Abstract
Myosin-X (Myo10) is an unconventional myosin with MyTH4-FERM domains that is best known for its striking localization to the tips of filopodia and its ability to induce filopodia. Although the head domain of Myo10 enables it to function as an actin-based motor, its tail contains binding sites for several molecules with central roles in cell biology, including phosphatidylinositol (3,4,5)-trisphosphate, microtubules and integrins. Myo10 also undergoes fascinating long-range movements within filopodia, which appear to represent a newly recognized system of transport. Myo10 is also unusual in that it is a myosin with important roles in the spindle, a microtubule-based structure. Exciting new studies have begun to reveal the structure and single-molecule properties of this intriguing myosin, as well as its mechanisms of regulation and induction of filopodia. At the cellular and organismal level, growing evidence demonstrates that Myo10 has crucial functions in numerous processes ranging from invadopodia formation to cell migration.
Key words: Myosin-X, Myo10, Filopodia, MyTH4-FERM, Intrafilopodial motility
Introduction
The myosins are a superfamily of actin-based motor proteins that have crucial roles in cellular and organismal physiology (Hartman et al., 2011). Myosins typically consist of a head domain that can bind to actin filaments and generate force, a neck domain that provides binding sites for myosin light chains, and a tail that endows specific properties, such as dimerization and binding to cargo. One particular group of myosins has tails that contain a myosin tail homology 4 (MyTH4) domain and a band 4.1, ezrin, radixin, moesin (FERM) domain. Members of this ‘MyTH-FERM’ superclass of myosins are expressed in organisms ranging from slime molds to humans (Breshears et al., 2010). Because the ciliate Tetrahymena thermophila also expresses myosins that contain MyTH4-FERM domains (Sugita et al., 2011; Williams and Gavin, 2005), myosins with these domains either arose very early in eukaryotic evolution or independently in two different lineages. Importantly, studies in Dictyostelium discoideum indicate that MyTH-FERM myosins have ancient and conserved roles in mediating membrane–cytoskeleton interactions in protrusive structures such as filopodia (Tuxworth et al., 2001). Of the four MyTH-FERM myosins that are expressed in humans – Myo7a, Myo7b, Myo10 and Myo15a – two are associated with human disease, with mutations in Myo15a causing deafness (Wang et al., 1998) and mutations in Myo7a resulting in both deafness and blindness (Weil et al., 1995).
Myo10 is the founding member of the class X myosins and is the only MyTH-FERM myosin with pleckstrin homology (PH) domains (Fig. 1). To follow the nomenclature convention for vertebrate myosins, which recommends that they be identified by the gene symbol for their heavy chain (Gillespie et al., 2001), we refer to myosin-X as Myo10. As the gene names for conventional myosins in vertebrates use ‘Myh’ as a prefix, whereas the gene names for unconventional myosins use ‘Myo’ as a prefix, it is very important to note that ‘Myh10’ corresponds to non-muscle myosin-IIb, not Myo10. Similarly, the Drosophila melanogaster myosin at chromosomal locus 10A, which is sometimes referred to as Drosophila Myo10A, corresponds to a MyTH-FERM myosin known as Myosin XV or Sisyphus.
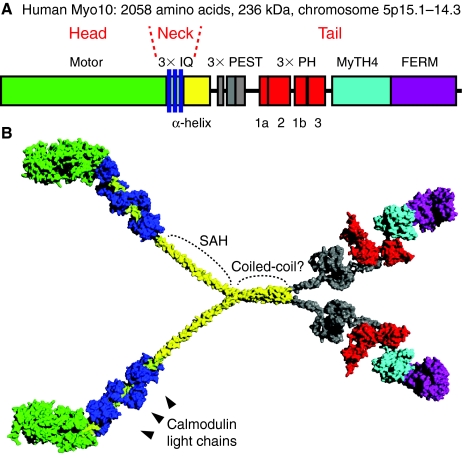
Fig. 1.
Structure of Myo10. (A) Diagram showing the domain structure of the Myo10 heavy chain. The cluster of three PH domains has a somewhat unusual arrangement, with PH2 located in a surface loop of PH1, splitting PH1 into two segments (1a and 1b). (B) Hypothetical model of Myo10 as a dimer with each heavy chain bound to three calmodulin light chains (dark blue). Note that other than the recently reported structures of the MyTH4-FERM region, the structure and overall organization of the Myo10 molecule remains unclear. The model here was constructed in PyMol on the basis of homology to domains of known structure to illustrate the relative size of the different domains. The neck is expected to have a length of ~11 nm. The beginning of the tail consists of a ~130-amino-acid α-helical region (yellow) that is depicted here consisting of a combination of an initial segment of single α-helix (SAH) and a distal segment of α-helical coiled-coil. Note that the precise length of the SAH is unclear and that the entire α-helical region is sometimes referred to as ‘CC’ because of initial predictions that it formed a coiled-coil. Although Myo10 is illustrated here as a parallel dimer, an antiparallel configuration has not been ruled out. The structure of the ~200-amino-acid region containing the three PEST sequences (gray) is completely unknown, but is depicted here as being relatively compact.
Myo10 is present in organisms ranging from humans to choanoflagellates (unicellular eukaryotes that are thought to be ancestral to metazoa), but Myo10 appears to have been lost in the lineages leading to Drosophila and Caenorhabditis elegans (Odronitz and Kollmar, 2007). Although humans and mice express only a single Myo10 gene, zebrafish have three Myo10-like genes (Sittaramane and Chandrasekhar, 2008). Myo10 was originally discovered in a screen for myosins in the inner ear (Solc et al., 1994), but it is now known to be expressed in most cells and tissues (Berg et al., 2000; Yonezawa et al., 2000). Myo10 appears to be expressed at quite low levels (Berg et al., 2000) and is thus likely to be several orders of magnitude less abundant than the major cellular myosins such as non-muscle myosin-II. Most importantly, growing evidence demonstrates that Myo10 is a molecular motor that has crucial functions in the slender actin-based extensions known as filopodia (Divito and Cheney, 2008).
Filopodia are cylindrical extensions of the plasma membrane that contain a bundle of parallel actin filaments at their core (Mattila and Lappalainen, 2008). Many cells appear to rely on these finger-like organelles to probe and interact with their surroundings, especially in processes such as axon guidance and angiogenesis (Eilken and Adams, 2010; Koleske, 2003). The precise mechanisms that underlie filopodia formation and function, however, remain unclear. The actin filaments in filopodia are known to have their barbed ends oriented towards the filopodial tip (Mattila and Lappalainen, 2008), and polymerization of actin monomers at the tip leads to a constant rearwards movement of actin filaments, which is known as retrograde flow or treadmilling (Medeiros et al., 2006). As discussed below, Myo10 is a MyTH-FERM myosin that localizes to filopodial tips, moves within filopodia and has potent filopodia-inducing activity (Berg et al., 2000; Bohil et al., 2006; Kerber et al., 2009). Here, we review key progress in understanding Myo10, from its domain structure and biophysical properties to its roles in cells and organisms.
Myo10 structure and biochemical properties
The Myo10 heavy chain has a molecular mass of ~237 kDa and can be divided into a head, neck and tail (Fig. 1) (Berg et al., 2000; Yonezawa et al., 2000). The head consists of a conserved myosin motor domain that can bind to F-actin, hydrolyze ATP and produce force. The Myo10 head is most similar to that of Myo7a, with which it shares 45% identity. The Myo10 neck consists of three IQ motifs, each of which can bind to a calmodulin or calmodulin-like light chain. The myosin neck domain is thought to act as a rigid lever that increases the myosin step size and is often involved in regulating motor activity. The Myo10 neck can bind to either calmodulin (Homma et al., 2001) or to an epithelia-specific protein known as calmodulin-like protein (CLP; encoded by CALML3) (Rogers and Strehler, 2001). CLP shares ~85% identity with calmodulin, but exhibits approximately eightfold weaker binding to Ca2+ (Rogers and Strehler, 2001). CLP binds preferentially to the third IQ motif of Myo10 (Caride et al., 2010), and overexpression of CLP increases Myo10 protein levels by several fold, apparently by stabilizing the Myo10 heavy chain during its translation (Bennett et al., 2008; Bennett et al., 2007; Bennett and Strehler, 2008).
The Myo10 tail begins with a short segment of ~130 amino acids that was initially predicted to form an α-helical coiled-coil by studies using computer programs such as PAIRCOIL and COILS. This suggested that Myo10 heavy chains can dimerize by way of a short stalk of coiled-coil (Berg et al., 2000). Detailed manual analysis of this α-helical region, however, revealed that it is unusually rich in charged residues and frequently lacks the hydrophobic residues that form the hydrophobic seam between the two α-helices in a coiled-coil (Knight et al., 2005). The work by Knight et al. also indicated that at least the initial portion of the α-helical region in Myo10 forms an intriguing structure known as either a single α-helix (SAH) (Knight et al., 2005) or an ER/K helix (Sivaramakrishnan et al., 2008). Although an individual α-helix was traditionally thought to be too compliant to stably span a large distance by itself, the alternating layers of positively and negatively charged residues found in a SAH endows greater stability and stiffness, and there is a growing recognition that a SAH can act as a structural element to lengthen the myosin lever arm and increase the step size (Baboolal et al., 2009; Knight et al., 2005; Sivaramakrishnan et al., 2008). Importantly, a baculovirus-expressed Myo10 construct consisting of the head, neck and entire α-helical region was largely monomeric when visualized by electron microscopy, although ~10% of the molecules appeared to form dimers (Knight et al., 2005). These results indicate that the proximal portion of the α-helical region forms a SAH, whereas the distal portion might undergo regulated dimerization by forming a coiled-coil or related structure. This also indicates that Myo10 constructs consisting of the head, neck and α-helical region, which are often referred to as ‘heavy meromyosin (HMM)-like’ because of their similarity to the dimeric HMM fragment of myosin-II, can exist as monomers, dimers or a mixture of both. Importantly, SAH domains are predicted in many proteins, including myosins such as Myo6 and Myo7a (Peckham and Knight, 2009; Sivaramakrishnan et al., 2008; Spink et al., 2008). Recent research with Myo6 indicates that its SAH uses electrostatic interactions to form parallel dimers that are offset by one turn of α-helix (Kim et al., 2010), and rearrangements of more proximal α-helical sequences might create additional calmodulin-binding sites (Liu et al., 2011). Given that the length of the SAH in Myo10 and the mechanism of dimerization are both unclear, an important goal for the future will be to determine the precise structure(s) formed by the α-helical region.
The next segment of the Myo10 tail contains three PEST regions. PEST regions are sequences that are enriched in proline, glutamate, serine and threonine residues and are often associated with proteins that rapidly turnover (Rechsteiner and Rogers, 1996). Myo10 can be cleaved by the calcium-dependent protease calpain at its PEST regions in vitro (Berg et al., 2000), but it is not yet known whether this occurs in vivo. Although the structure of the PEST regions in Myo10 is unknown, cleavage at the PEST regions would have the functionally interesting result of splitting a motorized HMM-like fragment away from the majority of the tail.
A cluster of three PH domains follows the PEST region (Berg et al., 2000; Yonezawa et al., 2003). The PH2 domain in Myo10 matches consensus sequences for binding to phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3] (Isakoff et al., 1998; Park et al., 2008; Tacon et al., 2004), a key cell signaling molecule that localizes primarily to the plasma membrane. Systematic screens of PH domains also demonstrate that the PH2 domain of Myo10 can be recruited to the plasma membrane by phosphoinositide 3-kinase (PI3K), the enzyme that generates PtdIns(3,4,5)P3 (Isakoff et al., 1998; Park et al., 2008). Although the PH2 domain of Myo10 preferentially binds to PtdIns(3,4,5)P3 in lipid-binding assays, the isolated PH1 or PH2 domains show little interaction with other phospholipids (Park et al., 2008; Plantard et al., 2010). Importantly, a construct consisting of all three PH domains is sufficient for targeting to the plasma membrane, where it has a mean residence time of ~20 seconds (Mashanov et al., 2004). Experiments with macrophages show that endogenous Myo10 is recruited to the phagocytic cup downstream of PI3K and that Myo10 is required for Fc-mediated phagocytosis (Cox et al., 2002). The PH domains of Myo10 appear to contribute to its localization to filopodia (Plantard et al., 2010; Umeki et al., 2011), and exciting new data discussed below show that binding to PtdIns(3,4,5)P3 activates Myo10 (Umeki et al., 2011).
The next domain in the Myo10 tail is a MyTH4 domain. MyTH4 domains have a well-conserved primary structure and are almost always located N-terminal to a FERM domain. MyTH4-FERM domains are found in several proteins other than myosins, including a family of plant kinesins in which they are implicated in targeting to microtubules (Narasimhulu and Reddy, 1998). The recently published crystal structures of the MyTH4-FERM domains in Myo7a (Wu et al., 2011) and Myo10 (Hirano et al., 2011; Wei et al., 2011) are a major advance and the first to reveal the structure of a MyTH4 domain. These papers show that the MyTH4 domain forms an ~200-amino-acid helical bundle, and that the MyTH4 and FERM domains form a supramodule with highly conserved contact residues. Functionally, the MyTH4 domain of Myo10 is sufficient for binding to microtubules, although the MyTH4-FERM supramodule showed stronger binding (Weber et al., 2004). Microtubule binding is mediated by a patch of positively charged residues in the MyTH4 domain that bind to the negatively charged E-hook region in the tails of α- and β-tubulin (Hirano et al., 2011). This ability to bind microtubules gives Myo10 the intriguing potential to act as a motorized link between actin filaments and microtubules.
The tail of Myo10 ends in a FERM domain that can bind to several β-integrins, a key family of cell surface receptors that are involved in cell adhesion and migration (Zhang et al., 2004). The Myo10 FERM domain shares ~28% identity with the FERM domain of talin, a major protein component of focal adhesions, which functions in ‘inside-out’ integrin signaling by binding to and activating integrins. Like the FERM domain in talin, the FERM domain in Myo10 binds to the NPXY motif that is present in the cytoplasmic tail of β-integrins. Unlike talin, however, Myo10 shows little or no localization to focal adhesions, the major sites of integrin localization in cells, and instead localizes to the tips of filopodia, where a small amount of integrin can be detected (Zhang et al., 2004). Together, these experiments indicate that Myo10 has important roles in the early steps of integrin-mediated adhesion, where it appears to function in the transport and/or tethering of integrins in filopodia. Important unanswered questions are whether Myo10 activates integrins and whether it competes or cooperates with other integrin-binding proteins such as talin.
The Myo10 FERM domain also binds to the cytoplasmic domains of the netrin receptors DCC (for deleted in colorectal cancer) and neogenin (Zhu et al., 2007). These guidance receptors have key roles in many processes, and Myo10 has been shown to be required for netrin-mediated axon guidance. The recently solved crystal structures of the Myo10 MyTH4-FERM supramodule bound to the P3 domain of DCC also provide the first views of an interaction between Myo10 and a candidate cargo (Hirano et al., 2011; Wei et al., 2011). Unlike the talin–integrin interaction, in which a groove on the F3 lobe binds to a β-strand formed by the NPXY motif, in the Myo10–DCC interaction the equivalent groove on the F3 lobe binds to an α-helix, thus revealing a new mode of binding for FERM domains. Biochemical experiments indicate that DCC, integrin and microtubules all compete for binding to the MyTH4-FERM supramodule, strongly suggesting that the supramodule can only bind to one of these candidate cargos at a time (Hirano et al., 2011).
Localization and dynamics of Myo10 in filopodia
Although the low abundance of endogenous Myo10 can make localization studies challenging, Myo10 clearly localizes to the tips of filopodia and retraction fibers (Fig. 2; supplementary material Movie 1) (Berg et al., 2000; Tokuo and Ikebe, 2004; Zhang et al., 2004). In addition to filopodial tips, Myo10 also shows some localization to other regions of dynamic actin, including lamellipodia (Berg et al., 2000), invadopodia (Schoumacher et al., 2010) and phagocytic cups (Cox et al., 2002). Because Myo10 constructs that lack the motor domain fail to localize to filopodial tips, it has been concluded that Myo10 uses its motor activity to transport itself along filopodial actin filaments to the tips of filopodia (Berg and Cheney, 2002). Moreover, as HMM-like constructs consisting of the head, neck and α-helical region are sufficient for tip localization, whereas a head–neck construct is not, tip localization was hypothesized to require both myosin motor activity and dimerization through some portion of the α-helical region. Consistent with this, forced dimerization of the head–neck–SAH is indeed sufficient to trigger tip localization (Kerber et al., 2009; Tokuo et al., 2007).
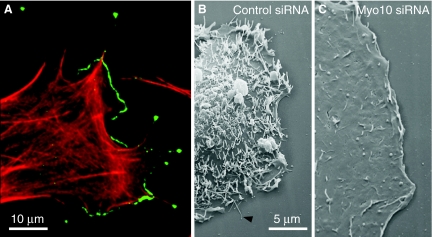
Fig. 2.
Myo10 localizes to filopodial tips and is required for filopodia formation. (A) Immunofluorescence image showing localization of F-actin (red) and endogenous Myo10 (green) at the tips of filopodia and along the leading edge of a bovine aortic endothelial cell. Image provided by Melinda DiVito, UNC Chapel Hill, NC. (B) Scanning electron microscopy (SEM) of a control HeLa cell showing a substrate-attached filopodium (arrowhead) and numerous dorsal filopodia. (C) SEM of a Myo10-knockdown cell at the same magnification showing a dramatic decrease in dorsal filopodia. SEM images provided by Aparna Bohil, UNC Chapel Hill, NC. siRNA, small interfering RNA.
Live-cell imaging shows that GFP–Myo10 is present at the tips of filopodia as soon as they form and remains there as the filopodia extend and retract (Berg and Cheney, 2002). Myo10 can also undergo several forms of movement within filopodia (Fig. 3). The bright puncta of GFP–Myo10 that are normally present at the tips of filopodia sometimes move slowly rearwards over a time course of several minutes at rates of 10–20 nm/second, leading to the hypothesis that the slow rearwards movements are due to Myo10 binding to filopodial actin filaments undergoing retrograde flow. This would provide a mechanism to allow Myo10 and any associated molecules to move rearwards, much like a package on a conveyor belt. Bright puncta of GFP–Myo10 also occasionally move forwards towards the filopodial tip at velocities on the order of ~80 nm/second, and this and other data indicate that Myo10 uses its motor activity to move forwards along filopodial actin (Berg and Cheney, 2002; Sousa and Cheney, 2005; Tokuo and Ikebe, 2004). Experiments with single-molecule sensitivity have revealed a faster and more frequent form of forwards motility, in which extremely faint particles of GFP–Myo10 can be observed moving towards the filopodial tip at ~600 nm/seconds, often along the entire length of a 5–10-μm filopodium (Kerber et al., 2009). These experiments also indicate that each filopodial tip contains in the order of 10–100 Myo10 molecules. Although it was unclear from these initial ‘single-molecule’ experiments whether the faint Myo10 particles corresponded to monomers or dimers, new data indicate that the faint particles can exhibit two-step photobleaching, as expected for a dimer (Watanabe et al., 2010). In addition, in vitro motility assays show that single molecules of HMM-like forced dimers are capable of moving along actin at velocities comparable to those of the faint particles in cells (Nagy et al., 2008; Sun et al., 2010). The in vivo movements of the faint particles of GFP–Myo10 provide one of the first examples of the direct visualization of long-range movement of individual myosin molecules in a living cell. These movements also raise the question of whether other motors in cells are busily shuttling to-and-fro at the single-molecule level.
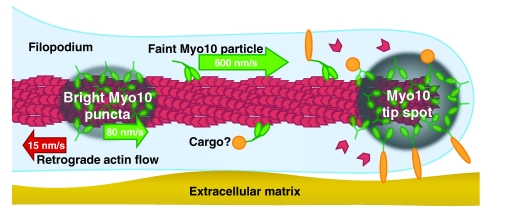
Fig. 3.
Myo10 undergoes several forms of intrafilopodial motility. Model of Myo10 movements in filopodia. In HeLa cells, bright puncta or clusters of Myo10 at tips of filopodia occasionally move rearwards within filopodia at the retrograde flow rate of ~15 nm/second. Bright puncta (areas of gray shading) have also been observed moving forwards within filopodia at ~80 nm/second. These puncta might include as-yet-undiscovered scaffolding components, actin polymerization machinery or cargos. Faint Myo10 particles (green), probably corresponding to dimers, move much faster (~600 nm/second) towards the filopodial tips. Forwards movements require Myo10 motor activity, whereas rearwards movements are hypothesized to be due to coupling to actin (red) undergoing retrograde flow. Myo10-interacting proteins (orange) include both cytoplasmic and membrane-associated proteins. See supplemental movie 5 of Kerber et al. for an animated model of Myo10's intrafilopodial motility (Kerber et al., 2009).
Is Myo10 a component of an intrafilopodial transport system?
As filopodia and related structures, such as microvilli and stereocilia, generally lack microtubules and intracellular vesicles, directed transport in these structures is likely to depend on actin and actin-based motors (Kerber et al., 2009; Nambiar et al., 2010; Salles et al., 2009). One candidate cargo for Myo10 is vasodilator-stimulated phosphoprotein (VASP), a cytoplasmic protein that that localizes to the tips of filopodia and lamellipodia. VASP has anti-capping activity and can stimulate filopodia formation (Applewhite et al., 2007), and it binds to the tail of Myo10 (Tokuo and Ikebe, 2004). Furthermore, in live-cell imaging experiments, bright puncta of VASP and Myo10 undergo co-transport in filopodia (Tokuo and Ikebe, 2004), and single-molecule-level imaging shows that faint particles of GFP–VASP move rapidly forwards in filopodia, much like faint particles of GFP–Myo10 (Kerber et al., 2009). Myo10 also binds to and is implicated in the transport and/or localization of several integral membrane proteins, including integrins (Zhang et al., 2004), the netrin receptors DCC and neogenin (Zhu et al., 2007), the bone morphogenetic protein (BMP) receptor activin receptor-like kinase 6 (ALK6) (Pi et al., 2007) and vascular endothelial (VE)-cadherin (Almagro et al., 2010).
Motor and single-molecule properties
The localization and intrafilopodial motility of Myo10 raise intriguing questions about its motor and biophysical properties. The initial studies with an HMM-like construct revealed that Myo10, like most myosins, moves towards the barbed end of the actin filament (Homma et al., 2001). Detailed kinetic studies of a head–3IQ construct reported a duty ratio of ~16% (Kovacs et al., 2005), and similar studies using a head–1IQ construct yielded a duty ratio of 60–70% (Homma and Ikebe, 2005). The duty ratio corresponds to the fraction of the ATPase cycle that the motor is tightly bound to the filament, so the value of 60–70% suggests that a Myo10 dimer can walk processively along a filament by alternating steps of its two heads. The kinetic studies also revealed that the ‘weak’ binding state of Myo10 has an unusually strong affinity for actin filaments, a property that could potentially keep it associated with actin and enhance processivity. Given the limited space for diffusion within a filopodium, even a moderately processive myosin might eventually succeed in moving to the tip as long as it moves fast enough to overcome retrograde flow.
In an exciting recent advance, full-length Myo10 has been expressed in baculovirus (Umeki et al., 2011). This revealed that purified Myo10 forms a compact monomer in which the Myo10 head binds to the PH and FERM domains in the tail. An HMM-like construct that was lacking most of the tail had an actin-activated ATPase activity of 20 ATP molecules per second, fivefold higher than that of full-length Myo10. Although the ATPase activity of full-length Myo10 was found not to be regulated by Ca2+, it was activated by membranes containing either PtdIns(3,4,5)P3 or phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2], and crosslinking showed that binding to these membranes induced Myo10 to form dimers (Umeki et al., 2011). This suggests a general model of Myo10 regulation in which membranes enriched in PtdIns(3,4,5)P3 or PtdIns(4,5)P2 recruit Myo10 and convert it from a compact monomer into an active dimer (Fig. 4). This would be analogous to the microtubule motor UNC-104/KIF1a, which is thought to exist as a monomer until it is targeted to PtdIns(4,5)P2-enriched vesicles by its PH domains, leading to an increased local concentration and the formation of processive dimers (Tomishige et al., 2002). Conversion from a compact monomer into an active dimer appears to be a general regulatory mechanism that is used by many different motors, including Myo6 (Park et al., 2006; Yu et al., 2009) and Myo7a (Umeki et al., 2009; Yang et al., 2009). Intriguingly, recent studies with Myo7a indicate that binding to a cargo induces this MyTH-FERM myosin to form dimers and localize to the tips of filopodia (Sakai et al., 2011).
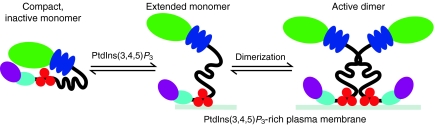
Fig. 4.
Model for Myo10 regulation. The tail of Myo10 binds to its head domain, forming a compact monomer that is enzymatically inactive (Umeki et al., 2011). Binding of the Myo10 tail to membranes containing PtdIns(3,4,5)P3 or PtdIns(4,5)P2 disrupts the intramolecular interactions between the head and tail, inducing the formation of an extended monomer. This, plus the increased local concentration of Myo10 resulting from its recruitment to membranes, is thought to lead to the formation of an active dimer that can move processively along actin.
A series of recent papers has begun to shed light, and controversy, on the single-molecule properties of Myo10 and the mechanisms that target it to filopodia. Using in vitro motility assays and an HMM–Myo10 construct (amino acids 1–920) that was forced to dimerize through a leucine zipper, single molecules were shown to move on individual actin filaments or actin bundles at velocities of 330–780 nm/second (Nagy et al., 2008). Interestingly, this construct also moved along filopodial actin bundles in detergent-permeabilized cells, although it appeared to run off the ends without pausing at the tip (Brawley and Rock, 2009). This construct also showed selectivity for actin bundles because its run length on bundles (630 nm) is fourfold longer than on single filaments, whereas Myo5a, a well-studied processive myosin, has similar run lengths (~600 nm) on either form of actin. This raised the possibility that Myo10 moves on bundles using a ‘straddle’ mechanism, in which the two heads bind to separate filaments (Nagy and Rock, 2010). The HMM–Myo10 forced dimer had an average step size of ~18 nm, too short to easily span the ~36 nm pseudohelical repeat of the actin filament (Nagy and Rock, 2010; Ricca and Rock, 2010). Unlike the ~36 nm step of Myo5a, which is ideal for stepping along one face of a single filament, an ~18 nm step is sterically awkward as it would force a myosin head to reach around to the back side of the filament. On a bundle, however, only the outer faces of actin filaments are available for binding and adjacent filaments provide additional binding sites, so an ~18 nm step would favor ‘straddling’ and might help direct Myo10 to filopodia.
Work from another group using a different HMM-like forced dimer, however, showed little or no selectivity for bundles (Sun and Goldman, 2011; Sun et al., 2010). The construct used in these experiments included amino acids 1–939 and was forced to dimerize through a coiled-coil sequence from Myo5a; similar results were also reported for a ‘native’ HMM-like Myo10 construct. Step-size measurements with the forced dimer showed that it took large steps of ~34 nm on single filaments. On bundles, this forced dimer exhibited both ~34 nm and ~20 nm steps, with the shorter steps attributed to stepping to adjacent filaments. These results challenge the straddling model, and suggest that other mechanisms might target Myo10 to filopodia. Given that tropomyosin regulates the binding of myosin to actin in muscle, it would be interesting to determine whether targeting of Myo10 to filopodia involves tropomyosins or other actin-binding proteins. Although the basis for the differences in step size and selectivity reported by the two groups are unclear, it is possible that differences in the sites and methods of forced dimerization lead to differences in the structure of the α-helical region or the precise positioning of the head domains. Sun and colleagues also used sophisticated biophysical approaches to show that their forced-dimer construct uses a hand-over-hand mechanism to move processively at ~200 nm/seconds with a run length of ~1000 nm (Sun et al., 2010). These authors also report the important finding that binding to actin can induce their ‘native’ HMM-like Myo10 to form processive dimers (Sun et al., 2010), a result remarkably similar to the proximity-induced dimerization seen with Myo6 (Park et al., 2006).
Myo10 induces filopodia by multiple mechanisms
In addition to its abilities to move on actin filaments, Myo10 exhibits potent filopodia-inducing activity. Light microscopy was initially used to show that overexpressing full-length Myo10, but not an HMM-like construct or the tail alone, increased the number and length of substrate-attached filopodia (Berg and Cheney, 2002). The ability of Myo10 to promote substrate-attached filopodia depends largely on its ability to bind to integrins through its FERM domain (Zhang et al., 2004), indicating that Myo10 can act by using a ‘sticky fingers’ mechanism that promotes adhesion of filopodia to the substrate and thus stabilizes them. Consistent with this, an analysis of the dynamics of filopodia in cells transfected with either full-length Myo10 or a FERM deletion construct shows that the full-length Myo10 increases the ability of filopodia that are extending along the substrate to undergo a second round of extension by approximately fivefold (Watanabe et al., 2010).
To test whether Myo10 also promotes filopodia induction by mechanisms that are independent of adhesion, scanning electron microscopy (SEM) was used to visualize dorsal filopodia (i.e. filopodia that are not attached to the substrate) in cells transfected with Myo10 or with a FERM deletion construct. Both constructs led to massive increases in dorsal filopodia, demonstrating that Myo10 can promote filopodia independently of adhesion or integrin binding (Bohil et al., 2006). Myo10 also appeared to act downstream of the Rho GTPase Cdc42 and independently of VASP proteins. Because an HMM-like construct failed to induce dorsal filopodia, elements of the Myo10 tail upstream of the FERM domain, such as the MyTH4 domain, appear to be important for the formation or stabilization of dorsal filopodia (Bohil et al., 2006), and recent data indicate roles for the PH domains (Umeki et al., 2011). The Myo10 tail could potentially stimulate filopodia formation through a variety of mechanisms, including regulating motor activity, inducing dimerization and/or transporting proteins that are important for filopodia formation.
Importantly, the induced dimerization of a Myo10 construct consisting of the head, neck and SAH is sufficient to trigger both tip localization and a pulse of short-lived filopodia (Tokuo et al., 2007). This result suggests that Myo10 dimers initiate filopodia formation by using their two heads to link the barbed ends of lamellipodial actin filaments together, thus triggering the formation of filopodial actin bundles. Consistent with this role, GFP–Myo10 has been observed in structures that resemble Λ-precursors (the Λ-shaped precursors to filopodial actin bundles) (Sousa et al., 2006), and GFP–Myo10 accumulates rapidly at sites of filopodial initiation (Watanabe et al., 2010). Importantly, endogenous Myo10 is required for filopodia formation because knockdown of Myo10 dramatically decreases the otherwise numerous dorsal filopodia in HeLa cells (Bohil et al., 2006) (Fig. 2). New data show that Myo10 is also required for the formation of invadopodia, actin-based protrusions that share several similarities with filopodia and that function in the digestion of extracellular matrix and metastasis of cancer cells (Schoumacher et al., 2010). Myo10 function is also required for the patterning of podosomes (McMichael et al., 2010), invadopodia-like structures that are involved in integrin-dependent adhesion in cells such as osteoclasts.
Myo10 is required for proper spindle assembly and orientation
In addition to its functions in filopodia and related structures, Myo10 has crucial functions in the spindle. In Xenopus laevis oocytes, an anti-Myo10 antibody labels microtubules and the meiotic spindle, especially where the spindle contacts the actin-rich cortex. Nuclear anchoring and spindle assembly are disrupted when Myo10 function is inhibited using an anti-Myo10 antibody or a dominant-negative tail construct (Weber et al., 2004). Myo10 and actin are also necessary in mitotic spindles, with morpholino-mediated knockdown of Myo10 resulting in multipolar spindles, increased spindle length and defects in spindle anchoring (Fig. 5) (Woolner et al., 2008). Woolner et al. also report that the MyTH4-FERM domain of Myo10 binds TPX2, a protein that is enriched in the spindle pole, and they propose that Myo10 acts at the spindle pole to suppress fragmentation and exert forces that oppose spindle lengthening (Woolner et al., 2008). Moreover, Myo10 is required for proper spindle orientation in HeLa cells (Toyoshima and Nishida, 2007), which normally orient their spindles parallel to the substrate in an integrin- and PtdIns(3,4,5)P3-dependent manner (Toyoshima et al., 2007). In addition, knockdown of Myo10 increases the number of multipolar spindles in MDA-231 cancer cells (Kwon et al., 2008), and these authors suggest that retraction fibers convey forces from the substrate to the spindle to orient the spindle and suppress fragmentation. Together, these results clearly demonstrate that Myo10 is required for spindle function, although the molecular mechanisms by which it acts will be an important area for future research (Wuhr et al., 2008).
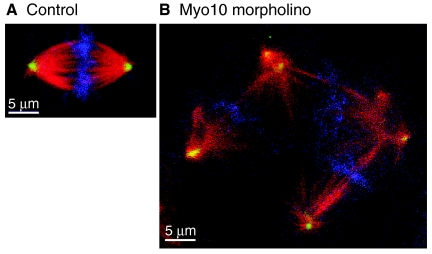
Fig. 5.
Knockdown of Myo10 results in spindle defects. (A) Normal mitotic spindle from a Xenopus embryo treated with a control morpholino. (B) An abnormal mitotic spindle from an embryo treated with a morpholino against Myo10. As described by Woolner and colleagues, knockdown of Myo10 leads to increased numbers of multipolar spindles and increased spindle length (Woolner et al., 2008). Spindles are displayed at the same magnification and were stained for β-tubulin to label microtubules (red), γ-tubulin to label spindle poles (green), and DAPI to label DNA (blue). Images provided by Josh Sandquist and William Bement, University of Wisconsin, Madison, WI.
Myo10 is required for angiogenic signaling in endothelial cells
Endothelial tip cells elaborate numerous filopodia during angiogenesis, and Myo10 is rapidly upregulated by BMP6, an angiogenic factor of the transforming growth factor-β (TGF-β) family (Pi et al., 2007). Knockdown of Myo10 in endothelial cells inhibits BMP6-mediated increases in filopodia number, cell orientation to BMP gradients, cell migration and endothelial tube formation (Pi et al., 2007). Myo10 also interacted with the BMP receptor ALK6 in pull-down assays, and is involved in early events in BMP signaling, possibly by localizing receptors to filopodia. Filopodia have also been hypothesized to help establish cell–cell junctions in endothelial cells, which are held together by a cell–cell adhesion receptor, VE-cadherin. Live-cell imaging shows that puncta of GFP–Myo10 and VE-cadherin move coordinately at rates of ~600 nm/s in endothelial-cell filopodia, and that they transiently colocalize at the tips of filopodia during early stages of cell–cell contact (Almagro et al., 2010). In pull-down assays the Myo10 FERM domain co-purified with VE-cadherin and other junction proteins, indicating that Myo10 and filopodia are likely to have important roles in junction formation.
Myo10 functions in melanosome transfer in melanocytes
In addition to acting as cellular sensors and contact points, filopodia can also serve as passageways for the transport of materials between cells. For example, melanocytes generate filopodia-like extensions that allow Myo5a to transport melanosomes towards their tips, and in a striking example of inter-cellular organelle transfer, the surrounding keratinocytes internalize the filopodia by a process that appears to be similar to phagocytosis. Myo10 was recently reported to function in melanosome transfer (Singh et al., 2010), with exposure to UV light increasing Myo10 expression, the number of filopodia and melanosome transfer, whereas knockdown of Myo10 decreased the number of filopodia and melanosome transfer. Although Myo10 was required for induction of filopodia in melanocytes, it might also have a role in ‘phagocytosis’ by the keratinocytes. Viruses such as HIV can also move from cell to cell using filopodia-like structures (Lehmann et al., 2005; Sherer et al., 2007; Sherer and Mothes, 2008), although the role of Myo10 in this process has not yet been tested.
Functions of Myo10 in neurons
Filopodia are thought to be especially important in neurons for growth cone guidance and synapse formation. Myo10 is developmentally regulated in brain, with relatively high levels in mouse cerebrum during the first two weeks after birth (Sousa et al., 2006), a peak period of synapse formation, when neurons elaborate an estimated 50,000 filopodia per day (Portera-Cailliau et al., 2003). Myo10 expression then drops to much lower levels in adult cerebrum. Brain also expresses a ‘headless’ form of Myo10 that lacks most of the head domain and thus is unable to act as a motor, localize to filopodial tips or induce filopodia (Sousa et al., 2006). Headless Myo10 has been hypothesized to act either as a scaffolding molecule or as a naturally occurring dominant negative (Sousa et al., 2006). Although the numerous binding partners of Myo10, such as integrin, can make interpretation of dominant-negative approaches challenging, motorless Myo10 constructs clearly inhibit netrin-dependent axon outgrowth (Zhu et al., 2007) as well as formation of focal adhesions and chemotaxis in a neuronal cell line (Wang et al., 2009). Axon guidance depends on both actin and microtubules, with filopodia being especially important for axonal path-finding (Dent and Gertler, 2003). Although cultures of cortical neurons that lack all three members of the VASP family fail to form filopodia or generate neurites, expressing full-length Myo10 rescues both filopodia formation and neuritogenesis (Dent et al., 2007). Myo10 is also upregulated approximately sevenfold following nerve injury, suggesting that it functions in nerve regeneration.
Myo10 is required for early development and neural crest migration
Although there are currently no Myo10-knockout animals, studies with morpholino-mediated knockdown in Xenopus embryos has revealed that Myo10 has important functions in development (Hwang et al., 2009; Nie et al., 2009). During early development, Myo10 is detected primarily in neural crest and paraxial mesoderm. Neural crest cells are highly migratory cells that give rise to several cell types, including craniofacial cartilage and bone, peripheral neurons and melanocytes. Although loss of maternal Myo10 appeared to be lethal, knockdown of zygotic Myo10 inhibited neural crest cell migration and resulted in a dramatic decrease in cranium size (Hwang et al., 2009). Consistent with the ability of Myo10 to bind integrins and promote filopodia, plating the knockdown cells on fibronectin revealed defects in adhesion, cell spreading, filopodia formation and cell migration (Hwang et al., 2009; Nie et al., 2009).
Conclusion and perspectives
Research over the past decade clearly demonstrates that Myo10 is a MyTH-FERM myosin with central roles in filopodia. The localization of Myo10 at the filopodial tip indicates that there is a largely uncharacterized filopodial tip complex that is likely to function in polymerization, adhesion and cell signaling. Much has been revealed about the basic properties of Myo10, and the mechanisms by which it targets to and induces filopodia are beginning to be unraveled. Exciting new theoretical studies indicate that the lengths of filopodia should be limited by the diffusion of proteins, such as actin, that are consumed during filopodial growth; motor proteins, such as Myo10, could overcome these limits if they actively transport such proteins to the tip (Zhuravlev et al., 2010; Zhuravlev and Papoian, 2009). The intrafilopodial movements of Myo10 raise intriguing questions about this form of motility and the identity of Myo10 cargos. In addition, these results raise the larger question of whether structures such as filopodia, microvilli and stereocilia use myosin motors to power transport systems that are analogous to intraflagellar transport, a transport system that is central to the formation of cilia and that is defective in a large number of human diseases. Biophysical studies of Myo10 are revealing fundamental mechanisms of motor regulation, such as induced dimerization and the roles of a new structural element, the SAH. Work with Myo10 also raises the question of the stepping patterns myosins use when moving on an array of filaments, an issue that could be investigated by tagging the two heads of a Myo10 dimer with different colors. The ability of Myo10 to bind to microtubules, and its functions in spindle orientation, also raise fundamental questions about the roles of actin and myosin in the spindle. It also remains to be seen whether Myo10 interacts with microtubules in interphase cells. As a myosin that binds to integrins, acts downstream of PI3K and is required for invadopodia formation, it will be important to investigate the functions of Myo10 in cell signaling and in disease processes such as cancer. Although morpholino studies in Xenopus embryos demonstrate important functions for Myo10 in developmental processes, such as neural crest migration, the generation of knockout or conditionally null animal models would greatly facilitate studies of the physiological functions of Myo10. Finally, research with Myo10 might be just the tip of the iceberg, as a growing number of unconventional myosins, including Myo3a (Salles et al., 2009), Myo7a (Sakai et al., 2011) and Myo15a (Belyantseva et al., 2005), appear to act as barbed-end-tracking proteins and have important functions in bundle-based protrusions.
Supplementary Material
Footnotes
Note added in proof
The structure of the PH1–PH2 region has recently been reported, and this region forms a rigid supramodule whose lipid-binding pockets are positioned side-by-side, allowing Myo10 to bind to phosphatidylinositol (3,4,5)-trisphosphate with high specificity and avidity (Lu et al., 2011).
Funding
Our work is supported by grants from the National Institutes of Heath, National Institute on Deafness and Other Communication Disorders [grant number R01-DC03299 to R.E.C.]; and National Institutes of Heath, National Heart, Lung, and Blood Institute [grant number P01-HL080166 to R.E.C.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.023549/-/DC1
References
- Almagro S., Durmort C., Chervin-Petinot A., Heyraud S., Dubois M., Lambert O., Maillefaud C., Hewat E., Schaal J. P., Huber P., et al. (2010). The motor protein myosin-X transports VE-cadherin along filopodia to allow the formation of early endothelial cell-cell contacts. Mol. Cell. Biol. 30, 1703-1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite D. A., Barzik M., Kojima S., Svitkina T. M., Gertler F. B., Borisy G. G. (2007). Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol. Biol. Cell 18, 2579-2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baboolal T. G., Sakamoto T., Forgacs E., White H. D., Jackson S. M., Takagi Y., Farrow R. E., Molloy J. E., Knight P. J., Sellers J. R., et al. (2009). The SAH domain extends the functional length of the myosin lever. Proc. Natl. Acad. Sci. USA 106, 22193-22198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyantseva I. A., Boger E. T., Naz S., Frolenkov G. I., Sellers J. R., Ahmed Z. M., Griffith A. J., Friedman T. B. (2005). Myosin-XVa is required for tip localization of whirlin and differential elongation of hair-cell stereocilia. Nat. Cell Biol. 7, 148-156 [DOI] [PubMed] [Google Scholar]
- Bennett R. D., Strehler E. E. (2008). Calmodulin-like protein enhances myosin-10 translation. Biochem. Biophys. Res. Commun. 369, 654-659 [DOI] [PubMed] [Google Scholar]
- Bennett R. D., Mauer A. S., Strehler E. E. (2007). Calmodulin-like protein increases filopodia-dependent cell motility via up-regulation of myosin-10. J. Biol. Chem. 282, 3205-3212 [DOI] [PubMed] [Google Scholar]
- Bennett R. D., Caride A. J., Mauer A. S., Strehler E. E. (2008). Interaction with the IQ3 motif of myosin-10 is required for calmodulin-like protein-dependent filopodial extension. FEBS Lett. 582, 2377-2381 [DOI] [PubMed] [Google Scholar]
- Berg J. S., Cheney R. E. (2002). Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 4, 246-250 [DOI] [PubMed] [Google Scholar]
- Berg J. S., Derfler B. H., Pennisi C. M., Corey D. P., Cheney R. E. (2000). Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J. Cell Sci. 113, 3439-3451 [DOI] [PubMed] [Google Scholar]
- Bohil A. B., Robertson B. W., Cheney R. E. (2006). Myosin-X is a molecular motor that functions in filopodia formation. Proc. Natl. Acad. Sci. USA 103, 12411-12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley C. M., Rock R. S. (2009). Unconventional myosin traffic in cells reveals a selective actin cytoskeleton. Proc. Natl. Acad. Sci. USA 106, 9685-9690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breshears L. M., Wessels D., Soll D. R., Titus M. A. (2010). An unconventional myosin required for cell polarization and chemotaxis. Proc. Natl. Acad. Sci. USA 107, 6918-6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caride A. J., Bennett R. D., Strehler E. E. (2010). Kinetic analysis reveals differences in the binding mechanism of calmodulin and calmodulin-like protein to the IQ motifs of myosin-10. Biochemistry 49, 8105-8116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D., Berg J. S., Cammer M., Chinegwundoh J. O., Dale B. M., Cheney R. E., Greenberg S. (2002). Myosin X is a downstream effector of PI(3)K during phagocytosis. Nat. Cell Biol. 4, 469-477 [DOI] [PubMed] [Google Scholar]
- Dent E. W., Gertler F. B. (2003). Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron 40, 209-227 [DOI] [PubMed] [Google Scholar]
- Dent E. W., Kwiatkowski A. V., Mebane L. M., Philippar U., Barzik M., Rubinson D. A., Gupton S., Van Veen J. E., Furman C., Zhang J., et al. (2007). Filopodia are required for cortical neurite initiation. Nat. Cell Biol. 9, 1347-1359 [DOI] [PubMed] [Google Scholar]
- Divito M. M., Cheney R. E. (2008). Myosin10. In Myosins: A Superfamily of Molecular Motors (ed. Coluccio L. M.), pp. 403-419 Dordrecht, The Netherlands: Springer; [Google Scholar]
- Eilken H. M., Adams R. H. (2010). Dynamics of endothelial cell behavior in sprouting angiogenesis. Curr. Opin. Cell Biol. 22, 617-625 [DOI] [PubMed] [Google Scholar]
- Gillespie P. G., Albanesi J. P., Bahler M., Bement W. M., Berg J. S., Burgess D. R., Burnside B., Cheney R. E., Corey D. P., Coudrier E., et al. (2001). Myosin-I nomenclature. J. Cell Biol. 155, 703-704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman M. A., Finan D., Sivaramakrishnan S., Spudich J. A. (2011). Principles of unconventional myosin function and targeting. Annu. Rev. Cell Dev. Biol. 27, 133-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y., Hatano T., Takahashi A., Toriyama M., Inagaki N., Hakoshima T. (2011). Structural basis of cargo recognition by the myosin-X MyTH4-FERM domain. EMBO J. 30, 2734-2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma K., Ikebe M. (2005). Myosin X is a high duty ratio motor. J. Biol. Chem. 280, 29381-29391 [DOI] [PubMed] [Google Scholar]
- Homma K., Saito J., Ikebe R., Ikebe M. (2001). Motor function and regulation of myosin X. J. Biol. Chem. 276, 34348-34354 [DOI] [PubMed] [Google Scholar]
- Hwang Y. S., Luo T., Xu Y., Sargent T. D. (2009). Myosin-X is required for cranial neural crest cell migration in Xenopus laevis. Dev. Dyn. 238, 2522-2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff S. J., Cardozo T., Andreev J., Li Z., Ferguson K. M., Abagyan R., Lemmon M. A., Aronheim A., Skolnik E. Y. (1998). Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 17, 5374-5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber M. L., Jacobs D. T., Campagnola L., Dunn B. D., Yin T., Sousa A. D., Quintero O. A., Cheney R. E. (2009). A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr. Biol. 9, 967-973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Hsin J., Liu Y., Selvin P. R., Schulten K. (2010). Formation of salt bridges mediates internal dimerization of myosin VI medial tail domain. Structure 18, 1443-1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight P. J., Thirumurugan K., Xu Y., Wang F., Kalverda A. P., Stafford W. F., 3rd, Sellers J. R., Peckham M. (2005). The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 280, 34702-34708 [DOI] [PubMed] [Google Scholar]
- Koleske A. J. (2003). Do filopodia enable the growth cone to find its way? Sci. STKE 2003, pe20 [DOI] [PubMed] [Google Scholar]
- Kovacs M., Wang F., Sellers J. R. (2005). Mechanism of action of myosin X, a membrane-associated molecular motor. J. Biol. Chem. 280, 15071-15083 [DOI] [PubMed] [Google Scholar]
- Kwon M., Godinho S. A., Chandhok N. S., Ganem N. J., Azioune A., Thery M., Pellman D. (2008). Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22, 2189-2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M. J., Sherer N. M., Marks C. B., Pypaert M., Mothes W. (2005). Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J. Cell Biol. 170, 317-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hsin J., Kim H., Selvin P. R., Schulten K. (2011). Extension of a three-helix bundle domain of myosin VI and key role of calmodulins. Biophys. J. 100, 2964-2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Yu J., Yan J., Wei Z., Zhang M. (2011). Structural basis of the myosin-X PH1N-PH2-PH1C tandem as a specific and acute cellular PI(3,4,5)P3 sensor. Mol. Biol. Cell (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashanov G. I., Tacon D., Peckham M., Molloy J. E. (2004). The spatial and temporal dynamics of pleckstrin homology domain binding at the plasma membrane measured by imaging single molecules in live mouse myoblasts. J. Biol. Chem. 279, 15274-15280 [DOI] [PubMed] [Google Scholar]
- Mattila P. K., Lappalainen P. (2008). Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9, 446-454 [DOI] [PubMed] [Google Scholar]
- McMichael B. K., Cheney R. E., Lee B. S. (2010). Myosin X regulates sealing zone patterning in osteoclasts through linkage of podosomes and microtubules. J. Biol. Chem. 285, 9506-9515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros N. A., Burnette D. T., Forscher P. (2006). Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 8, 215-226 [DOI] [PubMed] [Google Scholar]
- Nagy S., Rock R. S. (2010). Structured post-IQ domain governs selectivity of myosin X for fascin-actin bundles. J. Biol. Chem. 285, 26608-26617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy S., Ricca B. L., Norstrom M. F., Courson D. S., Brawley C. M., Smithback P. A., Rock R. S. (2008). A myosin motor that selects bundled actin for motility. Proc. Natl. Acad. Sci. USA 105, 9616-9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar R., McConnell R. E., Tyska M. J. (2010). Myosin motor function: the ins and outs of actin-based membrane protrusions. Cell. Mol. Life Sci. 67, 1239-1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhulu S. B., Reddy A. S. (1998). Characterization of microtubule binding domains in the Arabidopsis kinesin-like calmodulin binding protein. Plant Cell 10, 957-965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S., Kee Y., Bronner-Fraser M. (2009). Myosin-X is critical for migratory ability of Xenopus cranial neural crest cells. Dev. Biol. 335, 132-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F., Kollmar M. (2007). Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8, R196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Ramamurthy B., Travaglia M., Safer D., Chen L. Q., Franzini-Armstrong C., Selvin P. R., Sweeney H. L. (2006). Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol. Cell 21, 331-336 [DOI] [PubMed] [Google Scholar]
- Park W. S., Heo W. D., Whalen J. H., O'Rourke N. A., Bryan H. M., Meyer T., Teruel M. N. (2008). Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol. Cell 30, 381-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham M., Knight P. J. (2009). When a predicted coiled coil is really a single α-helix, in myosins and other proteins. Soft Matter 5, 2493-2503 [Google Scholar]
- Pi X., Ren R., Kelley R., Zhang C., Moser M., Bohil A. B., Divito M., Cheney R. E., Patterson C. (2007). Sequential roles for myosin-X in BMP6-dependent filopodial extension, migration, and activation of BMP receptors. J. Cell Biol. 179, 1569-1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantard L., Arjonen A., Lock J. G., Nurani G., Ivaska J., Stromblad S. (2010). PtdIns(3,4,5)P is a regulator of myosin-X localization and filopodia formation. J. Cell Sci. 123, 3525-3534 [DOI] [PubMed] [Google Scholar]
- Portera-Cailliau C., Pan D. T., Yuste R. (2003). Activity-regulated dynamic behavior of early dendritic protrusions: evidence for different types of dendritic filopodia. J. Neurosci. 23, 7129-7142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M., Rogers S. W. (1996). PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21, 267-271 [PubMed] [Google Scholar]
- Ricca B. L., Rock R. S. (2010). The stepping pattern of myosin X is adapted for processive motility on bundled actin. Biophys. J. 99, 1818-1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. S., Strehler E. E. (2001). The tumor-sensitive calmodulin-like protein is a specific light chain of human unconventional myosin X. J. Biol. Chem. 276, 12182-12189 [DOI] [PubMed] [Google Scholar]
- Sakai T., Umeki N., Ikebe R., Ikebe M. (2011). Cargo binding activates myosin VIIA motor function in cells. Proc. Natl. Acad. Sci. USA 108, 7028-7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles F. T., Merritt R. C., Jr, Manor U., Dougherty G. W., Sousa A. D., Moore J. E., Yengo C. M., Dose A. C., Kachar B. (2009). Myosin IIIa boosts elongation of stereocilia by transporting espin 1 to the plus ends of actin filaments. Nat. Cell Biol. 11, 443-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M., Goldman R. D., Louvard D., Vignjevic D. M. (2010). Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189, 541-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer N. M., Mothes W. (2008). Cytonemes and tunneling nanotubules in cell-cell communication and viral pathogenesis. Trends Cell Biol. 18, 414-420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer N. M., Lehmann M. J., Jimenez-Soto L. F., Horensavitz C., Pypaert M., Mothes W. (2007). Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat. Cell Biol. 9, 310-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K., Kurfurst R., Nizard C., Schnebert S., Perrier E., Tobin D. J. (2010). Melanin transfer in human skin cells is mediated by filopodia-a model for homotypic and heterotypic lysosome-related organelle transfer. FASEB J. 24, 3756-3769 [DOI] [PubMed] [Google Scholar]
- Sittaramane V., Chandrasekhar A. (2008). Expression of unconventional myosin genes during neuronal development in zebrafish. Gene Expr. Patterns 8, 161-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaramakrishnan S., Spink B. J., Sim A. Y., Doniach S., Spudich J. A. (2008). Dynamic charge interactions create surprising rigidity in the ER/K alpha-helical protein motif. Proc. Natl. Acad. Sci. USA 105, 13356-13361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solc C. K., Derfler B. H., Duyk G. M., Corey D. P. (1994). Molecular cloning of myosins from the bullfrog saccular macula: a candidate for the hair cell adaptation motor. Auditory Neurosci. 1, 63-75 [Google Scholar]
- Sousa A. D., Cheney R. E. (2005). Myosin-X: a molecular motor at the cell's fingertips. Trends Cell Biol. 15, 533-539 [DOI] [PubMed] [Google Scholar]
- Sousa A. D., Berg J. S., Robertson B. W., Meeker R. B., Cheney R. E. (2006). Myo10 in brain: developmental regulation, identification of a headless isoform and dynamics in neurons. J. Cell Sci. 119, 184-194 [DOI] [PubMed] [Google Scholar]
- Spink B. J., Sivaramakrishnan S., Lipfert J., Doniach S., Spudich J. A. (2008). Long single alpha-helical tail domains bridge the gap between structure and function of myosin VI. Nat. Struct. Mol. Biol. 15, 591-597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M., Iwataki Y., Nakano K., Numata O. (2011). Unique sequences and predicted functions of myosins in Tetrahymena thermophila. Gene 480, 10-20 [DOI] [PubMed] [Google Scholar]
- Sun Y., Goldman Y. E. (2011). Lever-arm mechanics of processive myosins. Biophys. J. 101, 1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Sato O., Ruhnow F., Arsenault M. E., Ikebe M., Goldman Y. E. (2010). Single-molecule stepping and structural dynamics of myosin X. Nat. Struct. Mol. Biol. 17, 485-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacon D., Knight P. J., Peckham M. (2004). Imaging myosin 10 in cells. Biochem. Soc. Trans. 32, 689-693 [DOI] [PubMed] [Google Scholar]
- Tokuo H., Ikebe M. (2004). Myosin X transports Mena/VASP to the tip of filopodia. Biochem. Biophys. Res. Commun. 319, 214-220 [DOI] [PubMed] [Google Scholar]
- Tokuo H., Mabuchi K., Ikebe M. (2007). The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J. Cell Biol. 179, 229-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M., Klopfenstein D. R., Vale R. D. (2002). Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science 297, 2263-2267 [DOI] [PubMed] [Google Scholar]
- Toyoshima F., Nishida E. (2007). Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 26, 1487-1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F., Matsumura S., Morimoto H., Mitsushima M., Nishida E. (2007). PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Dev. Cell 13, 796-811 [DOI] [PubMed] [Google Scholar]
- Tuxworth R. I., Weber I., Wessels D., Addicks G. C., Soll D. R., Gerisch G., Titus M. A. (2001). A role for myosin VII in dynamic cell adhesion. Curr. Biol. 11, 318-329 [DOI] [PubMed] [Google Scholar]
- Umeki N., Jung H. S., Watanabe S., Sakai T., Li X. D., Ikebe R., Craig R., Ikebe M. (2009). The tail binds to the head-neck domain, inhibiting ATPase activity of myosin VIIA. Proc. Natl. Acad. Sci. USA 106, 8483-8488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeki N., Jung H. S., Sakai T., Sato O., Ikebe R., Ikebe M. (2011). Phospholipid-dependent regulation of the motor activity of myosin X. Nat. Struct. Mol. Biol. 18, 783-788 [DOI] [PubMed] [Google Scholar]
- Wang A., Liang Y., Fridell R. A., Probst F. J., Wilcox E. R., Touchman J. W., Morton C. C., Morell R. J., Noben-Trauth K., Camper S. A., et al. (1998). Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science 280, 1447-1451 [DOI] [PubMed] [Google Scholar]
- Wang J. J., Fu X. Q., Guo Y. G., Yuan L., Gao Q. Q., Yu H. L., Shi H. L., Wang X. Z., Xiong W. C., Zhu X. J. (2009). Involvement of headless myosin X in the motility of immortalized gonadotropin-releasing hormone neuronal cells. Cell Biol. Int. 33, 578-585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T. M., Tokuo H., Gonda K., Higuchi H., Ikebe M. (2010). Myosin-X induces filopodia by multiple elongation mechanism. J. Biol. Chem. 285, 19605-19614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K. L., Sokac A. M., Berg J. S., Cheney R. E., Bement W. M. (2004). A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature 431, 325-329 [DOI] [PubMed] [Google Scholar]
- Wei Z., Yan J., Lu Q., Pan L., Zhang M. (2011). Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain. Proc. Natl. Acad. Sci. USA 108, 3572-3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D., Blanchard S., Kaplan J., Guilford P., Gibson F., Walsh J., Mburu P., Varela A., Levilliers J., Weston M. D., et al. (1995). Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374, 60-61 [DOI] [PubMed] [Google Scholar]
- Williams S. A., Gavin R. H. (2005). Myosin genes in Tetrahymena. Cell Motil. Cytoskeleton 61, 237-243 [DOI] [PubMed] [Google Scholar]
- Woolner S., O'Brien L. L., Wiese C., Bement W. M. (2008). Myosin-10 and actin filaments are essential for mitotic spindle function. J. Cell Biol. 182, 77-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Pan L., Wei Z., Zhang M. (2011). Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo. Science 331, 757-760 [DOI] [PubMed] [Google Scholar]
- Wuhr M., Mitchison T. J., Field C. M. (2008). Mitosis: new roles for myosin-X and actin at the spindle. Curr. Biol. 18, R912-R914 [DOI] [PubMed] [Google Scholar]
- Yang Y., Baboolal T. G., Siththanandan V., Chen M., Walker M. L., Knight P. J., Peckham M., Sellers J. R. (2009). A FERM domain autoregulates Drosophila myosin 7a activity. Proc. Natl. Acad. Sci. USA 106, 4189-4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa S., Kimura A., Koshiba S., Masaki S., Ono T., Hanai A., Sonta S., Kageyama T., Takahashi T., Moriyama A. (2000). Mouse myosin X: molecular architecture and tissue expression as revealed by northern blot and in situ hybridization analyses. Biochem. Biophys. Res. Commun. 271, 526-533 [DOI] [PubMed] [Google Scholar]
- Yonezawa S., Yoshizaki N., Sano M., Hanai A., Masaki S., Takizawa T., Kageyama T., Moriyama A. (2003). Possible involvement of myosin-X in intercellular adhesion: importance of serial pleckstrin homology regions for intracellular localization. Dev. Growth Differ. 45, 175-185 [DOI] [PubMed] [Google Scholar]
- Yu C., Feng W., Wei Z., Miyanoiri Y., Wen W., Zhao Y., Zhang M. (2009). Myosin VI undergoes cargo-mediated dimerization. Cell 138, 537-548 [DOI] [PubMed] [Google Scholar]
- Zhang H., Berg J. S., Li Z., Wang Y., Lang P., Sousa A. D., Bhaskar A., Cheney R. E., Stromblad S. (2004). Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat. Cell Biol. 6, 523-531 [DOI] [PubMed] [Google Scholar]
- Zhu X. J., Wang C. Z., Dai P. G., Xie Y., Song N. N., Liu Y., Du Q. S., Mei L., Ding Y. Q., Xiong W. C. (2007). Myosin X regulates netrin receptors and functions in axonal path-finding. Nat. Cell Biol. 9, 184-192 [DOI] [PubMed] [Google Scholar]
- Zhuravlev P. I., Papoian G. A. (2009). Molecular noise of capping protein binding induces macroscopic instability in filopodial dynamics. Proc. Natl. Acad. Sci. USA 106, 11570-11575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuravlev P. I., Der B. S., Papoian G. A. (2010). Design of active transport must be highly intricate: a possible role of myosin and Ena/VASP for G-actin transport in filopodia. Biophys. J. 98, 1439-1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.