Abstract
The Akt kinase is a critical effector in growth factor signaling. Activation of Akt driven by the growth factor dependent PI3K (phosphatidylinositol-3-OH kinase) is coupled to the plasma membrane translocation and phosphorylation of Akt on two sites by PDK1 (phosphoinositide-dependent protein kinase-1) on Thr-308 and by mTORC2 (mammalian Target Of Rapamycin Complex 2) on Ser-473. In our study we examined the sub-cellular localization of mTORC2 and identified that this kinase complex predominantly resides on endoplasmic reticulum (ER). Our immunostaining analysis did not show a substantial co-localization of the mTORC2 component rictor with Golgi, lysosome, clathrin-coated vesicles, early endosomes, or plasma membrane but indicated a strong co-localization of rictor with ribosomal protein S6 and ER marker. Our biochemical study also identified the mTORC2 components rictor, SIN1, and mTOR as the highly abundant proteins in the ER fraction, whereas only small amount of these proteins are detected in the plasma membrane and cytosolic fractions. We found that growth factor signaling does not alter the ER localization of mTORC2 and also does not induce its translocation to the plasma membrane. Based on our study we suggest that the mTORC2-dependent phosphorylation of Akt on Ser-473 takes place on the surface of ER.
Keywords: mTOR (mammalian Target Of Rapamycin), mTORC2 (mTOR Complex 2), rictor, cell signaling, endoplasmic reticulum
1. Introduction
Growth factor peptides regulate cell growth, proliferation, survival, and metabolism by activation of the phosphatidylinositol-3-OH kinase (PI3K)/Akt pathway [1]. PI3K nucleates growth factor signaling on the plasma membrane by functioning as a kinase of phosphatidylinositol-4,5-diphosphates and generating phosphatidylinositol-3,4,5-triphosphates (PIP3s). The plekstrin homology (PH) domain of Akt binds to PIP3s and recruits Akt to plasma membrane. This translocation step is critical in regulation of Akt because at the plasma membrane its phosphorylated on Thr-308 and Ser-473 sites required for its full activation [2]. The second Ser-473 site known as the regulatory hydrophobic motif site of Akt is phosphorylarted by the kinase complex known as mTORC2 (mammalian Target of Rapamycin Complex 2) [3].
Mammalian target of rapamycin (mTOR) is a central component of the essential and highly conserved signaling pathway that controls anabolic processes in cells [3]. The biochemical studies revealed that yeast TOR [4] or mammalian TOR (mTOR) with its interacting proteins mLST8 and DEPTOR exists at least in two distinct complexes [5]. Its first complex mTORC1 (mTOR Complex 1) assembled by mTOR and its binding proteins raptor. It functions as a nutrient-sensing complex and controls cell growth and cell size. The second complex mTORC2 is assembled by mTOR and its interacting proteins rictor and SIN1. The functional studies defined mTORC2 as the regulatory kinase of the distinct members of AGC (protein kinase A, G, and C) family including Akt, PKCa, and SGK1 [5].
mTORC2 regulates Akt by phosphorylation of its two different sites. The mTORC2-dependent phosphorylation of Akt on the regulatory Ser-473 site is dependent on growth factor signaling, whereas a basal activity of mTORC2 maintains the constitutive phosphorylation of Akt on its turn motif Thr-450 site [2]. This difference indicates that phosphorylation of the Thr-450 and Ser-473 sites on Akt by mTORC2 are separate events and might take place at different locations. It has been proposed that translocation of Akt to the plasma membrane coupled with its phosphorylation on Thr-308 and Ser-473 is a critical step in activation of Akt by growth factor signaling [2]. The functional localization of mTORC2 has been supported by the recent finding that in yeast TORC2 has been identified at the plasma membrane [6]. It has been also reported that mTORC2 in association with ribosomes promotes cotranslational phosphorylation of Akt on Thr-450 [7]. Importantly, activation of mTORC2 by association with ribosome has been demonstrated by Hall group [8]. Currently, the functional localization of mTORC2 remains elusive. In our present study we have addressed this problem by examining the sub-cellular localization of mTORC2.
2. Materials and methods
2.1 Materials
Reagents were obtained from the following sources: DMEM/F12 from Life Technologies; the Fetal Bovine Serum (FBS) from Hyclone, Fugene 6 transfection reagent and the complete protease inhibitor cocktail from Roche; for immunoblotting we used the following antibodies: rictor, HRP-labeled anti-rabbit/anti-mouse/anti-goat secondary antibodies, and tubulin from Santa Cruz Biotechnology and mTOR from Cell Signaling. The Alexa-coupled anti-mouse/anti-rabbit secondary antibodies from Invitrogen. For immunostaining we used the following set of antibodies: rictor (Santa Cruz #sc-50678), mTOR (Strategic Diagnostics Inc. #28130002, presently distributed by Novus Biologicals by the same catalog number), LAPM2 (BD Bioscience #555803) EEA1 (BD Bioscience #610456), Clathrin (Cell Signaling #2410), GM130 (Cell Signaling #2296), Calreticulin (Abcam #ab22683).
2.2 Cell lines and culture
MDA-MB-435 and A549 were obtained from American Type Culture Collection. MDA-MB-435 and A549 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) / F12 with 10% FBS. The cells reached 70–80% confluency were serum starved for 24 hours and stimulated by incubation with insulin-like growth factor I (IGFI) 50 ng/ml for 20 min.
2.3 Lentivirus production, and infection
Lentiviral shRNAs targeting human mTOR and rictor were generated and used as described previously [9]. Lentiviruses were harvested 48 hours after transfection and spinned at 3000 g for 15 min in order to eliminate any remaining 293T cells. The viral supernatant was added at a ratio of 1:1 to the culture medium in the presence of polybrene (8 µg/ml) and the cells were spinned at 1200 g for 45 min at 32°C, in order to increase the infection efficency. Cells incubated with the retroviruses for the following 24 hours. After an additional 24 hours of recovery in normal medium, infected cells were passaged and selected with puromycin (2.5 mg/ml for 3–4 days). To express ER-GFP the cells were infected by the Organelle Lights™ ER-GFP virus (Invitrogen #O36212).
2.4 Cell lysis and immunoblotting
Cells were rinsed with ice-cold PBS before lysis in buffer containing as described previously [9]. The scraped lysates were incubated for 20 min at 4°C to complete lysis. The soluble fractions of cell lysates were isolated by centrifugation at 13,000 rpm (17500g) for 10 min in a microcentrifuge. The protein concentration was measured using the 5X Biorad protein assay and the samples were denaturated in Laemmli buffer. 10 µg of protein were resolved by SDS-PAGE (7.5% acrylamide) and transferred to PVDF membrane. Proteins were analyzed by immunoblotting and detected by enhanced chemoluminescence (ECL).
2.5 Immunofluorescence
For endoplasmic reticulum (ER) staining, cells were pre-treated, two days before the experiment, with the Organelle Lights™ Reagents (Invitrogen) according to the manufacturer’s instruction. This technique is based on expression, via baculovirus delivery system, of fluorescent protein-signal peptide fusions for accurate and specific targeting to subcellular compartments and structures. Chamber slide were coated with fibronectin 10 µg/ml in PBS (3 µg/cm2) and incubated at 37°C C. After 2 hours, extra fibronectin was removed and cells were plated: 30,000 cells per dish for A549 and 100,000 cells per dish for MDA-MB-435. Twenty-four hours after plating, cultured cells were fixed with 3.7% paraformaldehyde for 15 min. After three washes with PBS, cells were permeabilized with 0.1% Saponin in PBS for 20 min. After another three PBS washes, free aldehydes were blocked with 50 mM ammonium chloride for 20 min followed by blocking of non specific sites with 10% horse serum diluted in PBS for 1 hour. The cells were then incubated with the appropriate primary antibodies overnight at 4°C in a humid chamber. The labeled proteins were detected using the appropriate Alexa 488 and/or Alexa 594-conjugated secondary antibodies for 1 hour at room temperature. The cells were then incubated with 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI) for 5 minutes. After three last PBS washes, the slides were quickly rinsed into water and mounted in Mowiol based mounting medium. Finally, slides were observed and pictures were taken using Olympus IX71FV500 Laser Scanning Confocal Microscope with UV excitation laser. The resulting images were analyzed using the software Fluoview 5.0, according to the manufacturer’s instructions (http://www.olympusfluoview.com/java/colocalization/index.html).
2.6 The biochemical sub-cellular fractionation
The cytosolic and endoplasmic reticulum (ER) fractions have been purified as described previously [10]. Briefly, we utilized the discontinuous sucrose gradients to purify ER. Initially, cells are lysed mechanically with sonication and, then, a low-speed centrifugation (700 × g) is used to remove large cellular debris. Supernatant from this step is collected as a total lysed protein fraction. A subsequent 15,000 × g centrifugation crudely pellets mitochondria and separates it from ER and other organelles. The supernatant is loaded onto a three-layered sucrose gradient and purified ER is banded by centrifugation at 152,000 × g. ER band accumulates at the interface of 1.3M Sucrose. The plasma membrane fraction was isolated by using BioVision kit (#K268-50) as recommended by the manufacture.
3. Results
3.1 Detection of mTORC2 by the co-localization of rictor and mTOR staining
Rictor is an essential component of mTORC2 [3]. To initiate our sub-cellular localization study of mTORC2, we pursued the rictor localization by immunostaining. First, we optimized the immunostaining of rictor by screening a set of the rictor antibodies and cellular fixation/permeabilization conditions in MDA-MB-435 cells. The permeabilization of cells following the fixation step with a low concentration of saponin has been selected as the most effective in detection of rictor. A specificity of the rictor detection in this condition has been validated by the knock down of rictor by the lentiviral expression of its targeting shRNA. The efficiency of the rictor knock down and deficiency of the mTORC2 signaling has been examined by immunoblotting (Fig. S1 A and B). Following the knock down of rictor expression as indicated by the immunoblotting of cellular lysates, in the absence of rictor we detected only a low background signal by immunostaining. The rictor staining shows a perinuclear and cytoplasmic dotted pattern implying that rictor is co-localized with cellular organelles. In our next step by a similar approach we have validated a specificity of the mTOR immunostaining (Fig. S2 A and B). We observed a similar dotted pattern of the mTOR staining only in control but not in the mTOR knock down cells indicating a specific detection of mTOR by immunostaining.
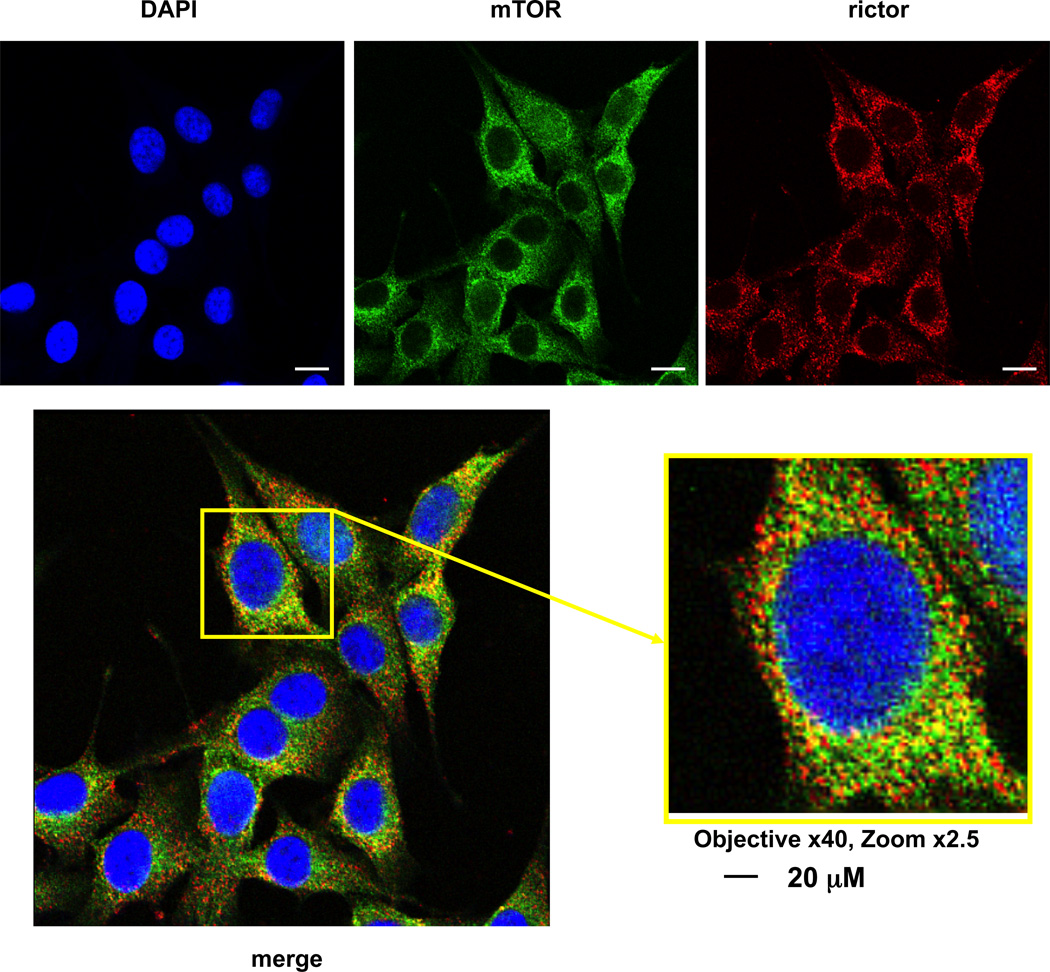
If we attained a specific immunostaining of rictor and mTOR, the mTORC2 complex will be revealed as the overlapping staining of both proteins. Following co-staining of rictor and mTOR we detected the dotted staining signals from each antibody and merging their images showed a strong overlapping (Pearson’s coefficient 0.74, Table 1) as we regard as a detection of mTORC2 (Fig. 1). It is remarkable that a substantial level of the mTOR staining shows no overlapping with rictor correlating with existence of mTOR in the rictor independent complex with raptor. Similarly, we observed some rictor staining without overlapping with mTOR supporting the recent finding that rictor also carries the mTOR independent function [11; 12]. Thus, our studies by co-localization of the rictor and mTOR immunostaining identified mTORC2 representing the substantial levels of rictor and mTOR as the coincident dotted pattern of both these proteins within perinuclear and cytoplasmic areas of cells.
Fig. 1.
Detection of mTORC2 by co-localization of rictor and mTOR. MDA-MB-435 cells growing in 10% serum were stained with the rictor and mTOR antibodies. The images of rictor, mTOR, and merged images are presented.
3.2 Rictor is poorly co-localized with the clathrin-coated vesicles, early endosomes, Golgi apparatus, and lysosomal markers
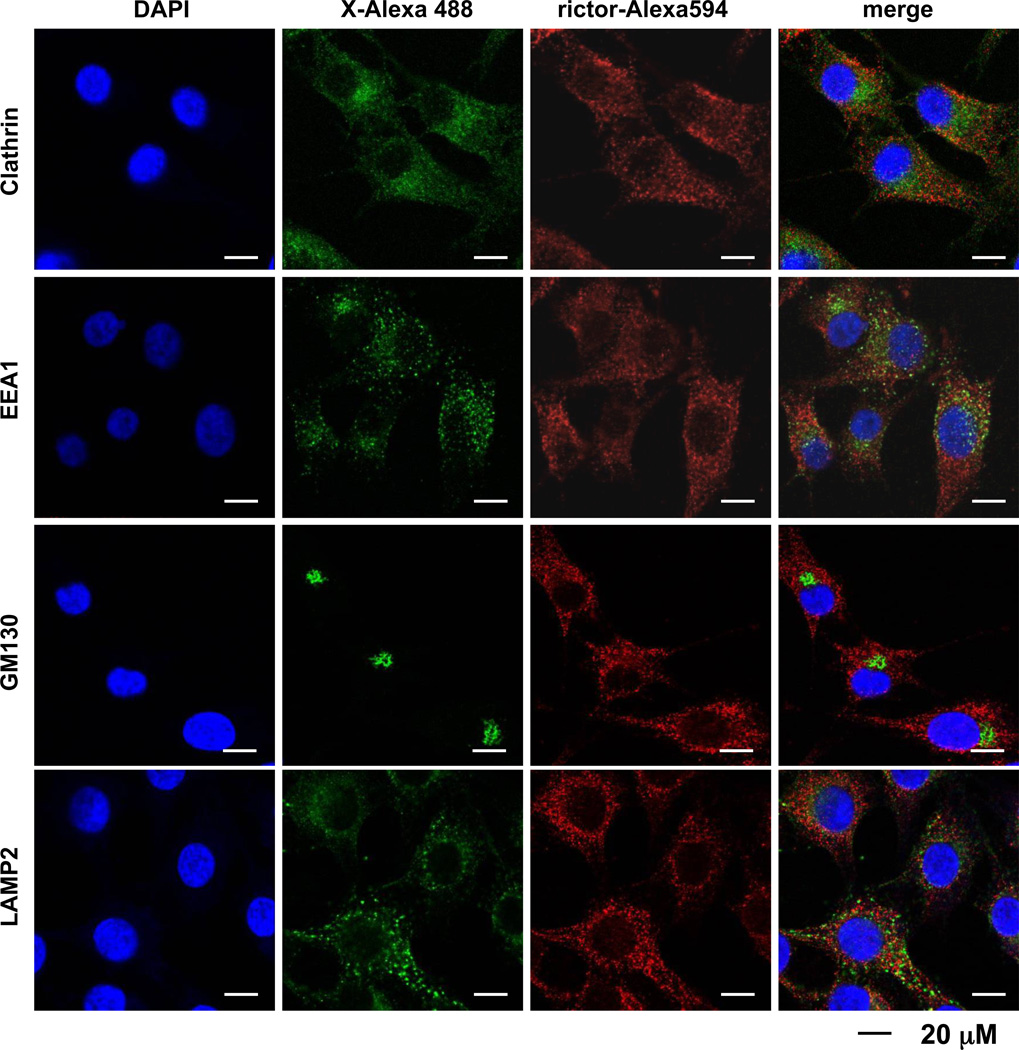
We found that a majority of the rictor staining overlaps with mTOR and this observation makes rictor as a valuable marker in pursuing the sub-cellular localization of mTORC2. We did not observe the localization of rictor or mTOR on the plasma membrane by immunostaining (Fig. 1) and in our next studies we studied the rictor localization with major intracellular organelles (Fig. 2). The recent studies have identified the early endosome vesicles compartment as an important site in regulation of Akt [13]. First, we studied if rictor might be located at this site because of the critical role of mTORC2 in regulation of Akt. We have detected the clathrin-coated endocytic vesicles by immunostaining cells with clathrin and the early endosomes were stained by EEA1 (Early Endosome Antigen 1) [14; 15]. The clathrin-coated endocytic vesicles stained as a particular dotted pattern but without a strong overlapping with the rictor staining (Pearson’s coefficient 0.52) (Fig. 2, the first panel). We also observed only a small fraction of rictor co-localized with early endosomes as detected by co-staining of rictor with EEA1 (Pearson’s coefficient 0.36) (Fig. 2, the second panel). Our data indicate that a bulk of rictor does reside in the clathrin-coated endocytic vesicles and early endosomes.
Fig. 2.
Bulk of rictor does not reside in endosomes, Golgi, and lysosomes. The immunofluorescence study has been performed to stain rictor with the specific markers of different organelles in MDA-MB-435. Clathrin was used as a specific marker for clathrin-coated vesicules, EEA1 was used as a specific marker for early endosomes, GM 130 was used as a specific marker for Golgi apparatus, and LAMP2 was used as a specific marker for lysosomes.
Golgi apparatus is an important sorting organelle in eukaryotic cells [16]. In our next study we analyzed the rictor localization in Golgi network by staining cells with the Golgi marker GM130 (Golgi Matrix protein of 130 kDa) [17]. By using this marker we detected the Golgi network as a distinct perinuclear structure very similar to the staining of Golgi in HeLa cells [18]. Interestingly, the rictor speckled staining pattern filling the cytoplasm shows a dim staining particular in the perinuclear location detected as the Golgi network area (Fig. 2, the third panel). As a result, we did not detect the rictor co-staining with the GM130 marker (with a lowest Pearson’s coefficient 0.19) indicating that the Golgi network is not a main location site of rictor.
The lysosomal surface has been identified as a critical site in the nutrient sensing mechanism of mTORC1 [19]. To address whether mTORC2 might also associate with lysosomes, we performed the co-staining of rictor with the lysosomal marker LAMP2 (Lysosomal-Associated Membrane Protein 2) [20]. We found that the staining of LAMP2 shows the speckled staining through the cytosole resembling the rictor staining, but we observed only a weak overlap of rictor and LAMP2 staining with a Pearson’s coefficient 0.52 (Fig. 2, the forth panel). Our analysis of the lysosomal marker also led to a conclusion that a bulk of rictor does not associate with cellular lysosomes.
3.3 Rictor is co-localized with the ribosomal protein S6 and ER marker
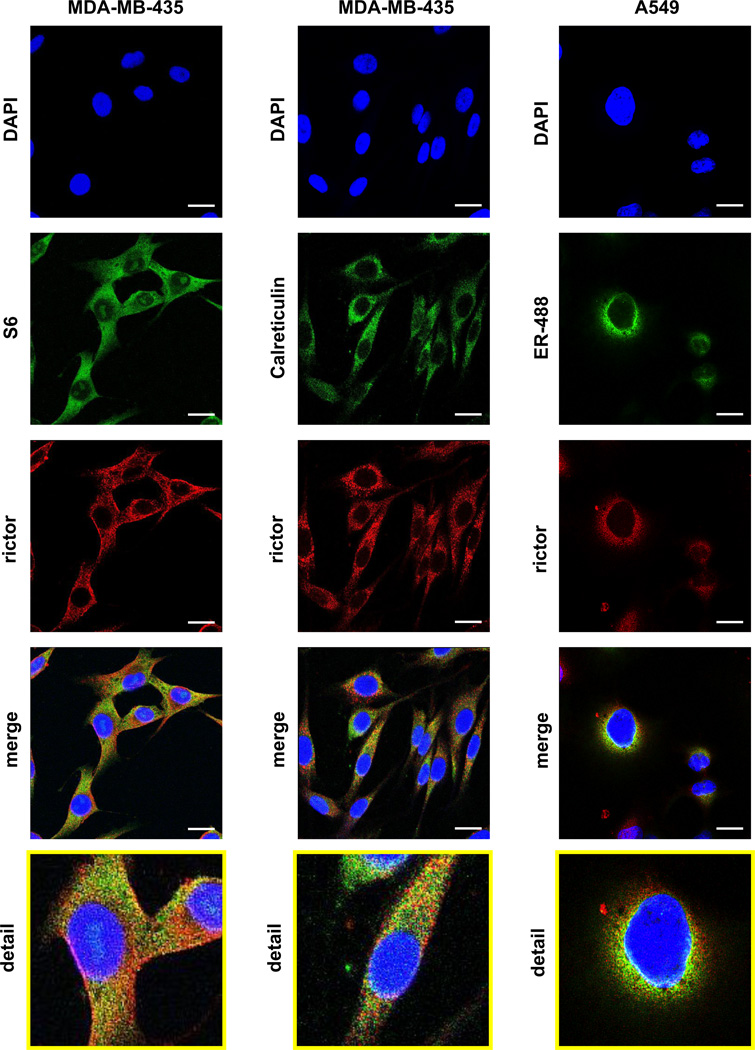
The recent functional studies of mTORC2 carried a new insight by connecting this kinase complex with ribosomes. It has been shown that mTORC2 in association with ribosome promotes the co-translational phosphorylation of Akt on its Thr-450 turn motif site [7]. Importantly, activation of mTORC2 in association with ribosome has been recently identified [8]. If a substantial portion of mTORC2 associates with ribosomes, in this case the co-localization of rictor with ribosomal proteins will be detectable by immunostaining. To pursue this lead, we co-stained rictor with the ribosomal protein S6 (Fig. 3, the left row). The ribosomal protein has been detected as a dense speckled staining through most of the cytoplasm. Importantly, among all markers we have analyzed in this study, the ribosomal S6 protein detection shows the most substantial overlap with rictor (Pearson’s coefficient 0.73). A considerably strong co-localization of rictor with the ribosomal protein implies that rough endoplasmic reticulum (ER), known as the highly enrich organelle with ribosomes, is a main localization site of mTORC2. It has been supported by our co-localization study of rictor with the different ER markers: calreticulin [21] and green fluorescent protein containing the ER localization and retention peptide sequences (ER-GFP) [22] as shown in Fig. 3B. We detected a strong co-localization of rictor with calreticulin in MDA-MB-435 cells (Fig. 3, the middle row, the Pearson’s coefficient 0.66). In addition, the ER-GFP protein expressed in A549 cells has indicated a substantial overlap with the rictor staining (Fig. 3, the right row, the Pearson’s coefficient 0.73). In the previous studies mTOR, the binding partner of rictor, has been identified as the protein located in ER [23] suggesting that mTORC2 might be located in ER. Thus, our immunostaining analysis demonstrates that rictor is localized mostly with ER.
Fig. 3.
Rictor is co-localized with the ribosomal protein S6 and ER markers. The immunofluorescence study has been performed in MDA-MB-435 cells to stain rictor with the ribosomal protein S6 (left panel) and the ER markers - calreticulin (middle panel) and ER-GFP (right panel).
3.4 The biochemical sub-cellular fractionation locates the mTORC2 components predominantly in ER
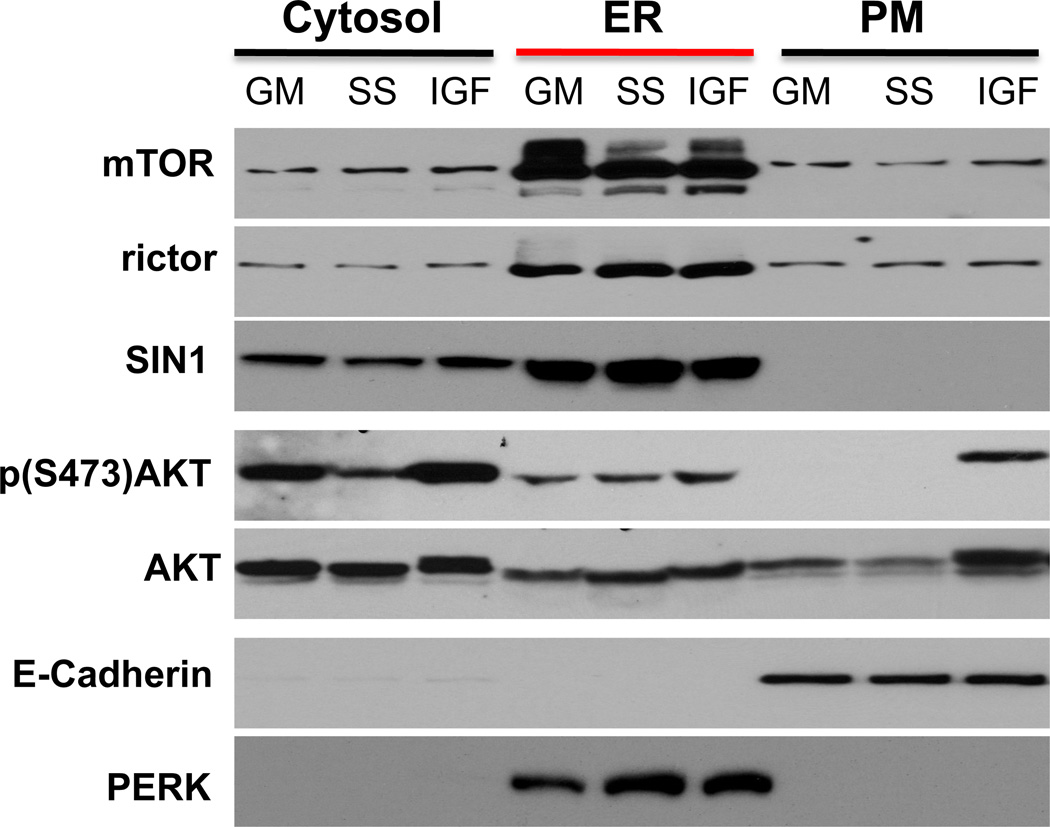
The co-localization data of rictor with the ribosomal protein S6 and ER markers obtained by immunostaining indicate that rictor might reside primarily in ER. If this finding is true, we anticipate that localization of mTORC2 as the protein complex defined by rictor also associates predominantly with ER. To address a sub-cellular localization of mTORC2 by an alternative approach, we have performed the biochemical fractionation of MDA-MB-435 cells by isolating the cytosolic, ER, and plasma membrane fractions (Fig. 4). First, we have validated a purity of the isolated sub-cellular fractions by detecting the ER marker PERK (protein kinase RNA-like endoplasmic reticulum kinase) [24] and the plasma membrane marker E-cadherin [25]. We have identified PERK only in the ER fraction, whereas E-cadherin has been detected in the plasma membrane fraction (Fig. 4).
Fig. 4.
The biochemical sub-cellular fractionation identified mTORC2 in ER. MDA-MB-435 cells incubated in the medium containing 10% serum (GM, growth medium), serum starved for 24 hrs (SS), and serum starved for 24 hrs and stimulated with IGFI (50 ng/ml) for 20 min (IGF) were analyzed by fractionation and isolation the cytosolic, ER (endoplasmic reticulum) and PM (plasma membrane) fractions. Each fraction representing different cell culture condition were lysed and analyzed by loading the equal protein amount on a gel by immunoblotting with the indicated antibodies.
Following validation of our biochemical sub-cellular fractionation approach, we analyzed abundance of Akt in the isolated fractions (Fig. 4). Similar to the initial localization study of c-Akt [26], we found that Akt is localized predominantly in cytoplasm. We have also detected Akt in the ER fraction as it has been reported previously [27]. The Akt localization within this fraction has not been altered by the IGFI stimulation indicating that Akt resides on ER independent of growth factor signaling. When the serum starved cells are stimulated by IGFI, we observed a substantial translocation of Akt to the plasma membrane and this step is known to be critical in the PI3K-dependent activation of Akt [2]. A high basal phosphorylation of Akt on the mTORC2-dependent Ser-473 site is detected in the cytosolic fraction in cells growing in the 10% serum cell culture condition (GM, growth medium) and under the serum starvation condition we observed a low phosphorylation of Akt. Following stimulation of the serum starved cells by IGFI, we detected a robust phosphorylation of Akt not only in the cytosolic but also in the plasma membrane fraction. It indicates that the growth factor stimulation induces activation of Akt is coupled with its translocation to the plasma membrane. Importantly, our fractionation study shows a high abundance of mTOR, rictor, and Sin1 in the ER fraction but only the weak signals of these proteins are detected in the cytosolic and plasma membrane fractions (Fig. 4). In contrary to Akt, we did not detect the growth factor dependent translocation of the mTORC2 components indicating that growth factor signaling does not regulate the localization of this kinase complex. Thus, our biochemical studies determine that mTORC2 is predominantly localized within the ER fraction that is consistent with our immunostaining analysis of rictor.
4. Discussion
The plasma membrane has been defined as a major activation site of Akt coupled to its phosphorylation on Thr-308 and Ser-473 [2]. A poor detection of mTORC2 on the plasma membrane and a high abundance of this kinase complex in the ER fraction suggest that the functional activity of mTORC2 as the regulatory kinase of Akt and its related kinases takes place on the ER surface. A basal activity of mTORC2 maintains a constitutive phosphorylation of Akt on its turn motif Thr-450 site, whereas growth factor signaling by regulating the mTORC2 kinase activity controls phosphorylation of Akt on its hydrophobic regulatory Ser-473 site.
Our finding on the sub-cellular localization of mTORC2 relates well with the recent reports indicating the functional relationship between mTORC2 and ribosomes. It has been shown that mTORC2 can associate with ribosomes to promote the cotranslational phosphorylation of Akt on Thr-450 site and stabilize a nascent Akt polypeptide [7]. In addition, the growth factor/PI3K dependent activation of mTORC2 in association with ribosomes has been recently reported [8]. It has been proposed that ribosome as an essential and rate-limiting component in protein synthesis by binding to mTORC2 coordinates activity of this kinase complex and consequently controls the mTORC2 dependent anabolic pathway. Our finding by defining ER as a major localization site of mTORC2 places this kinase complex with the cellular organelle highly enriched with ribosomes and is coherent with the findings linking ribosomes in regulation of mTORC2.
Based on our sub-cellular localization study of mTORC2, we believe that mTORC2 resides on the surface of ER and is accessible to bind and phosphorylate its substrates. At this location, mTORC2 is controlled by the functional activity of ER, the organelle playing a crucial role in anabolic processes including protein and lipid synthesis. It has been recently reported that ER stress inhibits the kinase activity of mTORC2 by a specific phosphorylation of its essential component rictor mediated by GSK-3b [28]. The mechanism how growth factor signaling by regulating association of mTORC2 with ribosomes [8] controls the kinase activity of this complex is the next important question to be addressed. Besides, mTORC2 as the highly conserved complex in all eykaryotes remains poorly characterized and most likely its essential function in ER is yet to be elucidated.
Highlights.
-
––
mTORC2 is predominantly localized in endoplasmic reticulum
-
––
Growth factor signaling does not alter the sub-cellular localization of mTORC2
-
––
Growth factor signaling has induced translocation of Akt but not mTORC2 to the plasma membrane
Supplementary Material
Acknowledgements
We are thankful to Jared K. Burks for supporting our imaging analysis at our institutional Flow Cytometry & Cell Imaging Core Facility, the core is funded by the NIH grant CA16672. This work was supported by the M. D. Anderson Trust Fellow Fund and NIH grant CA 133522 (D.D.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Delphine Boulbes, Email: dboulbes@mdanderson.org.
Tattym Shaiken, Email: tshaikenov@mdanderson.org.
Dos D. Sarbassov, Email: dsarbass@mdanderson.org.
References
- 1.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 2.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 3.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 5.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berchtold D, Walther TC. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol Biol Cell. 2009;20:1565–1575. doi: 10.1091/mbc.E08-10-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 10.Bozidis P, Williamson CD, Colberg-Poley AM. Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts. Curr Protoc Cell Biol Chapter. 2007;3 doi: 10.1002/0471143030.cb0327s37. Unit 3 27. [DOI] [PubMed] [Google Scholar]
- 11.Hagan GN, Lin Y, Magnuson MA, Avruch J, Czech MP. A Rictor-Myo1c complex participates in dynamic cortical actin events in 3T3-L1 adipocytes. Mol Cell Biol. 2008;28:4215–4226. doi: 10.1128/MCB.00867-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald PC, Oloumi A, Mills J, Dobreva I, Maidan M, Gray V, Wederell ED, Bally MB, Foster LJ, Dedhar S. Rictor and integrin-linked kinase interact and regulate Akt phosphorylation and cancer cell survival. Cancer Res. 2008;68:1618–1624. doi: 10.1158/0008-5472.CAN-07-5869. [DOI] [PubMed] [Google Scholar]
- 13.Schenck A, Goto-Silva L, Collinet C, Rhinn M, Giner A, Habermann B, Brand M, Zerial M. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 2008;133:486–497. doi: 10.1016/j.cell.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 14.Mu FT, Callaghan JM, Steele-Mortimer O, Stenmark H, Parton RG, Campbell PL, McCluskey J, Yeo JP, Tock EP, Toh BH. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine "fingers" and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 15.Bananis E, Murray JW, Stockert RJ, Satir P, Wolkoff AW. Regulation of early endocytic vesicle motility and fission in a reconstituted system. J Cell Sci. 2003;116:2749–2761. doi: 10.1242/jcs.00478. [DOI] [PubMed] [Google Scholar]
- 16.Polishchuk RS, Mironov AA. Structural aspects of Golgi function. Cell Mol Life Sci. 2004;61:146–158. doi: 10.1007/s00018-003-3353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nacak TG, Leptien K, Fellner D, Augustin HG, Kroll J. The BTB-kelch protein LZTR-1 is a novel Golgi protein that is degraded upon induction of apoptosis. J Biol Chem. 2006;281:5065–5071. doi: 10.1074/jbc.M509073200. [DOI] [PubMed] [Google Scholar]
- 19.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 21.Sonnichsen B, Fullekrug J, Nguyen Van P, Diekmann W, Robinson DG, Mieskes G. Retention and retrieval: both mechanisms cooperate to maintain calreticulin in the endoplasmic reticulum. J Cell Sci. 1994;107 (Pt 10):2705–2717. doi: 10.1242/jcs.107.10.2705. [DOI] [PubMed] [Google Scholar]
- 22.Roderick HL, Campbell AK, Llewellyn DH. Nuclear localisation of calreticulin in vivo is enhanced by its interaction with glucocorticoid receptors. FEBS Lett. 1997;405:181–185. doi: 10.1016/s0014-5793(97)00183-x. [DOI] [PubMed] [Google Scholar]
- 23.Drenan RM, Liu X, Bertram PG, Zheng XF. FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J Biol Chem. 2004;279:772–778. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- 24.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65:3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed NN, Franke TF, Bellacosa A, Datta K, Gonzalez-Portal ME, Taguchi T, Testa JR, Tsichlis PN. The proteins encoded by c-akt and v-akt differ in post-translational modification, subcellular localization and oncogenic potential. Oncogene. 1993;8:1957–1963. [PubMed] [Google Scholar]
- 27.Giorgi C, Ito K, Lin HK, Santangelo C, Wieckowski MR, Lebiedzinska M, Bononi A, Bonora M, Duszynski J, Bernardi R, Rizzuto R, Tacchetti C, Pinton P, Pandolfi PP. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science. 330:1247–1251. doi: 10.1126/science.1189157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CH, Shaikenov T, Peterson TR, Aimbetov R, Bissenbaev AK, Lee SW, Wu J, Lin HK, Sarbassov dos D. ER stress inhibits mTORC2 and Akt signaling through GSK-3beta-mediated phosphorylation of rictor. Sci Signal 4 ra10. doi: 10.1126/scisignal.2001731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.