural Research Program of the NIH at the NICHD and at the Center for Cancer Research, NCI.
Footnotes
This manuscript has been published online, prior to printing for Cell Cycle, Volume 5, Issue 19. Definitive page numbers have not been assigned. The current citation is: Cell Cycle 2006; 5(19): http://www.landesbioscience.com/journals/cc/abstract.php?id=3292
References
- 1.Hirano T. Condensins: Organizing and segregating the genome. Curr Biol. 2005;15:R265–75. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 2.Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol. 2000;149:811–24. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–21. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 4.Kimura K, Cuvier O, Hirano T. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J Biol Chem. 2001;276:5417–20. doi: 10.1074/jbc.C000873200. [DOI] [PubMed] [Google Scholar]
- 5.Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 1995;9:587–99. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- 6.Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell. 2003;5:323–36. doi: 10.1016/s1534-5807(03)00199-0. [DOI] [PubMed] [Google Scholar]
- 7.Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 2002;16:729–42. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouspenski II, Cabello OA, Brinkley BR. Chromosome condensation factor brn1p is required for chromatid separation in mitosis. Mol Biol Cell. 2000;11:1305–13. doi: 10.1091/mbc.11.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–21. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- 10.Cobbe N, Savvidou E, Heck MM. Diverse mitotic and interphase functions of condensins in Drosophila. Genetics. 2006;172:991–1008. doi: 10.1534/genetics.105.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bazett-Jones DP, Kimura K, Hirano T. Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol Cell. 2002;9:1183–90. doi: 10.1016/s1097-2765(02)00546-4. [DOI] [PubMed] [Google Scholar]
- 12.Ono T, Fang Y, Spector DL, Hirano T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell. 2004;15:3296–308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeshima K, Laemmli UK. A two-step scaffolding model for mitotic chromosome assembly. Dev Cell. 2003;4:467–80. doi: 10.1016/s1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 14.Wang BD, Eyre D, Basrai M, Lichten M, Strunnikov A. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol Cell Biol. 2005;25:7216–25. doi: 10.1128/MCB.25.16.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang BD, Yong-Gonzalez V, Strunnikov AV. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 2004:3. doi: 10.4161/cc.3.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan M, Higuchi T, Katis VL, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–82. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- 17.D’Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–69. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- 18.Machin F, Torres-Rosell J, Jarmuz A, Aragon L. Spindle-independent condensation-mediated segregation of yeast ribosomal DNA in late anaphase. J Cell Biol. 2005;168:209–19. doi: 10.1083/jcb.200408087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganley AR, Hayashi K, Horiuchi T, Kobayashi T. Identifying gene-independent noncoding functional elements in the yeast ribosomal DNA by phylogenetic footprinting. Proc Natl Acad Sci USA. 2005;102:11787–92. doi: 10.1073/pnas.0504905102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chernoff YO, Vincent A, Liebman SW. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. Embo J. 1994;13:906–13. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nierras CR, Liebman SW, Warner JR. Does Saccharomyces need an organized nucleolus? Chromosoma. 1997;105:444–51. [PubMed] [Google Scholar]
- 22.Wai HH, Vu L, Oakes M, Nomura M. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: Use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 2000;28:3524–34. doi: 10.1093/nar/28.18.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi T, Nomura M, Horiuchi T. Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:136–47. doi: 10.1128/MCB.21.1.136-147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nogi Y, Vu L, Nomura M. An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7026–30. doi: 10.1073/pnas.88.16.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venema J, Dirks-Mulder A, Faber AW, Raue HA. Development and application of an in vivo system to study yeast ribosomal RNA biogenesis and function. Yeast. 1995;11:145–56. doi: 10.1002/yea.320110206. [DOI] [PubMed] [Google Scholar]
- 27.Pasero P, Bensimon A, Schwob E. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 2002;16:2479–84. doi: 10.1101/gad.232902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–54. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 29.Rose MD, Winston F, Hieter P. Methods in yeast genetics, a laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1990. [Google Scholar]
- 30.Brown A, Tuite M, editors. Yeast Gene Analysis. San Diego, London: Academic Press; 1998. [Google Scholar]
- 31.Strunnikov AV, Larionov VL, Koshland D. SMC1: An essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. JCB. 1993;123:1635–48. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.San-Segundo PA, Roeder GS. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97:313–24. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- 33.Pearson CG, Maddox PS, Zarzar TR, Salmon ED, Bloom K. Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol Biol Cell. 2003;14:4181–95. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–8. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heix J, Vente A, Voit R, Budde A, Michaelidis TM, Grummt I. Mitotic silencing of human rRNA synthesis: Inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. Embo J. 1998;17:7373–81. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elliott SG, McLaughlin CS. Regulation of RNA synthesis in yeast. III Synthesis during the cell cycle. Mol Gen Genet. 1979;169:237–43. doi: 10.1007/BF00382269. [DOI] [PubMed] [Google Scholar]
- 37.Snyder M, Sapolsky RJ, Davis RW. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2184–94. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill A, Bloom K. Genetic manipulation of centromere function. Mol Cell Biol. 1987;7:2397–405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Studitsky VM, Walter W, Kireeva M, Kashlev M, Felsenfeld G. Chromatin remodeling by RNA polymerases. Trends Biochem Sci. 2004;29:127–35. doi: 10.1016/j.tibs.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Dammann R, Lucchini R, Koller T, Sogo JM. Transcription in the yeast rRNA gene locus: Distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol Cell Biol. 1995;15:5294–303. doi: 10.1128/mcb.15.10.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol. 2003;23:1558–68. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–30. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powers T, Dilova I, Chen CY, Wedaman K. Yeast TOR signaling: A mechanism for metabolic regulation. Curr Top Microbiol Immunol. 2004;279:39–51. doi: 10.1007/978-3-642-18930-2_3. [DOI] [PubMed] [Google Scholar]
- 44.Martin DE, Hall MN. The expanding TOR signaling network. Curr Opin Cell Biol. 2005;17:158–66. doi: 10.1016/j.ceb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Choi JH, Adames NR, Chan TF, Zeng C, Cooper JA, Zheng XF. TOR signaling regulates microtubule structure and function. Curr Biol. 2000;10:861–4. doi: 10.1016/s0960-9822(00)00599-6. [DOI] [PubMed] [Google Scholar]
- 46.Burkhalter MD, Sogo JM. rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol Cell. 2004;15:409–21. doi: 10.1016/j.molcel.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 47.Muller M, Lucchini R, Sogo JM. Replication of yeast rDNA initiates downstream of transcriptionally active genes. Mol Cell. 2000;5:767–77. doi: 10.1016/s1097-2765(00)80317-2. [DOI] [PubMed] [Google Scholar]
- 48.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 49.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lavoie BD, Hogan E, Koshland D. In vivo dissection of the chromosome condensation machinery: Reversibility of condensation distinguishes contributions of condensin and cohesin. J Cell Biol. 2002;156:805–15. doi: 10.1083/jcb.200109056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strunnikov AV, Aravind L, Koonin EV. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics. 2001;158:95–107. doi: 10.1093/genetics/158.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 53.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–3. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi T. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol Cell Biol. 2003;23:9178–88. doi: 10.1128/MCB.23.24.9178-9188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Di Felice F, Cioci F, Camilloni G. FOB1 affects DNA topoisomerase I in vivo cleavages in the enhancer region of the Saccharomyces cerevisiae ribosomal DNA locus. Nucleic Acids Res. 2005;33:6327–37. doi: 10.1093/nar/gki950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johzuka K, Horiuchi T. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells. 2002;7:99–113. doi: 10.1046/j.1356-9597.2001.00508.x. [DOI] [PubMed] [Google Scholar]
- 58.Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–76. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative real-time RT-PCR analysis of rDNA transcript levels in Fob1+ and Fob1− cells. RNA was prepared from asynchronous populations of two isogenic strains 532-W303 (FOB1) and 532-W303-1 (fob1 I) and analyzed as in Figure 1C.
Examples of time-lapse video microscopy of anaphase in 20rDNA (TAK300) and wild type rDNA (TAK300E) cells. Total nuclear chromatin is visualized by green fluorescence: ectopically expressed Scc3p-GFP fusion (under control of partially repressed MET15 promoter). Red fluorescence – nucleolar marker: Sik1-mRFP fusion expressed from integrated single copy SIK1:mRFP gene. (A) TAK300E (wild type rDNA length). (B) TAK300 (20 rDNA repeats).