Abstract
C19ORF5 is a sequence homologue of microtubule-associated proteins MAP1A/MAP1B of unknown function, except for its association with mitochondria-associated proteins and the paclitaxel-like microtubule stabilizer and candidate tumor suppressor RASSF1A. Here, we show that when overexpressed in mammalian cells the recombinant 393-amino acid residue COOH terminus of C19ORF5 (C19ORF5C) exhibited four types of distribution patterns proportional to expression level. Although normally distributed throughout the cytosol without microtubular association, C19ORF5C specifically accumulated on stabilized microtubules in paclitaxel-treated cells and interacted directly with paclitaxel-stabilized microtubules in vitro. The native 113-kDa full-length C19ORF5 and a shorter 56-kDa form similarly associated with stabilized microtubules in liver cells and stabilized microtubules from their lysates. As C19ORF5 accumulated, it appeared on mitochondria and progressively induced distinct perinuclear aggregates of mitochondria. C19ORF5 overlapped with cytochrome c-deficient mitochondria with reduced membrane potential. Mitochondrial aggregation resulted in gross degradation of DNA, a cell death–related process we refer to as mitochondrial aggregation and genome destruction (MAGD). Deletion muta-genesis revealed that the C19ORF5 hyperstabilized microtubule-binding domain resides in a highly basic sequence of <100 residues, whereas the MAGD activity resides further downstream in a distinct 25-residue sequence (F967–A991). Our results suggest that C19ORF5 mediates communication between the microtubular cytoskeleton and mitochondria in control of cell death and defective genome destruction through distinct bifunctional structural domains. The accumulation of C19ORF5 and resultant MAGD signaled by hyperstabilized microtubules may be involved in the tumor suppression activity of RASSF1A, a natural microtubule stabilizer and interaction partner with C19ORF5, and the taxoid drug family.
Introduction
Aneuploidy is thought to be the predominant source of genetic instability necessary for evolution of diverse malignant pheno-types of cancer cells (1–4). Elaborate quality control mechanisms insure orderly and symmetrical partitioning of chromosomes at mitosis where missegregation may result in aneuploidy (5, 6). The microtubule spindle complex is the single most important organelle underlying chromosomal segregation during cell division (7) and cellular factors that control microtubule dynamics by stabilization and destabilization are critical in the process (8–10). Mitotic spindle checkpoint mechanisms monitor quality of the spindle and symmetry of chromosome segregation and cause temporary arrest of mitosis to allow for repair and synchronization when the process is less than symmetrical (3, 5). When checkpoint repair mechanisms fail and symmetrical chromosome segregation is catastrophically stalled, cell death and destruction of the genome is necessary to prevent inheritable aneuploidy due to chromosome missegregation that may confer the genetic plasticity that underpins evolution of malignant cancer (5). Large gaps exist in our knowledge of how catastrophic defects in the mitotic spindle, particularly the microtubular structure, are monitored and coupled to genome destruction and cell death. Understanding these mechanisms, how they are bypassed in cancer cells and how to restore them are essential to understanding tumor suppression through prevention of aneuploidy.
Protein C19ORF5 is a homologue of microtubule-associated protein MAP1B that interacts with and colocalizes with leucine-rich PPR-motif containing protein (LRPPRC, AAA67549; ref. 11). LRPPRC is a nucleic acid binding (12, 13), mitochondria-associated protein composed of modular domains homologous to proteins involved in cytoskeletal dynamics, nucleocytosolic shuttling, and chromosome activity (14). Mutations in LRPPRC cause Leigh syndrome, French-Canadian type, a defect in oxidative metabolism that is thought to be related to a deficit in mitochondrial cytochrome c oxidase activity (15). A screen using the LRPPRC-interactive COOH terminus of C19ORF5 as the bait for interactive substrates revealed that, in addition to LRPPRC, C19ORF5 interacted with mitochondria-associated NADH dehydrogenase I and cytochrome c oxidase I and RASSF1A (11, 16, 17). RASSF1A is a microtubule-associated protein that is a powerful natural microtubule stabilizer and inhibitor of the anaphase-promoting complex (16–19). RASSF1A has been proposed as a tumor suppressor because it is silenced by hypermethylation in a large majority of human tumors (20–22). Recently, others have confirmed our report of the interaction of RASSF1A with C19ORF5 both in vitro and in vivo (19). Collectively, these studies suggest that both RASSF1A and C19ORF5 are interactive with microtubules, dramatically affect microtubule dynamics, and play a role in spindle dynamics and chromosome segregation at mitosis. If and how the two proteins play a role in tumor suppression through coupling defective microtubule spindle dynamics to cell death and genome destruction remains to be established.
In addition to their role in cellular energy production and metabolic homeostasis, mitochondria play a key role in some forms of cell death and are a source of factors that mediate cell death-related genome degradation (23–25). We hypothesized that C19ORF5 may be a potential link between microtubule dynamics and mitochondria based on its interaction with at least three mitochondria-associated proteins, its homology to known microtubule-associated proteins MAP1B and MAP1A, and its interaction with the microtubule stabilizer RASSF1A. Here, we show that both endogenous and overexpressed recombinant C19ORF5 normally does not decorate microtubules but is recruited to paclitaxel-stabilized microtubules both in vivo and in vitro. We show that increasing levels of the 393-amino acid residue COOH terminus of C19ORF5 in mammalian cells induced aggregation of cytochrome c-deficient mitochondria with reduced membrane potential around the nucleus and degradation of DNA within the aggregates. We refer to this process as mitochondrial aggregation and genome destruction (MAGD) to distinguish it from the plethora of markers and phenomena observed in diverse types of cell death short of complete genome destruction (26). Structure-function analysis indicated that distinct sequence domains within the C19ORF5 COOH terminus mediate the two functions and the activities occur independent of each other. The independent domains may link catastrophically stable spindle microtubules to cell death and genome destruction to suppress tumors by prevention of inheritable genomic abnormalities that threaten due to mitotic catastrophe.
Materials and Methods
Plasmid construction
GFP-C19ORF5C was a construct with an enhanced green fluorescent protein (GFP) fused at the NH2 terminus of a C19ORF5 fragment that represents the COOH-terminal 393 residue of C19ORF5 protein (11). We isolated a 1.25-kb cut fragment from C19ORF5-pACT2 (14) by NcoI-BglII double digestion, filled it into blunt ends, ligated into a 5-kb SmaI-cut pGEX-4T-1 vector (Amersham Biosciences, Stockholm, Sweden), and created GST-C19ORF5C with glutathione S-transferase (GST) at the NH2 terminus of C19ORF5C. Different fragments of C19ORF5 were generated by PCR using template C19ORF5C-pACT2 (14) and primers that target the exact beginning or end coding sequences of each construct. Four nucleotides (AATT) plus an EcoRI (GAATTC) or NotI (GCGGCCGC) site were added to the 5′ or 3′ end of the 15 nucleotide-long primers, respectively. The GST-fused D667–S766, S766–L866, A867–E966, S767–E966, and F967–F1059 were constructed by ligation of the EcoRI-NotI-cut 5-kb pGEX-4T-1 vector with the corresponding PCR fragments cut with the same enzymes. GFP-tagged D667–S766, S766–L866, A867–E966, S767–E966, F967–F1059, F967–S1041, F967–D1016, and F967–A991 are the ligation products of SmaI-cut pEGFP-C3 vector with the corresponding EcoRI-NotI-cut PCR fragments that were filled into blunt ends. Fidelity of constructs was verified by DNA sequence.
Cell transfection, morphologic analysis, and cellular localization analysis
COS7 cells were used and maintained and transiently transfected with GFP-C19ORF5C and other constructs, the medium was removed, and attached cells were washed with PBS, fixed with 4% paraformaldehyde at 37°C to preserve cytoskeletal architecture, stained with 4′,6-diamidino-2-phenylindole (DAPI), anti-β-tubulin, or anti–cytochrome c (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) antibodies, and visualized using Alexa Fluor 350 goat anti-mouse secondary antibody for cytochrome c (Molecular Probes, Inc., Eugene, OR) and Texas red–conjugated secondary antibody for β-tubulin as described (11). The mitochondria were labeled in the same way as described (27). Continuing observation of living cells was conducted in four-well Lab-Tek chambered cover glass slips (Nalge Nunc International, Rochester, NY). For controls, cells were transfected with the same amount of vector bearing GFP alone as indicated. The expression of GFP-C19ORF5C and GFP was determined by immunoblotting of transfected cell lysates.
Expression and purification of glutathione S-transferase–tagged C19ORF5 constructs
The GST fusion proteins indicated in the text were expressed in Escherichia coli BL21, the cells were lysed in buffer I (PBS with 1 mmol/L phenylmethylsulfonyl fluoride, 2 μg/mL pepstatin, 2 μg/mL leupeptin, 10 μg/mL aprotinin, 1 mmol/L EDTA, 0.1 mmol/L DTT) containing 0.5 mg/mL lysozyme, and the lysate was subjected to glutathione affinity chromatography according to the manufacturer's recommendations. The purified products were dialyzed in buffer II [50 mmol/L Tris-HCl (pH 8.0)] and then applied to DNA-agarose beads (Amersham Biosciences) at 4°C overnight. Beads were collected, packed into a column, washed thrice with buffer II, and eluted with 0.5 mol/L NaCl in buffer II. The eluate was dialyzed against buffer II. The 37-kDa NH2-terminal segments D667–S766 and S767–L866 purified by glutathione affinity failed to bind to the DNA affinity column. GST-C19ORF5C with multiple bands was purified to achieve homogeneity of the intact 68-kDa product assessed by SDS-PAGE. The product of the A867–E966 construct purified by both glutathione and DNA affinity yielded three bands on SDS-PAGE, the largest of which was 34 kDa instead of the predicted 37.5 kDa. This may be a consequence of conformation on SDS-PAGE but more likely a truncation at the COOH terminus in bacteria because the products all bind to immobilized glutathione. The GST-tagged E967–F1059 construct was insufficiently soluble to recover enough purified material for biochemical assessment in vitro. Protein concentration was determined using Micro-BCA Protein Assay Reagent kit (Pierce Biotechnology, Inc., Rockford, IL). The purified GST fusion products were concentrated at ≥2 mg/mL and stored in aliquots at −80°C until immediate use. Purified C19ORF5 undergoes severe fragmentation on dilution and freezing and thawing accompanied by loss of activities.
Validation of mAb4G1 and analysis of native C19ORF5 in HepG2 cells
A monoclonal anti-C19ORF5 antibody (mAb4G1) was generated in collaboration with A&G Pharmaceuticals, Inc. (Baltimore, MD) using purified GST-C19ORF5C as antigen. The authenticity of the antibody was confirmed by immunoblot of the purified GST-C19ORF5C protein and immunostain of GFP-C19ORF5C-transfected COS7 cells. GST-C19ORF5C or GST (~40 μg) was analyzed by SDS-PAGE and stained with Coomassie blue. About 400 ng was subjected to immunoblot with 1 μg/mL mAb4G1 and 0.1 μg/mL alkaline phosphatase-conjugated anti-mouse antibody. COS7 cells transfected with GFP-C19ORF5C were incubated with 50 μL of 1:20 diluted stock mAb4G1 (0.9 mg/mL) and visualized by treatment with Texas red–conjugated anti-mouse IgG antibody. The subcellular distribution of native C19ORF5 protein in normal or paclitaxel-treated cells was immunostained with the mAb4G1 antibody and FITC-conjugated anti-mouse antibody (Santa Cruz Biotechnology). The antibody mAb4G1 reacted specifically with the C19ORF5 portion of GST-C19ORF5C or its breakdown products after dilution. The mAb4G1-reactive antigen overlapped precisely with the GFP signal from GFP-C19ORF5C in transfected COS7 cells. The mAb4G1 epitope was further localized to amino acid sequence 667 to 766 using deletion mutants of C19ORF5C.
Interaction with microtubules in vitro
Assembly and stabilization of microtubules in vitro using pure proteins was done using modifications of previous reports (28). Phosphocellulose-purified α-tubulin and β-tubulin (Cytoskeleton, Inc., Denver, CO) and C19ORF5 proteins were coassembled into microtubules in G-PEM buffer [80 mmol/L Na-PIPES (pH 6.9), 0.5 mmol/L MgCl2, 1 mmol/L EGTA, 1 mmol/L GTP] in the presence of paclitaxel (Sigma-Aldrich Corp., St. Louis, MO). Association of native C19ORF5 with paclitaxel-stabilized microtubules in HepG2 cell lysates was tested by a described ultracentrifugation procedure with minor modification (29, 30).
Results
Progressive perinuclear accumulation of normally cytosolic C19ORF5C
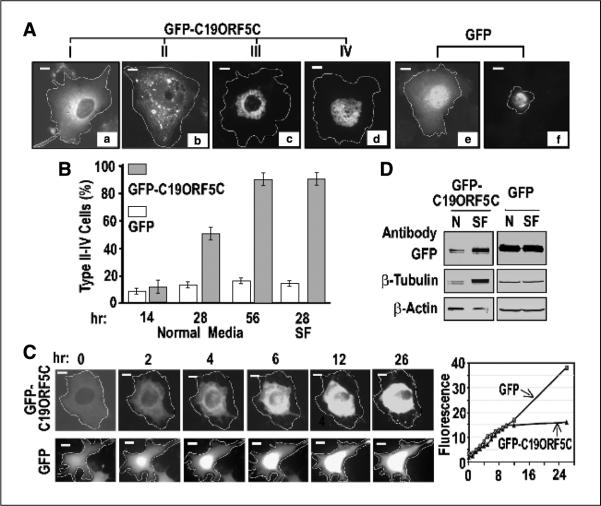
In the absence of inducers of microtubule hyperstabilization, both the full-length C19ORF5 (19) and the 393-residue COOH terminus of C19ORF5 fused to GFP at the NH2 terminus (GFP-C19ORF5C) exhibited a dispersed cytosolic distribution spreading out from a normal nucleus in >90% of COS7 cells after 12 to 14 hours post-transfection (type I cells; Fig. 1A, a). In the remaining 10% of cells, three additional distribution patterns of the GFP signal were noted that were not observed in cultures transfected with GFP alone (11). The distribution in type II cells was still largely distributed throughout the cytosol, but the GFPC19ORF5C also appeared in punctated areas of higher fluorescent intensity (Fig. 1A, b). A third type of morphologic distribution was apparent in type III cells that still exhibited well-defined intact nuclei, but the GFP signal intensified into a perinuclear punctate network (Fig. 1A, c). In a fourth type of distribution (type IV), an intact nucleus was not observed and only the punctate network of highest fluourescent intensity was predominant (Fig. 1A, d). In contrast to the dispersed cytosolic distribution of GFP-C19ORF5C that was excluded from the nucleus in the majority of transfected cells, GFP alone was most intense in the nucleus in addition to the uniform cytosolic distribution (Fig. 1A, e). The intense net-like GFP signal observed in GFP-C19ORF5C-expressing type III and IV cells was unique and in sharp contrast to the intensified but irregular signal from GFP alone that was observed in <10% of cells exhibiting a more conventional apoptotic morphology in GFP-transfected control cultures (Fig. 1A, f). Quantification by direct count of the diverse morphologic cell types indicated by distribution of GFP-C19ORF5C over time revealed that although collectively the type II to IV cells represented <10% of the transfected cell population 14 hours after replacement of the original transfection medium with fresh culture medium, at 28 hours, type II to IV cells represented ~50% of the transfected cell population and >90% by 56 hours (Fig. 1B). Serum-free medium, a condition commonly used to induce environmental stress and accelerate cell death (31), resulted in the increase of type II to IV cells to 90% of total GFP-C19ORF5C-expressing cells as early as 28 hours after transfection (Fig. 1B). This was in marked contrast to the cultures transfected with GFP alone in which type II to IV cells were not observed. Cells exhibiting other than the majority pattern (Fig. 1A, e), including cell morphologies typical of classic apoptosis (Fig. 1A, f), were never >20% of the GFP-positive cell population.
Figure 1.
Cellular distribution patterns of GFP-C19ORF5C. A, four general morphologic profiles of GFP distribution in COS7 cells transfected with GFP-C19ORF5C. Cells were classified as type I to IV according to increasing association with punctiform aggregates. Cell boundaries were located by light microscope and then determined by increasing exposure times to maximum to detect total GFP signal across the field. The transfected cell periphery was then outlined as indicated. Field exposure was then reduced to facilitate maximum resolution of the high-intensity punctate structures. Bars, 10 μm in all photomicrographs. B, time-dependent increase in type II to IV cells exhibiting punctiform aggregates of GFP-C19ORF5C. Columns, mean counts of type I to IV cell types from three independent experiments in which at least 500 total transfected cells among 50 microscopic fields were counted at the indicated times; bars, SD. Normal medium contained 5% fetal bovine serum. SF, serum-free medium. The number of cells exhibiting the conventional apoptotic morphology in (A, f) was scored in cultures transfected with GFP alone (open columns). C, time-dependent perinuclear clustering of GFP-C19ORF5C in single transfected cells at room temperature. Images were captured at the indicated times from single cells observed continuously. Representative cell of replicate observations and time-dependent increase in intensity of GFP fluorescence in single cells expressing GFP-C19ORF5C or GFP. Relative florescence intensity was determined by dividing the average fluorescence intensity in arbitrary units in a field with brightest florescence in the cells by that of a field of the same size outside the cell boundary. D, direct analysis of total GFP-C19ORF5 or GFP, β-tubulin, and β-actin in transfected cells. GFP-C19ORF5C or GFP transfected COS7 cells were cultured in 25 cm2 tissue culture flasks in medium with (N) or without 5% fetal bovine serum (SF) for 28 hours. Extracts were made from 8 × 105 cells collected by scraping into 200 μL buffer I. An equal amount of soluble protein (160 μg) was applied to each lane of SDS-PAGE, and after electrophoretic transfer, GFP-C19ORF5C, GFP, tubulins, and actin were detected with 1 μg/mL polyclonal antibody against GFP or monoclonal antibodies against β-tubulin or β-actin. Bands were visualized with 0.1 βg/mL alkaline phosphatase–conjugated anti-rabbit IgG or anti-mouse IgG antibodies.
To determine whether the increasingly intense network of C19ORF5C condensate at the nucleus reflected in type II to IV morphologies was temporally related and occurred progressively within a single cell, we tracked single cells expressing GFP-C19ORF5C or GFP in transfected cultures and plotted the increase in fluorescent intensity over a period of 26 hours (Fig. 1C). The results confirmed that a time-dependent increase in intensity of GFP signal from GFP-C19ORF5C was coincident with progressive aggregation around the nucleus within a single transfected cell (Fig. 1C). This was quite different from the control GFP signal of which a large part is nuclear that continued to increase in linear mode, whereas intensity of GFP-C19ORF5C that is excluded from nuclei reached a plateau by 12 hours (Fig. 1C). This further indicates a biological effect of C19ORF5C fused to GFP that limits its own synthesis or stability relative to GFP alone whose expression continues to increases linearly with time after 12 hours. Lastly, to confirm that the increase in fluorescent intensity of GFP-C19ORF5C in type II to IV cells reflected an overall increase in GFP-C19ORF5C protein in addition to perinuclear accumulation, we compared the total GFP-C19ORF5C from nonionic detergent extracts of cultures at 28 hours with or without serum. Separate analyses of total cell extracts with SDS indicated that >90% of total cellular antigens analyzed were soluble and in the nonionic detergent-soluble fraction shown here. Compared with β-actin that was assumed to reflect an unchanged standard cell protein, serum-free cultures in which predominantly the type III and IV phenotypes were present exhibited a net increase in both total GFP-C19ORF5C and h-tubulin proteins (Fig. 1D). Comparable changes were not observed in cultures expressing control GFP. This indicates that increasing levels of cellular C19ORF5C occur in the type II to IV morphologies coincident with the perinuclear translocation. As cellular levels of C19ORF5C increase, there is a progressive shift from a general distribution in the cytosol to punctate foci that continues to build into an intense perinuclear aggregate network that ends in apparent nuclear disintegration.
The purified recombinant C19ORF5 COOH terminus interacts with paclitaxel-stabilized microtubules assembled from tubulins in vitro
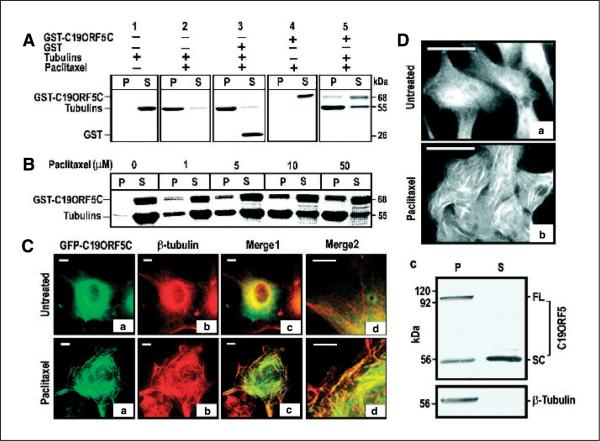
Although even in the type I cells GFP-C19ORF5C did not clearly decorate microtubules in intact transfected cells, we tested for a direct interaction of GST-C19ORF5C with microtubules reconstituted from soluble tubulins (55 kDa) in vitro (Fig. 2A, lanes 1 and 2). Similar to 26-kDa GST alone in the presence of microtubules, no 68-kDa GST-C19ORF5C was detectable in the pellet in absence of tubulin (Fig. 2A, lanes 3 and 4). However, significant levels of GST-C19ORF5C associated with the pelleted microtubules (Fig. 2A, lane 5). Figure 2B further shows that the amount of microtubule-associated GST-C19ORF5C was proportional to the microtubule yield, as yield in the pellet was enhanced with increasing paclitaxel concentration in the assay. This indicated that the COOH terminus of C19ORF5 exhibited a potential microtubule-binding domain that was not apparent in intact cells with arrays of microtubules in dynamic steady state.
Figure 2.
Specific association of the C19ORF5 with paclitaxel-stabilized microtubules. A, association of purified GST-C19ORF5C with paclitaxel-stabilized microtubules. Tubulins (10 μg) were incubated with 10 μg GST-C19ORF5C, GST, or other control GST fusion proteins in G-PEM buffer for 30 minutes at 37°C in the presence of 5 μmol/L paclitaxel. The 20 μL mixtures were layered on 10 μL of 10% sucrose solution in PEM buffer and centrifuged at 10,000 × g for 20 minutes at room temperature. Supernatant (S, 30 μL) containing soluble tubulins and other relatively low molecular weight components and pellet (P) resuspended in 30 μL G-PEM buffer containing stabilized microtubules and associated proteins were loaded side by side on 7.5% SDS-PAGE. Protein bands were visualized with Coomassie blue stain. B, paclitaxel concentration-dependent association of GST-C19ORF5C protein with microtubules. Microtubules were assembled in the presence of the same amount of GST-C19ORF5C and tubulin employed in (A) with the indicated amounts of paclitaxel. C, specific association of the C19ORF5 with paclitaxel-stabilized microtubules in vivo. COS7 cells treated with 10 μmol/L paclitaxel where indicated at the time of transfection with cDNA coding for GFP-C19ORF5C were stained with anti-β-tubulin and Texas red–conjugated anti-mouse antibody. The relationship of GFP (green) and anti-β-tubulin antibody (red) fluorescence was observed and colors merged to test for overlap of signals (yellow). Merge2, segments of cells in Merge1 magnified 2.5 times. Representative of 90% of transfected cells from three independent experiments. D, distribution of native C19ORF5 in normal (a) and paclitaxel-treated HepG2 cells (b) and its association with paclitaxel-stabilized microtubules from HepG2 cell lysate in vitro (c). Cells were treated with paclitaxel as indicated and analyzed with mAb4G1. Representative of the majority of cells in three independent experiments. For in vitro assembly of microtubules, ~1 × 106 HepG2 cells were lysed in 150 μL buffer and followed by sonication for 10 seconds. The cell lysate was clarified by centrifugation at 10,000 × g for 20 minutes and treated with 10 μmol/L paclitaxel at 37°C for 1 hour to stabilize microtubules and the reaction mixture was loaded on a cushion of 20 μL of 20% sucrose. Stabilized microtubules and associated proteins were sedimented by ultracentrifugation (Sorvall RC M120) at 100,000 × g for 1 hour. The pellet containing the stabilized microtubules was resuspended in 150 μL buffer. An equal volume of pellet and supernatant (25 μL) fractions were analyzed on SDS-PAGE and immunoblotted with antibodies against C19ORF5 (mAb4G1) or β-tubulin, respectively. FL, full-length C19ORF5 predicted from cDNA; SC, short chain.
GFP-C19ORF5C associates with paclitaxel-stabilized microtubules in transiently transfected COS7 cells
Because the yield of stable sedimentable microtubules formed from tubulins in vitro necessarily required stabilization with paclitaxel, we reexamined the distribution of C19ORF5 in cells transfected with cDNA coding for a GFP-C19ORF5C after treatment with paclitaxel. Remarkably, the GFP-C19ORF5C that was normally distributed throughout the cytosol among normal microtubular arrays with no clear association with them in untreated cells was concentrated most often in distinct foci on microtubule bundles induced by the paclitaxel treatment (Fig. 2C). This indicated that C19ORF5 accumulated on specifically hyperstabilized microtubules from its dispersed cytosolic location and a microtubule association domain resides within the 393–amino acid residue COOH terminus of C19ORF5. The association seems to be direct and independent of other cofactors as RASSF1.
Association of native C19ORF5 with paclitaxel-stabilized microtubules in differentiated liver cells
The distribution of endogenous native C19ORF5 was analyzed in well-differentiated human hepatoma cells (HepG2) using a monoclonal antibody (mAb4G1). Similar to the overexpressed C19ORF5C in COS cells, native C19ORF5 was distributed diffusely in the cytosol with an increasing intensity around the nucleus and not distinctly associated with microtubules (Fig. 2D, a). However, similar to cells expressing recombinant C19ORF5C, native C19ORF5 was concentrated on the bundled microtubular structures in paclitaxel-treated cells (Fig. 2D, b). Recovery and immunochemical analysis of the paclitaxel-stabilized microtubules from lysates of HepG2 cells revealed that all of the 113-kDa band representing the full-length C19ORF5 was associated with the pelleted microtubule fraction, whereas a 56-kDa short chain was equally distributed between pellet and supernatant (Fig. 2D, c). These results show that the recruitment of transfected recombinant 393-residue COOH terminus of C19ORF5 accurately reflects the recruitment of native full-length C19ORF5 to specifically hyperstabilized microtubules.
Punctate aggregates of C19ORF5C reflect association with mitochondrial aggregates
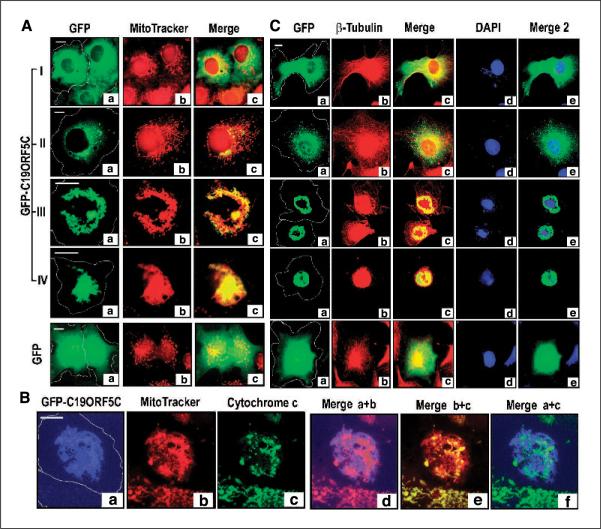
The punctate pattern of GFPC19ORF5C observed in type II cells and particularly the net-like nuclear-associated aggregates observed in type III and IV cells (Fig. 1A, c and d) were suggestive of reticular networks of mitochondria and more specifically the mitochondrial clustering that is associated with some forms of cell death (23, 32). Because of this, the association of C19ORF5C with mitochondria-associated protein LRPPRC and other mitochondrial proteins by yeast two-hybrid analysis (14), we examined the relationship of C19ORF5 in the four types of cells described in Fig. 1A with MitoTracker dye. MitoTracker is a selective mitochondrial indicator stain that requires an intact mitochondrial membrane potential to concentrate it for retention (33). In type I cells exhibiting the diffuse cytosolic distribution of C19ORF5C, mitochondria were evenly distributed in their characteristic punctate pattern and neither exhibited a particularly characteristic association indicated by overlap analysis with GFP-C19ORF5C (Fig. 3A). In type II cells, the majority of mitochondria were still largely normal in the expected dispersed cytosolic arrays. However, punctate areas of increased GFP intensity from GFP-C19ORF5C clearly overlapped with clusters of mitochondria that were out in the cytosol and most intense at the nucleus that were no longer characteristic of the distinctive reticular network of normal mitochondria (Fig. 3A, II). The intense perinuclear aggregates of C19ORF5C in type III cells were associated with apparently fused aggregates of perinuclear mitochondria that have completely lost the distinct reticular pattern of cytosolic mitochondria distributed along the microtubular cytoskeleton (Fig. 3A, III). The overlap of GFP-C19ORF5C with aggregates of mitochondria indicated by MitoTracker (yellow in the merge) instead of normally distributed networks of mitochondria (red in the merge) among the majority population of C19ORF5C-associated aggregates indicates that C19ORF5 is selectively associated with aggregated mitochondria. In the type IV cells, very few mitochondria that do not overlap with C19ORF5C are visible (Fig. 3A, IV). Intense fused aggregates of overlapping GFP-C19ORF5C and mitochondria penetrated and completely enveloped what was the nucleus. In contrast to the type II to IV morphologies caused by transfection with GFP-C19ORF5C, cells transfected with only GFP exhibited the normal punctate cytosolic networks of mitochondria (Fig. 3A). In sum, these results show that the time-dependent increase in intensity and punctate pattern of C19ORF5C distribution with time and accumulation of C19ORF5 represents an increasing degree of mitochondrial aggregation with which C19ORF5 is associated relative to its normal uniform cytosolic distribution.
Figure 3.
MAGD caused by C19ORF5. A, relationship of GFP-C19ORF5C and mitochondria in type I to IV cells. Cell boundaries were determined and then outlined as described in Fig. 1. The green GFP signal (a) was merged with that of red MitoTracker (b) in Merge (c). Yellow, overlap of the GFP signal with mitochondria. B, relation of GFP-C19ORF5C-associated mitochondria with cytochrome c. To achieve maximum color contrast, the strong GFP signal from GFP-C19ORF5C was artificially assigned blue. The cytochrome c signal was assigned green. The overlap of blue GFP-C19ORF5C with red MitoTracker, red MitoTracker with green cytochrome c, or blue GFP-C19ORF5C with green cytochrome c is indicated by purple, yellow, or cyan in Merge d, e, or f, respectively. C, coincidence of microtubular collapse and DNA degradation with C19ORFC-associated mitochondrial aggregation. The green GFP signal from GFP-C19ORF5 (a) was analyzed with that of red β-tubulin immunochemical stain (b) in Merge (c) and blue DAPI DNA stain (d) in Merge 2 (e). Yellow in Merge is indicative of overlap of the GFP signal with tubulin and cyan in Merge 2 is indicative of overlap of the GFP signal with DAPI. Microtubules were visualized by immunoanalysis as described in Materials and Methods. Control cells transfected with GFP alone are indicated.
C19ORF5C-associated mitochondrial aggregates are cytochrome c–deficient and exhibit reduced membrane integrity
We noted that the mitochondria associated with GFP-C19ORF5C exhibited a diminished MitoTracker signal compared with normal adjacent reticular networks (Fig. 3A, III). Because retention of dye is dependent on mitochondrial membrane potential, this was suggestive of a reduction in potential in specifically the mitochondrial aggregates. Cytochrome c release from mitochondria is a common measure of loss of mitochondrial integrity associated with cell death and delivery of genome destructive elements from permeabilized mitochondria (34–36). Therefore, we compared the distribution of cytochrome c in the C19ORF5-induced mitochondrial aggregates with which C19ORF5 was associated to normally dispersed mitochondria that exhibited little overlap with C19ORF5C. As shown in Fig. 3B, a and b, areas of weakest MitoTracker (green) intensity exhibited maximum overlap with GFP-C19ORF5C (blue) indicated by purple in merged Fig. 3B, d. Consistent with a loss of mitochondrial integrity indicated by the diminished MitoTracker signal, areas of weakest cytochrome c staining (green; Fig. 3B, c) overlapped with areas of weakest MitoTracker retention (Fig. 3B, e). In contrast, normal mitochondria exhibited maximum overlap of MitoTracker and cytochrome c (yellow; Fig. 3B, e) as illustrated most effectively in the neighboring untransfected cell at the bottom of the panels. GFP-C19ORF5C-associated mitochondria were largely depleted of cytochrome c (Fig. 3B, f).
Microtubular cytoskeletal collapse and DNA degradation occurs with perinuclear aggregation of C19ORF5C-associated mitochondria
Because distribution of mitochondria is normally dependent on the microtubular cytoskeleton and the transition from type I to type III and IV cells culminated in severe nuclear perturbation, we examined the status of microtubules with anti-β-tubulin and nuclear DNA with DAPI stain in type I to IV cell phenotypes. As reported previously (11, 19), C19ORF5C is dispersed in the cytosol with no particular association with normal microtubular arrays in the majority of cells (type I) at early times after transfection (Fig. 3C). Type I cells exhibited a normal nuclear morphology and DNA content. The cytoskeleton and nuclei of type II cells exhibiting punctate foci of mitochondria-associated GFP-C19ORF5C were still intact. However, in type III and IV cells exhibiting extensive perinuclear aggregation of GFP-C19ORF5C with mitochondria, coincident perinuclear collapse of the microtubular cytoskeleton was apparent. The DNA stain confirmed that the nuclear morphology was severely perturbed in type III and IV cells and a significant loss of DNA within the nuclear condensates was evident. The DNA decondensation in areas with C19ORF5 aggregates represented an inverse phenomenon of the DNA condensation observed in typical apoptosis. In type IV cells in which GFP-C19ORF5C, mitochondria, and β-tubulin were most intense, intact nuclei could not be distinguished and only traces of DNA remained in the spherical cellular remnants. These phenotypes were in sharp contrast to cells transfected with GFP alone that exhibited a normally dispersed microtubular cytoskeleton and an intact nucleus with normal DNA content unquenched by the strong nuclear GFP signal in >90% of cells at all times. These results show that collapse of the microtubular cytoskeleton accompanies the accumulation of C19ORF5C and perinuclear aggregation of C19ORF5C-associated mitochondria. The fact that association of C19ORF5C with mitochondria and some aggregation was evident in type II cells before collapse of microtubular arrays suggested that cytoskeletal collapse may be the consequence rather than cause of the mitochondrial aggregation. Disintegration of the nuclear structure and global DNA degradation seems to be an end consequence of progressive accumulation of C19ORF5 on mitochondria and the aggregation of C19ORF5-associated mitochondria. We subsequently refer to the process as MAGD.
Localization of the hyperstabilized microtubule-binding activity to a highly basic sequence domain in the C19ORF5 COOH terminus
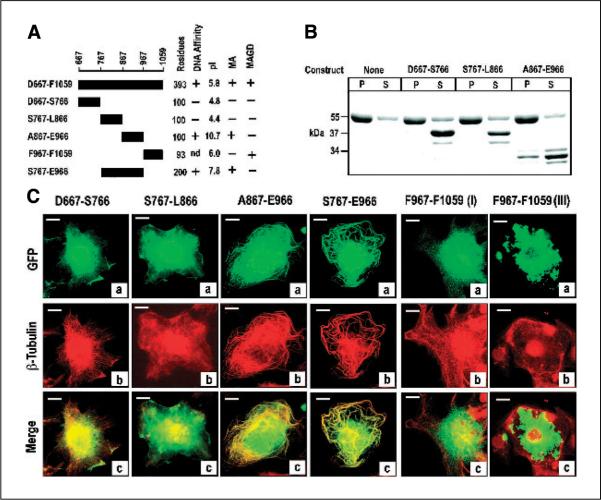
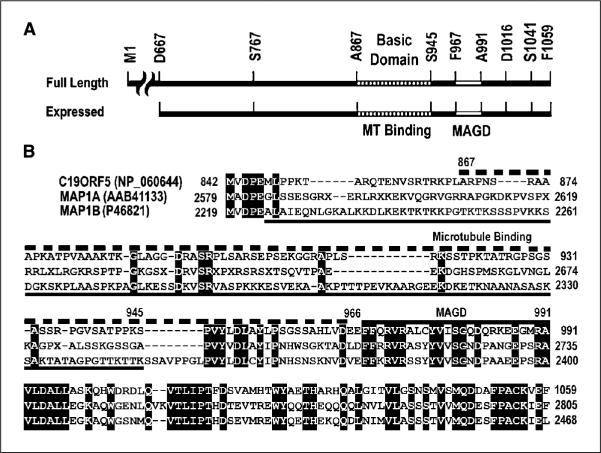
To determine whether a distinct microtubule-binding domain could be shown within C19ORF5C, we constructed, expressed in bacteria, and purified a series of fragments tagged with GST (Fig. 4A). The GST fusion constructs were then examined for their association with paclitaxel-stabilized microtubules in vitro as described for GST-C19ORF5C in Fig. 2A. Only the ~33-kDa truncated products of A867-E966 were observed associated with pelleted stabilized microtubules (Fig. 4B). We then examined the microtubular association of products from the constructs listed in Fig. 4A, with a GFP tag substituted for the GST in transfected COS7 cells. Similar to the parent 393-residue C19ORF5C (Fig. 2C), none of the green GFP-tagged products exhibited association with the microtubular cytoskeleton in steady state indicated by overlap with β-tubulin (red; data not shown). Therefore, we examined distribution of the GFP signal in transfected cells treated with paclitaxel. Only GFP-tagged A867-E966 and S767-E966 associated with the hyperstabilized microtubules induced by the paclitaxel treatment (Fig. 4C). Notably, a significant amount of GFP signal from all partial constructs appeared in the nucleus in addition to decorating arrays of stabilized microtubules in the cytosol in contrast to 393-residue C19ORF5C. This may indicate that flanking sequences in C19ORF5C underlie exclusion of the GFP from the nucleus or that partial degradation of constructs down to or near GFP that appears in the nucleus when expressed as a single product. In addition to a lack of association with microtubules, construct S767-L866 seemed to cause disorganization of microtubular arrays relative to the other constructs (Fig. 4C). In contrast, only construct F967-F1059 was cytotoxic and caused the time-dependent aggregation of mitochondria characteristic of the C19ORF5C (Fig. 3A, III). Together, these results indicate that hyperstabilized microtubule-binding activity of C19ORF5 resides within the highly basic 100 amino acid residues within A867-E966. Because an apparent COOH-terminally truncated product that may be as few as 60 to 80 residues from A867, the microtubule-binding domain may reside in the highly basic sequence A867 to K945 or less. Theoretical calculated pI values for all constructs examined are summarized in Fig. 4A.
Figure 4.
Identification of the hyperstabilized microtubule-binding domain within the C19ORF5 COOH terminus in vitro. A, sequence and properties of constructs. The indicated constructs were fused with GST or GFP at their NH2 terminus. The GST fusions were expressed, recovered, and purified using glutathione and DNA affinity as described in Materials and Methods. Numbering is according to full-length cDNA of C19ORF5, single letter code for amino acids at the NH2- and COOH-terminal residue of the product, and the total residues are indicated. Physicochemical and functional properties of the construct products are summarized on the right. Theoretical pI values were calculated using the Web-based tool from http://us.expasy.org/tools/pi_tool.html. MA, microtubule association activity; MAGD, cytotoxic MAGD activity; nd, not determined. B, binding to paclitaxel-stabilized microtubules in vitro. Association of the indicated constructs with paclitaxel-stabilized microtubules assembled from tubulins was determined as described in Fig. 1A. The 55-kDa band is tubulin and the 37-kDa or smaller bands are the GST fusions and their lower molecular weight truncates. As indicated in Materials and Methods, purified products of the A867-E966 construct were ≤34 kDa. Insufficient recovery of purified GST-tagged F967-F1059 from bacteria prevented assessment in vitro. The purified GST-S767-E966 with predicted molecular weight of 47 kDa overlapped tubulin on SDS-PAGE and is not shown. P, pellet containing microtubules; S, supernatant containing unpolymerized tubulin. C, confirmation of the hyperstabilized microtubule-binding domain in C19ORF5. COS cells transfected with the indicated constructs with GFP at the NH2 terminus were treated with paclitaxel and cells were examined for association of the green GFP signal with hyperstabilized microtubules as well as overlap with red anti-β-tubulin as in Fig. 2. Representative of near 100% of transfected cells with the first four constructs, 90% and 10% of transfected cells with F967-F1059, respectively, in at least three independent experiments in which >2,000 cells were examined.
A distinct sequence independent of the microtubule-binding domain supports C19ORF5 mitochondrial aggregation and genome destruction activity
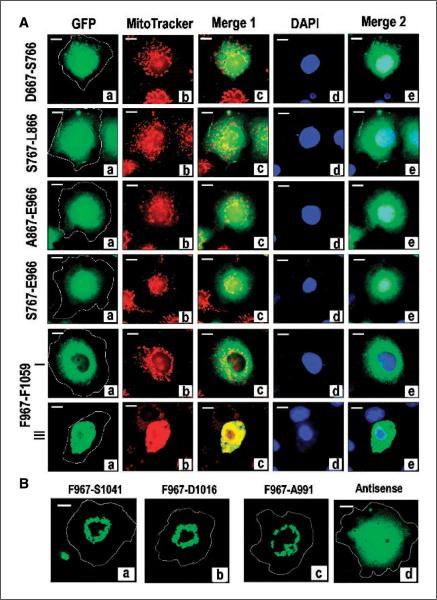
To determine whether the sequence required for MAGD activity was overlapping or distinct and independent, we compared effect, distribution, and overlap with mitochondria of products from the same GFP-tagged constructs. The expression levels and solubility of full-length products of the indicated constructs in the transfected COS cells was verified by comparison of extracts of the indicated transfected cultures with nonionic detergent to that with SDS and subsequent analysis on SDS-PAGE (data not shown). Cells exhibiting the type II to IV morphology when transfected with GFP-tagged D667–S766, S767–L866, A867–E966, or S767–E966 could not be detected. Treatment with MitoTracker and DAPI indicated that all transfected cells exhibited the normal reticular distribution of mitochondria in the cytosol and normal nuclear morphology and DNA content (Fig. 5A). In contrast, the product of GFP-labeled F967–F1059 that is devoid of the hyperstabilized microtubule-binding domain was excluded from the nucleus similar to the full-length 393-residue C19ORF5 COOH terminus (Fig. 5A). Although ~90% of cells exhibited the normal type I morphology, similar to GFP-C19ORF5C, ~10% of cells collectively exhibited the type II to IV phenotype that increased with increasing time after transfection. This indicated that the MAGD domain is distinct and downstream of the microtubule-binding domain.
Figure 5.
Identification of an independent MAGD domain. A, analysis of GFP-tagged constructs coding for subdomains of C19ORF5C. COS cells were transfected with the indicated constructs with GFP at the NH2 terminus and cells were stained with MitoTracker and DAPI dyes. Cultures were examined for type I to IV phenotypes 24 hours after transfection in the same way as described for GFP-C19ORF5C. Yellow in Merge 1 and cyan in Merge 2 indicate overlap of green GFP-C19ORF5C with mitochondria (red) or DNA (blue), respectively. Representative cell from each transfected culture. Type II to IV cell morphologies were only observed in cultures transfected with F967-F1059 at the relative frequencies described for GFP-C19ORF5C in Fig. 1. Representative type I and III cell. B, reduction of the MAGD domain to a 25-residue sequence. The indicated constructs of MAGD domain-containing GFP-F967-F1059 with sequential deletions at the COOH terminus were tested for induction of the type II to IV phenotypes indicative of MAGD activity. Antisense, coding sequence for GFP fused to the in-frame antisense coding sequence for F967-A991.
Lastly, we tested whether the MAGD-promoting sequence domain could be further reduced by construction and observation of three progressive deletions at the COOH terminus of GFP-F967–F1059 (Fig. 5B). All exhibited MAGD activity indicated by the presence of type II to IV cell morphologies. This indicated that as few as 25 amino acid residues encoded in construct F967–A991 underlie the MAGD activity. An antisense construct of F967–A991 encoding an unrelated GFP-tagged 25–amino acid peptide exhibited no MAGD activity and a distribution characteristic of GFP. This indicated that the GFP and the linker sequence between GFP and constructs had no contribution to the MAGD activity and cellular distribution of the C19ORF5 constructs. These results show that distinct and independent sequence domains within C19ORF5C support the hyperstabilized microtubule-binding and mitochondrial aggregation activities. The distinct sequence domains of C19ORF5 and a comparison to homologous regions of MAP1A and MAP1B are summarized in Fig. 6.
Figure 6.
Comparison of functional sequence domains in the COOH terminus of C19ORF5 to sequence of the light chains of MAP1A and MAP1B. A, sequence domain structure of C19ORF5. Full-length C19ORF5 and the 393–amino acid residue C19ORF5C (D667-F1059) with residues flanking constructs used in this study are indicated. A867-S945 indicates the highly basic microtubule-binding domain. F967-A991 is the MAGD domain. B, sequence alignment of human C19ORF5 with the light chains of MAP1A (MAP1A-LC2) and MAP1B (MAP1B-LC1). Identical residues in all three sequences are in black. Dashed bar (top), C19ORF5 sequence A867-E966 containing the microtubule-binding domain; bottom solid line, microtubule-binding domain of MAP1B (45); top solid bar, MAGD domain of C19ORF5.
Discussion
Here, we show that C19ORF5 normally distributes in the cytosol but is recruited to stabilized microtubules and as it builds up with time causes aggregation of mitochondria resulting in cell death and destruction of genomic DNA (MAGD). Association with stabilized microtubules and MAGD activity are distinct and independent activities determined by separate sequence domains within the last 200 residues of the COOH terminus of C19ORF5. Aggregation and perinuclear clustering of mitochondria has been observed associated with some forms of cell death, although the role of mitochondrial aggregation as an active participant or consequential end point in each type remains to be established (23, 34, 36, 37). Diverse forms of perinuclear aggregation of mitochondria have been reported to be associated with activation of tumor necrosis factor (34, 36, 37), treatment with staurosporine (38), expression of hepatitis B virus X protein (39), the Bcl-2-related proapoptotic protein Bax (23, 40), Bim (29), and Bid (41), and disruption of the microtubule-associated protein Tau (42) and motor protein kinesin (23, 32). The net-like tangles of C19ORF5C-induced C19ORF5C-associated mitochondria and microtubules culminating with complete engulfment of the nucleus and gross destruction of genomic DNA are distinct from prior reports where perinuclear-associated mitochondria were reported as largely punctiform organelle fragments surrounding intact nuclei (23). The C19ORF5-induced phenotypes were also in sharp contrast to the occasional dying cells showing condensates of nuclear DNA in cultures transfected with GFP alone or those induced by another pro–cell death partner of LRPPRC, cat eye syndrome chromosome region candidate 2 (11). Notably in C19ORF5C-transfected cells, nuclear DNA appeared progressively decondensed until its complete degradation within perinuclear aggregates of C19ORF5 and mitochondria. Degradation of genomic DNA was sufficiently severe to preclude detection of partial DNA fragmentation using conventional DNA ladder analysis of cell extracts or terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling analysis in situ in the relevant cell morphologies. This is consistent with the massive engulfment of nuclei by the mitochondrial aggregates, their weaker staining with MitoTracker dye whose intensity is proportional to intact membrane potential and extensive cytochrome c deficiency. Permeabilized mitochondria are a source of nuclease activity as well as the trigger for other nonmitochondrial nucleases involved in cell death (43, 44). We refer to this unconventional mode of cell death as MAGD because of its potential importance in prevention of propagation of a defective genome caused by catastrophic mitotic microtubule spindle defects.
A hallmark of C19ORF5 is its homology to microtubule-associated proteins MAP1A and MAP1B (14). Unlike C19ORF5, MAP1A and MAP1B seem to independently associate with microtubular arrays under steady-state conditions and overexpression results in stabilization and resistance of microtubules to nocodazole (45). Moreover, the light chains of MAP1A and MAP1B also interacted with actin stress fibers when overexpressed in mammalian cells (46). Here, we show in agreement with others (19) that even at high levels achieved by overexpression, C19ORF5 neither associates with normal steady-state arrays of microtubules nor causes microtubule stabilization. Moreover, neither the 393-residue COOH terminus of C19ORF5 (11) nor full-length C19ORF53 exhibits interaction with the actin cytoskeleton. Instead, C19ORF5 is specifically recruited to hyperstabilized microtubules. The association seems direct, occurs in vitro with microtubules reconstituted and stabilized with taxol from purified tubulins, and is independent of cellular cofactors, as its interaction partners the RASSF1 isoforms that at sufficient levels cause paclitaxel-like microtubule stabilization (16–19, 47).
C19ORF5 plays a pivotal role in specificity of the RASSF1A isoform of products of the RASSF1 gene for association with microtubules and promotion of microtubule hyperstabilization (11, 27). Although the A and C isoforms of RASSF1 are expressed in most tissues, specifically the A isoform is silenced in a variety of human tumors by epigenetic hypermethylation of CpG islands in specifically the RASSF1A promoter, whereas the RASSF1C isoforms remain constant (20–22). Both RASSF1A and RASSF1C alone exhibit identical properties with respect to cellular location, trafficking from mitochondria to microtubules to cause hyperstabilization, interference with cell cycling, and tumor suppression properties in model tumor cells and tissues (11, 16–19, 27). However, only complexes of the COOH terminus of C19ORF5 and RASSF1A associate with and hyperstabilize microtubules (27). Complexes of C19ORF5C and RASSF1C fail to associate with microtubules. This indirectly confers specificity on the hypermethylated suppressed RASSF1A isoform for association with microtubules and induction of microtubule hyperstabilization required for accumulation of C19ORF5. Accumulation on hyperstabilized microtubules may increase the local concentration of C19ORF5 to levels mimicked by overexpression of C19ORF5 that result in initiation of MAGD. This differential interaction of C19ORF5 with RASSF1 isoforms may underlie specific tumor suppression activity of only RASSF1A. Because of its dual functional microtubule-binding and MAGD domains, C19ORF5 is well endowed to sense prolonged and potentially catastrophic stabilization of spindle microtubules that threatens cells with chromosome missegregation at mitosis and thus to play a role in prevention of inheritance of the resultant genomic defect. Aneuploidy that could arise at a single mitosis is the single most common abnormality associated with all cancers and thought to underlie the genetic instability that allows their plasticity (1–3). C19ORF5 is a candidate for mediation of the tumor suppression activity of the apparent natural paclitaxel-like microtubule stabilizer RASSF1A because of its particular affinity for it. However, because of its independent affinity for hyperstabilized microtubules, it may also play a role in tumor suppression by the taxoid class of chemotherapeutic agents.
The COOH-terminal most 200 residues of the 393–amino acid residue recombinant COOH terminus of C19ORF5 employed in this study exhibited all activities of full-length C19ORF5 described to date (19). By homology, this domain corresponds to the light chains of microtubule-associated proteins MAP1A and MAP1B (Fig. 6). The full-length translation products of MAP1A and MAP1B are processed into heavy chains composed of the NH2-terminal 2,607 and 2,210 residues, respectively, and light chains composed of the COOH-terminal 222 and 250 residues, respectively. The heavy and light chains oligomerize and associate with microtubules in a 1:2 ratio (48). Here, we showed that the 113-kDa full-length native C19ORF5 is a major product associated with hyperstabilized microtubules in lysates of human hepatoma cells, making it unlikely that C19ORF5 requires post-translational processing into the heavy and light chain counterparts of MAP1A/MAP1B to interact with microtubules. More work is needed to determine whether proteolytic processing or domain-specific splice variants of C19ORF5 play a role in linking microtubule status to mitochondria-associated cell death and genome destruction in various tumor tissues and cells. Recently, an alternative splice variant from human neuroblastoma cells (CR610661) has been reported as a full-length cDNA in Genbank whose predicted product is the light chain counterpart of C19ORF5 that comprises the last 218 residues within the 393-residue COOH terminus employed in our study.
A surprising and major finding of our study is the definition of a minimal and independent 25–amino acid residue sequence within the C19ORF5 COOH terminus that was responsible for the MAGD activity of C19ORF5 (Fig. 6). The sequence domain is distinct from the microtubule association domain that is spatially homologous to microtubule association domains in the light chains of MAP1A/MAP1B. Except for the characteristic enrichment of basic residues, the microtubule-binding domains among C19ORF5, MAP1A, MAP1B, and other microtubule-associated proteins, such as MAP2 and Tau, are unique to each factor (49). This likely reflects the unique way in which each interacts with microtubules and their effect on the microtubular cytoskeleton. In contrast to the weak homology among the microtubule association domains of C19ORF5 and MAP1A/MAP1B that does not exceed 3% in identity, both MAP1A and MAP1B exhibit MAGD sequence domains that are 52% homologous to that in C19ORF5. This suggests an unrecognized potential role of MAP1A/MAP1B in mitochondria-associated cell death similar to that we have described for C19ORF5. In contrast to C19ORF5 that is widely expressed in tissues (14), expression of MAP1A and MAP1B is restricted to adult and developing neuronal tissue, respectively, where it plays important roles in neuronal development and differentiation (45, 46). MAP1B expression is specific to the neural system, the earliest microtubule-associated protein expressed in the developing nervous system, and becomes silent in adult neurons (49). It becomes reactivated in neurofibrillary tangles and senile plaques in neurites and has been associated with neuronal cell death in Alzheimer's disease (45). Our results suggest a reexamination of a potential role of the light chains of MAP1A and MAP1B in quality control of microtubules and cell death in the neural system is warranted. The results of Mei et al. (48) have suggested roles of light chains beyond their association with microtubules and showed that independently expressed MAP1B light chain appeared in granular filamentous structures suggestive of aggregated mitochondria. Lastly, Dallol et al. reported that candidate tumor suppressor RASSF1 isoforms interact with MAP1B in addition to C19ORF5. Because the expression of MAP1B is largely limited to the nervous system, the MAP1B-RASSF1 partnership, both of which associate with and stabilize microtubules at sufficient levels, may play a specialized role in microtubule quality in differentiation, function, and homeostasis in neural tissues. The wide expression of C19ORF5 in tissues, its recognition of specifically hyperstabilized microtubules, and its determination of specificity of hypermethylation-suppressed RASSF1A in vivo make it a well-suited partner for mediation of RASSF1A tumor suppression in a wide range of tissues.
Taken together, our results suggest an intimate communication among mitochondria and microtubules mediated by multiple but independent functional domains within RASSF1A and C19ORF5, with potential roles in normal microtubule dynamics, monitoring microtubule spindle quality, and triggering tumor suppression through MAGD if needed.
Acknowledgments
Grant support: National Institutes of Diabetes, Digestive and Kidney Diseases USPHS grants DK35310 and DK47039, National Cancer Institute, NIH grant CA59971, and Distinguished Visiting Scholar grant from the Office of the Vice-President of Research, Texas A&M University (G. Liu).
We thank Dr. Serafín Piñol-Roma (Mount Sinai School of Medicine at New York, New York, NY) for mouse monoclonal antibody against LRPPRC and Dr. Le Sun and Joe Corvera (A&G Pharmaceutical) for collaboration in preparation of the mouse monoclonal antibody (mAb4G1) against C19ORF5.
Footnotes
Unpublished results.
References
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 2.Duesberg P, Rasnick D. Aneuploidy, the somatic mutation that makes cancer a species of its own. Cell Motil Cytoskeleton. 2000;47:81–107. doi: 10.1002/1097-0169(200010)47:2<81::AID-CM1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 3.Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer. 2001;1:109–17. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- 4.Schneider BL, Kulesz-Martin M. Destructive cycles: the role of genomic instability and adaptation in carcino-genesis. Carcinogenesis. 2004;25:2033–44. doi: 10.1093/carcin/bgh204. [DOI] [PubMed] [Google Scholar]
- 5.Sorger PK, Dobles M, Tournebize R, Hyman AA. Coupling cell division and cell death to microtubule dynamics. Curr Opin Cell Biol. 1997;9:807–14. doi: 10.1016/s0955-0674(97)80081-6. [DOI] [PubMed] [Google Scholar]
- 6.Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–27. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- 7.Pihan GA, Doxsey SJ. The mitotic machinery as a source of genetic instability in cancer. Semin Cancer Biol. 1999;9:289–302. doi: 10.1006/scbi.1999.0131. [DOI] [PubMed] [Google Scholar]
- 8.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 9.Downing KH. Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu Rev Cell Dev Biol. 2000;16:89–111. doi: 10.1146/annurev.cellbio.16.1.89. [DOI] [PubMed] [Google Scholar]
- 10.Nogales E. Structural insight into microtubule function. Annu Rev Biophys Biomol Struct. 2001;30:397–420. doi: 10.1146/annurev.biophys.30.1.397. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Vo A, Liu G, McKeehan WL. Novel complex integrating mitochondria and the microtubular cytoskeleton with chromosome remodeling and tumor suppressor RASSF1 deduced by in silico homology analysis, interaction cloning in yeast, and colocalization in cultured cells. In Vitro Cell Dev Biol Anim. 2002;38:582–94. doi: 10.1290/1543-706x(2002)38<582:ncimat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mili S, Pinol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol Cell Biol. 2003;23:4972–82. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuchiya N, Fukuda H, Sugimura T, Nagao M, Nakagama H. LRP130, a protein containing nine pentatricopeptide repeat motifs, interacts with a single-stranded cytosine-rich sequence of mouse hyper-variable minisatellite Pc-1. Eur J Biochem. 2002;269:2927–33. doi: 10.1046/j.1432-1033.2002.02966.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, McKeehan WL. Sequence analysis of LRPPRC and its SEC1 domain interaction partners suggest roles in cytoskeletal organization, vesicular trafficking, nucleocytosolic shuttling and chromosome activity. Genomics. 2002;79:124–36. doi: 10.1006/geno.2001.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mootha VK, Lepage P, Miller K, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci U S A. 2003;100:605–10. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Tommasi S, Lee DH, Dammann R, Pfeifer GP. Control of microtubule stability by the RASSF1A tumor suppressor. Oncogene. 2003;22:8125–36. doi: 10.1038/sj.onc.1206984. [DOI] [PubMed] [Google Scholar]
- 17.Song MS, Song SJ, Ayad NG, et al. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat Cell Biol. 2004;6:129–37. doi: 10.1038/ncb1091. [DOI] [PubMed] [Google Scholar]
- 18.Vos MD, Martinez A, Elam C, et al. A role for the RASSF1A tumor suppressor in the regulation of tubulin polymerization and genomic stability. Cancer Res. 2004;64:4244–50. doi: 10.1158/0008-5472.CAN-04-0339. [DOI] [PubMed] [Google Scholar]
- 19.Dallol A, Agathanggelou A, Fenton SL, et al. RASSF1A interacts with microtubule-associated proteins and modulates microtubule dynamics. Cancer Res. 2004;64:4112–6. doi: 10.1158/0008-5472.CAN-04-0267. [DOI] [PubMed] [Google Scholar]
- 20.Burbee DG, Forgacs E, Zochbauer-Muller S, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–9. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–9. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 22.Dammann R, Schagdarsurengin U, Strunnikova M, et al. Epigenetic inactivation of the Ras-association domain family 1 (RASSF1A) gene and its function in human carcinogenesis. Histol Histopathol. 2003;18:665–77. doi: 10.14670/HH-18.665. [DOI] [PubMed] [Google Scholar]
- 23.Desagher S, Martinou JC. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 2000;10:369–77. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 24.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–33. [PubMed] [Google Scholar]
- 25.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–6. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 26.Leist M, Jaattela M. Four deaths and a funeral: from caspases to alternative mechanisms. Nat Rev Mol Cell Biol. 2001;2:589–98. doi: 10.1038/35085008. [DOI] [PubMed] [Google Scholar]
- 27.Liu L, Vo A, McKeehan WL. Specificity of the methylation-suppressed A isoform of candidate tumor suppressor RASSF1 for microtubule hyperstabilization is determined by cell death inducer C19ORF5. Cancer Res. 2005;65:1830–8. doi: 10.1158/0008-5472.CAN-04-3896. [DOI] [PubMed] [Google Scholar]
- 28.Eichenmuller B, Everley P, Palange J, Lepley D, Suprenant KA. The human EMAP-like protein-70 (ELP70) is a microtubule destabilizer that localizes to the mitotic apparatus. J Biol Chem. 2002;277:1301–9. doi: 10.1074/jbc.M106628200. [DOI] [PubMed] [Google Scholar]
- 29.Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell. 1999;3:287–96. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- 30.Chen D, Wang M, Zhou S, Zhou Q. HIV-1 Tat targets microtubules to induce apoptosis, a process promoted by the pro-apoptotic Bcl-2 relative Bim. EMBO J. 2002;21:6801–10. doi: 10.1093/emboj/cdf683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui XS, Jeong YJ, Lee HY, Cheon SH, Kim NH. Fetal bovine serum influences apoptosis and apoptosis-related gene expression in porcine parthenotes developing in vitro. Reproduction. 2004;127:125–30. doi: 10.1530/rep.1.00039. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka Y, Kanai Y, Okada Y, et al. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–58. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- 33.Pendergrass W, Wolf N, Poot M. Efficacy of MitoTracker Green trade mark and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry. 2004;61A:162. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- 34.Thomas WD, Zhang XD, Franco AV, Nguyen T, Hersey P. TNF-related apoptosis-inducing ligand-induced apoptosis of melanoma is associated with changes in mitochondrial membrane potential and perinuclear clustering of mitochondria. J Immunol. 2000;165:5612–20. doi: 10.4049/jimmunol.165.10.5612. [DOI] [PubMed] [Google Scholar]
- 35.Haga N, Fujita N, Tsuruo T. Mitochondrial aggregation precedes cytochrome c release from mitochondria during apoptosis. Oncogene. 2003;22:5579–85. doi: 10.1038/sj.onc.1206576. [DOI] [PubMed] [Google Scholar]
- 36.De Vos K, Goossens V, Boone E, et al. The 55-kDa tumor necrosis factor receptor induces clustering of mitochondria through its membrane-proximal region. J Biol Chem. 1998;273:9673–80. doi: 10.1074/jbc.273.16.9673. [DOI] [PubMed] [Google Scholar]
- 37.Suen YK, Fung KP, Choy YM, Lee CY, Chan CW, Kong SK. Concanavalin A induced apoptosis in murine macrophage PU5-1.8 cells through clustering of mitochondria and release of cytochrome c. Apoptosis. 2000;5:369–77. doi: 10.1023/a:1009691727077. [DOI] [PubMed] [Google Scholar]
- 38.Frank S, Gaume B, Bergmann-Leitner ES, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–25. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 39.Takada S, Shirakata Y, Kaneniwa N, Koike K. Association of hepatitis B virus X protein with mitochondria causes mitochondrial aggregation at the nuclear periphery, leading to cell death. Oncogene. 1999;18:6965–73. doi: 10.1038/sj.onc.1203188. [DOI] [PubMed] [Google Scholar]
- 40.Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. Movement of Bax from the cytosol to mitochondria during apoptosis. J Cell Biol. 1997;139:1281–92. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 42.Sato S, Tatebayashi Y, Akagi T, et al. Aberrant τ phosphorylation by glycogen synthase kinase-3β and JNK3 induces oligomeric τ fibrils in COS-7 cells. J Biol Chem. 2002;277:42060–5. doi: 10.1074/jbc.M202241200. [DOI] [PubMed] [Google Scholar]
- 43.Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–9. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 44.Widlak P, Li P, Wang X, Garrard WT. Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J Biol Chem. 2000;275:8226–32. doi: 10.1074/jbc.275.11.8226. [DOI] [PubMed] [Google Scholar]
- 45.Togel M, Wiche G, Propst F. Novel features of the light chain of microtubule-associated protein MAP1B: microtubule stabilization, self interaction, actin filament binding, and regulation by the heavy chain. J Cell Biol. 1998;143:695–707. doi: 10.1083/jcb.143.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noiges R, Eichinger R, Kutschera W, et al. Microtubule-associated protein 1A (MAP1A) and MAP1B: light chains determine distinct functional properties. J Neurosci. 2002;22:2106–14. doi: 10.1523/JNEUROSCI.22-06-02106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rong R, Jin W, Zhang J, Saeed Sheikh M, Huang Y. Tumor suppressor RASSF1A is a microtubule-binding protein that stabilizes microtubules and induces G(2)/M arrest. Oncogene. 2004;23:8216–30. doi: 10.1038/sj.onc.1207901. [DOI] [PubMed] [Google Scholar]
- 48.Mei X, Sweatt AJ, Hammarback JA. Microtubule-associated protein 1 subunit expression in primary cultures of rat brain. Brain Res Bull. 2000;53:801–6. doi: 10.1016/s0361-9230(00)00416-0. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez-Billault C, Jimenez-Mateos EM, Caceres A, Diaz-Nido J, Wandosell F, Avila J. Microtubule-associated protein 1B function during normal development, regeneration, and pathological conditions in the nervous system. J Neurobiol. 2004;58:48–59. doi: 10.1002/neu.10283. [DOI] [PubMed] [Google Scholar]