Abstract
Accurate chromosome segregation requires the assembly of kinetochores, multiprotein complexes that assemble on the centromere of each sister chromatid. A key step in this process involves binding of the constitutive centromere-associated network (CCAN) to CENP-A, the histone H3 variant that constitutes centromeric nucleosomes. This network is proposed to operate as a persistent structural scaffold for assembly of the outer kinetochore during mitosis. Here, we show by fluorescence resonance energy transfer (FRET) that the N-terminus of CENP-N lies in close proximity to the N-terminus of CENP-A in vivo, consistent with in vitro data showing direct binding of CENP-N to CENP-A. Furthermore, we demonstrate in living cells that CENP-N is bound to kinetochores during S phase and G2, but is largely absent from kinetochores during mitosis and G1. By measuring the dynamics of kinetochore binding, we reveal that CENP-N undergoes rapid exchange in G1 until the middle of S phase when it becomes stably associated with kinetochores. The majority of CENP-N is loaded during S phase and dissociates again during G2. We propose a model in which CENP-N functions as a fidelity factor during centromeric replication and reveal that the CCAN network is considerably more dynamic than previously appreciated.
Key words: Mitosis, Centromere, Kinetochore, CCAN complex, CENP-N, Cell cycle
Introduction
The kinetochore is a multiprotein complex that assembles at the centromeric DNA (‘centromere’) of each chromosome to permit proper segregation of sister chromatids during cell division. Although kinetochores are composed of over 100 different subunits, at their structural core they contain two conserved protein networks, the KMN (for KNL1/Blinkin/Spc105p, MIND/MIS12/Mtw1 and NDC80/Hec1) (De Wulf et al., 2003; Nekrasov et al., 2003; Cheeseman et al., 2004; Cheeseman et al., 2006; Obuse et al., 2004; Liu et al., 2005; Meraldi et al., 2006) and the constitutive centromere-associated network (CCAN) or CENP-A–NAC/CAD kinetochore complex (Foltz et al., 2006; Okada et al., 2006; Meraldi et al., 2006; McClelland et al., 2007; Hori et al., 2008a; Hori et al., 2008b; Amano et al., 2009) (reviewed by Santaguida and Musacchio, 2009). The KMN network is essential for kinetochore–microtubule binding (Tanaka and Desai, 2008; Cheeseman and Desai, 2008), whereas the CCAN network is associated to the centromeric nucleosomes (Carroll et al., 2009; Foltz et al., 2006). The centromeric nucleosomes, which contain the histone H3 variant CENP-A, assemble on repetitive α-satellite DNA. The structural core of the CENP-A-containing nucleosome is key to marking kinetochore position on the chromosome (Black et al., 2007a; Black et al., 2007b; Dalal et al., 2007; Sekulic and Black, 2009; Furuyama and Henikoff, 2009; Sekulic et al., 2010; Dimitriadis et al., 2010; Cho and Harrison, 2011; Zhou et al., 2011; Hu et al., 2011; Dechassa et al., 2011). Functionally, the CCAN network is required for the efficient recruitment of CENP-A into centromeric nucleosomes at the end of mitosis (Okada et al., 2006; Carroll et al., 2009; Carroll et al., 2010) and the maintenance of centromeric chromatin, but it is also involved in regulation of kinetochore fibre stability and dynamics, to control chromosome alignment and bipolar spindle assembly (Fukagawa et al., 2001; Foltz et al., 2006; Okada et al., 2006; McAinsh et al., 2006; McClelland et al., 2007; Toso et al., 2009; Amaro et al., 2010). Because several of the CCAN proteins are constitutively present at centromeres, this network has been proposed to act as a structural scaffold that is stably associated with the centromeric nucleosomes (Cheeseman and Desai, 2008). One key subunit of the CCAN is CENP-N, which binds directly to CENP-A-containing nucleosomes. It is required for the loading of all other CCAN subunits onto kinetochores (Okada et al., 2006; McClelland et al., 2007; Carroll et al., 2009). CENP-N directly binds CENP-L within the CCAN network, thus providing a possible link between the CENP-A nucleosomes and the rest of the network (Carroll et al., 2009).
In contrast to the static scaffold model, immunofluorescence experiments suggest that certain components of CCAN are not constitutively associated with the centromeres, but rather are dynamic and only associate with centromeres during certain phases of the cell cycle. In particular, it has been reported that the kinetochore-bound levels of CENP-N decrease when cells enter mitosis, with low binding detectable during metaphase (McClelland et al., 2007). This would suggest that the CCAN network is not a constitutive scaffold, but dynamically changes during cell cycle progression.
Here, we applied a broad range of methodologies to characterise the dynamic or static nature of CENP-N in a cellular context. Using fluorescence resonance energy transfer (FRET), we demonstrate that the N-terminus of EGFP–CENP-N is in direct proximity to the N-terminus of CENP-A in vivo, consistent with a close association to CENP-A-containing nucleosomes. We further show by several independent methods that CENP-N binds to the kinetochore at the end of G1 and during S phase, and is released during G2. Detailed FRAP analysis indicates that EGFP–CENP-N exchanges fast at kinetochores in G1 and is loaded to kinetochores by fast exchange in early S phase, before binding stably with very slow loading dynamics in middle and late S phase. These dynamics result in maximal CENP-N protein levels at kinetochores in S phase, reduced levels in G2, but low values in M phase and G1. We therefore conclude that the CCAN is not constitutive, but rather has a dynamic composition at the kinetochore during the cell cycle.
Results
CENP-N binds in close proximity to CENP-A in vivo
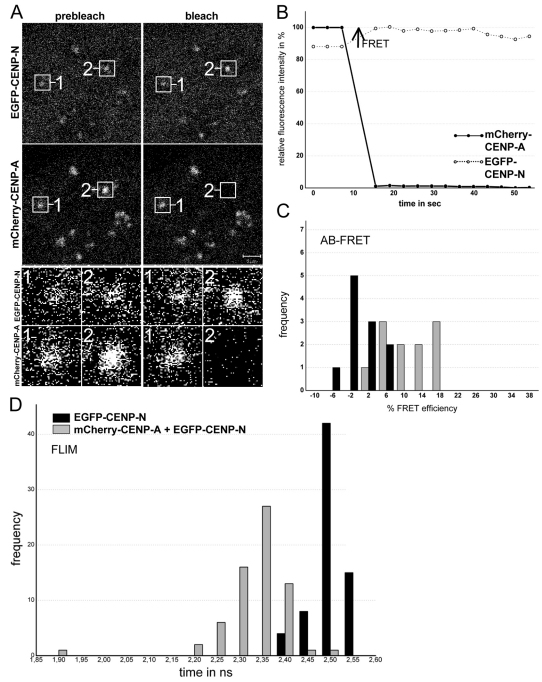
The CCAN subunit CENP-N binds in vitro to CENP-A-containing nucleosomes via a folded region that spans the CENP-N protein (Carroll et al., 2009). However, whether CENP-A and CENP-N are closely associated in vivo is not known. To analyse the binding of cellular CENP-N to the CENP-A-containing nucleosome in centromeric chromatin, we performed a FRET-based approach, which we have previously utilised to study protein proximities and interactions in cells (Hellwig et al., 2009; Orthaus et al., 2008; Orthaus et al., 2009). We expressed a full-length human CENP-N fusion with EGFP and confirmed that the EGFP–CENP-N colocalised to kinetochores with CENP-A in human HEp-2 cells (Fig. 1A). Next, we measured the FRET efficiency between the donor fluorophore (EGFP–CENP-N) and the acceptor fluorophore (mCherry–CENP-A), a method that can only generate a positive result when the distance between donor and acceptor is less than ~10 nm (see Fig. 1). In our live cell experiments, the orientation of the fluorophore dipole moment of the acceptor relative to that of the donor is not known. Thus, we did not deduce defined distance values from our measured FRET efficiencies but interpreted the appearance of FRET as an indication that donor and acceptor chromophores are close to one another within 10 nm.
Fig. 1.
FRET analysis. (A) AB-FRET of the protein pair EGFP–CENP-N and mCherry–CENP-A. Human HEp-2 cells were co-transfected with both fusion proteins. Typical cell nucleus, showing colocalisation at specific (centromeric) locations. Two of these locations, 1 and 2, were selected for fluorescence intensity analysis before and after acceptor bleaching (see enlargement below). Spot 1 served as control and showed no detectable intensity change. At spot 2, the acceptor fluorophore mCherry was bleached (compare prebleach and postbleach images). (B) Time course of the fluorescence intensity of the donor and the acceptor. The acceptor intensities in the ROI (region of interest; black circles) were averaged and normalized to the mean intensity measured at the three time points before bleaching. The donor intensities in the ROI were averaged and normalized to the intensity measured at the first time point after bleaching. Bleaching of the acceptor resulted in a fluorescence intensity increase of the donor (open circles) indicating the presence of FRET (see arrow). (C) Donor fluorescence intensity variation observed during acceptor bleaching was normalized to the intensity measured at the first time point after bleaching to give FRET efficiency (E). For spot 1 (acceptor not bleached, control, 11 kinetochores), this yielded Evar (black bars) and for spot 2 (acceptor bleached, 10 kinetochores) EFRET (grey bars). Frequency values are the number of observed single cases (grouped into Evar or EFRET value ranges of 4%) and are plotted against the values of Evar or EFRET. Only some EFRET values coincided with the distribution of Evar values, however, most EFRET values exceeded the Evar values, indicating the presence of FRET. (D) Histogram of the donor fluorescence lifetimes obtained in FLIM experiments. HEp-2 cells were transfected with EGFP–CENP-N alone (donor only, black bars, control, 69 kinetochores) or co-transfected with EGFP–CENP-N and mCherry–CENP-A. In these living cells, the donor fluorescence lifetimes were measured by TCSPC. The histograms display the fitted lifetime values of all single kinetochores. The heights of the bars represent the numbers of kinetochores (frequency) whose lifetimes fall within the indicated 0.3 nanosecond range (time). When the acceptor is present (grey bars, 64 kinetochores), the donor fluorescent lifetimes were shorter than the control values for donor only (black bars), clearly indicating the presence of FRET. The weak binding to kinetochores of the CENP-NΔC mutant lacking the CENP-N C-terminus (Carroll et al., 2009) resulted in low fluorescence intensities and did not allow for FRET measurements.
Our measurements were based on two independent, but complementary, experimental approaches: first, we measured the FRET donor fluorescence intensity with or without photo-inactivation of the acceptor (acceptor-photobleaching FRET; AB-FRET) and, second, the donor fluorescence lifetime (FLIM), which can yield more detailed information and is less error-prone. In AB-FRET, the acceptor chromophore is destroyed by photobleaching, thereby preventing FRET from the donor to the acceptor. If the donor and acceptor are in close enough proximity for energy transfer, photobleaching the acceptor results in an observable increase in donor fluorescence. In our experiments, two separate kinetochore locations were identified in each image (Fig. 1A, spots 1 and 2). In spot 2, the acceptor (mCherry–CENP-A) was photobleached, whereas spot 1 was not photobleached and served as an internal control for any non-FRET effects (Fig. 1A, and black bars in Fig. 1C). During bleaching of spot 2, the donor fluorescence intensity significantly increased (Fig. 1B, black arrow) indicating that FRET occurs between EGFP–CENP-N and mCherry–CENP-A (Hellwig et al., 2009). Careful quantification indicated that such FRET transfer occurred in 64% of the cases (P<0.001; Fig. 1C, grey bars), even though the FRET efficiency values were rather low. These data imply that the N-terminus of CENP-N is in close proximity to the N-terminus of CENP-A. By contrast, we detected no FRET between EGFP–CENP-N and mCherry–CENP-C (Hellwig et al., 2009) and between EYFP–CENP-C and Cerulean–CENP-A (S. Orthaus, Towards the architecture of the human kinetochore, PhD thesis, Friedrich Schiller University, Jena, Germany, 2006). These results are consistent with recent findings on the kinetochore binding of CENP-C (Przewloka et al., 2011; Screpanti et al., 2011). Additionally, we observed indications for very weak FRET between the C-terminus of CENP-C and the flexible N-terminus of CENP-A; these data however require further intensive experimentation. Recently, we observed that CENP-C binds directly and specifically to CENP-A-containing nucleosomes, requiring the extreme C-terminus of CENP-A for binding (Carroll et al., 2010).
We next confirmed our proximity results by measuring FRET in the lifetime domain (FLIM) with the same FRET pair (mCherry–CENP-A and EGFP–CENP-N) and time-correlated single photon counting (TCSPC) at kinetochore locations identified by mCherry–CENP-A. When FRET occurs in such an experiment, the donor fluorescence lifetime is decreased due to energy transfer to the acceptor. The mean donor lifetime in the absence of an acceptor was 2.49±0.01 nanoseconds (69 kinetochores from 10 cells; see Fig. 1D, black bars). In the presence of the acceptor, however, the donor lifetime distribution significantly shifted to shorter values with a mean value of 2.33±0.03 nanoseconds (67 kinetochores in 11 cells; see Fig. 1D, grey bars) with a mean FRET efficiency of 7% (P<0.001). These combined results indicate the presence of FRET between the fluorophores positioned at the N-termini of CENP-A and CENP-N, demonstrating that these entities are close to one another in vivo, consistent with previous in vitro biochemical data (Carroll et al., 2009).
CENP-N binds to kinetochores mainly in S phase and G2
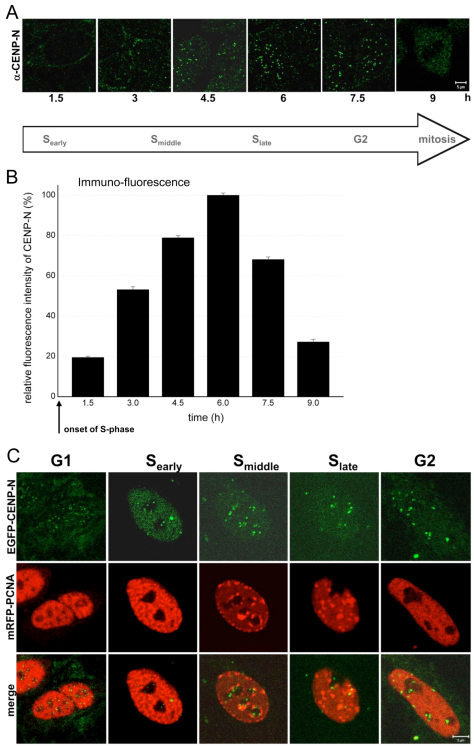
Previous immunofluorescence experiments in asynchronous cell populations indicated that CENP-N levels at kinetochores decrease during mitosis (McClelland et al., 2007) suggesting that the CENP-N kinetochore binding varies during the cell cycle. To substantiate this result, we synchronised cells and quantified the levels of endogenous CENP-N at kinetochores at different phases of the cell cycle, using the CENP-N antibody (McClelland et al., 2007), from early S phase until mitosis (Fig. 2A). Cell cycle phases were identified either by PCNA (S phase), DAPI (chromosome condensation in mitosis) or CENP-F staining (G2, mitosis). After double-thymidine block release, the fluorescence intensity profile versus time shows a continuous increase at kinetochores with a clear maximum after 6 hours in the second half of S phase (Fig. 2B). Relative to this maximal value, the fluorescence intensity values after 1.5 hours (early S phase) and 9 hours (mitosis) were 19% and 27%, respectively (Fig. 2B). The low abundance of CENP-N at kinetochores during mitosis thus agreed with previous findings (McClelland et al., 2007).
Fig. 2.
Amount of endogenous immunostained and EGFP-labelled CENP-N at kinetochores in early, middle and late S phase, G2 and mitosis. (A) CENP-N labelled by an anti-CENP-N primary and a fluorescently labelled secondary antibody in HEp-2 nuclei fixed 1.5, 3.0, 4.5, 6.0, 7.5 and 9.0 hours after double-thymidine block release. The amount of antibody-marked endogenous CENP-N at kinetochores increased during S phase and was low in mitosis. Scale bar: 5 μm. A G1 image is not included due to the low CENP-N levels in G1. (B) For each kinetochore, the fluorescence intensity per area was determined and compared. The mean secondary antibody fluorescence intensity per area of 70–250 kinetochores per time point (10–20 kinetochores per cell) was determined at kinetochores within cells at different time points after double-thymidine block release. The fluorescence intensity values are plotted relative to the value at 6.0 hours (100%). Endogenous CENP-N at kinetochores increased during S phase and was depleted from kinetochores during G2. (C) Analysis of EGFP–CENP-N at kinetochores. Detection of EGFP–CENP-N (upper row), mRFP-PCNA as replication marker (middle row) and merge (lower row) in HEp-2 cell nuclei. The fluorescence intensity of EGFP-labelled CENP-N increased from G1 into S phase. Scale bar: 5 μm.
Immunofluorescence-based quantifications, however, have the disadvantage that epitope-masking effects cannot be excluded. Therefore, for comparison, we determined whether EGFP–CENP-N is also differentially bound to kinetochores in living human cells. We analysed the localisation of EGFP–CENP-N by live-cell imaging throughout G1, early, middle and late S phase, and G2 using mRFP-PCNA localisation as a readout of the cell cycle phase (Madsen and Celis, 1985). PCNA is diffuse in G1 nuclei, coalesces into many small foci in early S phase, migrates to the inner nuclear membrane and nucleolar surface in middle S phase, grows to large foci in late S phase followed by diffuse redistribution in G2, similar to G1 localisation (Somanathan et al., 2001; Sporbert et al., 2005). Furthermore, G1 and G2 nuclei could be distinguished by determining whether mitotic condensed chromosomes and cytokinesis, or nuclear PCNA foci preceded the cell cycle stage (Hemmerich et al., 2008). This analysis showed low levels of CENP-N bound to kinetochores during G1, and an increase in kinetochore binding during S phase (Fig. 2C). By measuring the fluorescence intensities, the cell cycle dependent levels of EGFP–CENP-N were quantified at kinetochores, in the interphase nucleus and in the whole cell. The obtained cell cycle fluctuations were very similar to those of the endogenous protein. To quantify in more detail the removal of CENP-N from the kinetochores, we measured the decrease of EGFP–CENP-N fluorescence intensity at kinetochores after S phase at particular time points in living G2 cells and observed a continuous disappearance of CENP-N during the first 3 hours after entry into G2.
Thus, CENP-N immunofluorescence and EGFP–CENP-N measurements indicated that CENP-N is recruited to kinetochores at the onset of S phase, reaches a maximal abundance during late S phase, and is continuously displaced from kinetochores during G2.
Cellular CENP-N protein levels are maximal during S phase
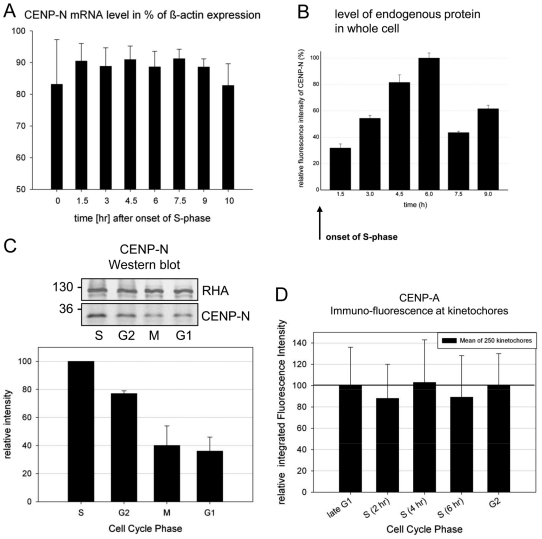
The S phase- and G2-specific accumulation of CENP-N at kinetochores could reflect either the general cellular abundance of CENP-N or a specific binding of CENP-N to kinetochores during S phase and G2. To distinguish between these possibilities, we determined CENPN mRNA and protein expression levels in the whole cell at different cell cycle phases (identified as described above). First, we measured the CENPN mRNA level of double-thymidine block synchronised cells over a period of 10 hours after release, well into G2 and mitosis. We observed stable CENPN mRNA values of 80–90% relative to β-actin (Fig. 3A), indicating that CENPN mRNA levels remain constant throughout the cell cycle. We next determined endogenous CENP-N protein levels in whole synchronised cells by quantitative immunofluorescence microscopy. We observed maximum fluorescence intensity in the whole cell after 6 hours in the second half of S phase (Fig. 3B). This maximum intensity was three times higher than the level during early S phase and ~1.6 times higher than the level in mitosis. These data were confirmed by an immunoblotting analysis (Fig. 3C). Three hours after double-thymidine block release we found maximal amounts of CENP-N (S phase), which decreased to 77±2% after 7 hours (G2), 40±14% after 9 hours (mitosis) and 36±10% after 12 hours (G1). Finally, we also tested whether CENP-N binding to kinetochores might be influenced by the cell-cycle dependent changes in CENP-A levels. We thus quantified CENP-A levels at kinetochores by immunofluorescence and found constant CENP-A levels over S phase and G2 (Fig. 3D).
Fig. 3.
CENPN mRNA and whole cell protein levels. (A) mRNA levels (mean of three independent experiments) of CENPN are plotted relative to the value of β-actin in the cells. Human HEp-2 cells were synchronised by a double-thymidine block and analysed after release in time intervals of 90 minutes for 11 hours. (B) After immunostaining of endogenous CENP-N, the whole cell fluorescence was measured and integrated at 1.5, 3.0, 4.5, 6.0, 7.5 and 9.0 hours after double-thymine block release. Whole cell fluorescence intensity values are plotted relative to the maximal value at 6.0 hours (100%). Although the mRNA levels were rather constant, the amount of CENP-N in the cell increased in S phase (with a maximum in late S phase) and decreased afterwards. (C) Western blot analysis of whole cell CENP-N protein levels at 3 (S phase), 7 (G2), 9 (M phase) and 12 (G1) hours after double-thymidine block release relative to the house-keeping gene product RHA. CENP-N was identified by a CENP-N-specific antibody (molecular weight marker values are shown on the left). CENP-N amounts were quantified by the ODYSSEY Infrared Imaging System and are shown in the histogram below. CENP-N levels were maximal in S phase and low in mitosis and G1. (D) CENP-A was antibody stained at 0 (G1–S), 2 (S), 4 (S), 6 (S) and 8 (G2) hours after HEp-2 cells double-thymidine block release and its amounts quantified by integrating the secondary antibody fluorescence intensity at kinetochores. CENP-A levels at centromeres remained constant during S phase and G2. Error bars indicate s.d.
We conclude that CENP-N is present in the cell during the whole cell cycle. However, protein levels vary within a factor of three, with highest values during second half of S phase. The binding of CENP-N to kinetochores varied by a factor of five in S phase compared with mitosis–G1. This implies that during S phase, increasing cellular CENP-N protein concentrations drive CENP-N into a complex with CENP-A-containing nucleosomes. In G2, however, CENP-N is released from kinetochores. This reduction of CENP-N levels at CENP-A-containing nucleosomes in G2 is not explained by a potential reduction of CENP-A levels, but coincides with the degradation of CENP-N.
Kinetochore-binding dynamics of CENP-N
The varying presence of CENP-N at kinetochores suggests a dynamic binding behaviour. The binding of CENP-N to the kinetochores might either be stable, or transient, with fast exchange of CENP-N at the binding sites. Recently, we detected stable binding to kinetochores during the whole cell cycle for CENP-A and CENP-I, whereas hMis12 stably bound to the kinetochores only during mitosis but showed fast exchange during interphase (Hemmerich et al., 2008). To monitor the behaviour of CENP-N, we applied two independent, complementary methods, the SNAP-tag and fluorescence recovery after photobleaching (FRAP).
SNAP-tag
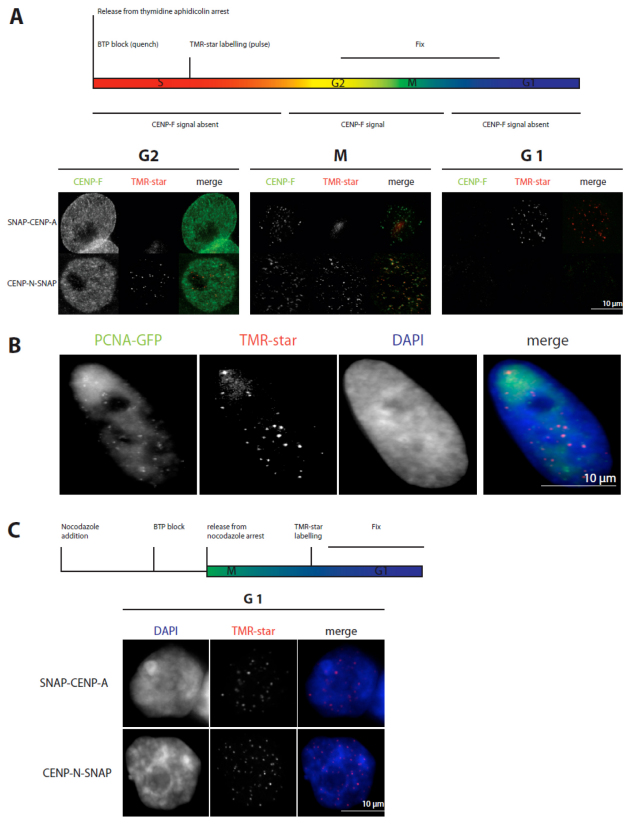
The SNAP-tag is a versatile protein tag that can catalyse the formation of a covalent bond to a benzyl-guanine moiety coupled to different fluorescent or non-fluorescent membrane-permeable reagents (Keppler et al., 2003). Importantly, this tag allows pulse-chase experiments at a single protein level, because no other protein in a cell will react with membrane-permeable benzyl-guanines. The tag has previously been used to show that CENP-A is loaded on centromeres exclusively in early G1 (Jansen et al., 2007). We therefore used the same technique to ask at which cell cycle stage CENP-N-SNAP is incorporated into kinetochores. First, to validate the method in our hands, we monitored the centromere assembly of CENP-A in HeLa cells (Fig. 4). Cells were transiently transfected with SNAP–CENP-A and arrested in early S phase with a double-thymidine and aphidicolin arrest. Cellular SNAP–CENP-A was quenched with BTP (a benzyl guanine not coupled to a fluorophore) for 30 minutes. Cells were released from the arrest, labelled in vivo 4 hours later with the fluorophore TMR-star, and fixed for immunofluorescence analysis 12 hours after labelling. This resulted in a mix of G2, mitotic, and early G1 cells, which could be distinguished by CENP-F staining, which labels the whole nucleus in G2 and specifically binds to kinetochores during M phase, but shows no specific localisation in early G1 cells (Liao et al., 1995). Consistent with previous data, TMR-star fluorescence on SNAP–CENP-A was only observed in G1 cells (Fig. 4A), confirming that CENP-A is specifically loaded in G1 (Jansen et al., 2007; Hemmerich et al., 2008). By contrast, when transfecting a CENP-N–SNAP construct, after double-thymidine and aphidicolin block release and applying the same protocol, we found CENP-N–SNAP present at kinetochores of G2 cells, but not of early G1 cells (Fig. 4A). This suggested that CENP-N is loaded onto kinetochores in or before G2, that it is released as cells progress through M phase, and that it is not reloaded in early G1. To extend our temporal analysis to further phases of the cell cycle, we repeated these experiments in U2OS because these cells have a longer cell cycle. Thus, again 12 hours after release and following the same experimental procedure, U2OS cells can be analysed in late-S phase. Here, TMR-star fluorescence for CENP-N–SNAP was already detected in late S phase as judged by PCNA–GFP fluorescence. Thus, CENP-N assembles at kinetochores already in late S phase or earlier (Fig. 4B). Finally, to measure the earliest time point at which CENP-N can assemble into kinetochores, CENP-N–SNAP-transfected HeLa cells were arrested in mitosis for 12 hours by a nocodazole block and quenched with BTP for 30 minutes (Fig. 4C). At 4 hours after quenching, the cells were released from nocodazole arrest. After another 5 hours, SNAP-tagged CENP-N was fluorescently labelled with TMR-star for 30 minutes and fixed for examination. TMR-star fluorescence was detected (Fig. 4C) in roughly half the cells examined, indicating that CENP-N–SNAP can be loaded during the first 5 hours after M phase in G1. Overall, these experiments suggest a time window of middle G1 to G2 for CENP-N binding and loading to kinetochores.
Fig. 4.
CENP-N loading to kinetochores measured by the SNAP-tag technology. (A) Above: Scheme of the performed experiment. Below: Representative images of cells showing TMR-star fluorescence for SNAP–CENP-A only in G1 and for CENP-N–SNAP in G2 and M phase. Cell cycle phases G2 (CENP-F staining of the whole nucleus) and mitosis (specific kinetochore binding of CENP-F) are clearly identified. (B) The same experiment as in A was performed with U2OS cells stably expressing PCNA–GFP. CENP-N–SNAP fluorescence appeared at kinetochores in late S phase as judged from cellular PCNA distributions. (C) Top: Scheme of the performed experiment. Below: Representative images of cells expressing SNAP–CENP-A and CENP-N–SNAP showing fluorescence at kinetochores during G1. CENP-N was loaded to kinetochores in G1 and S phase.
FRAP
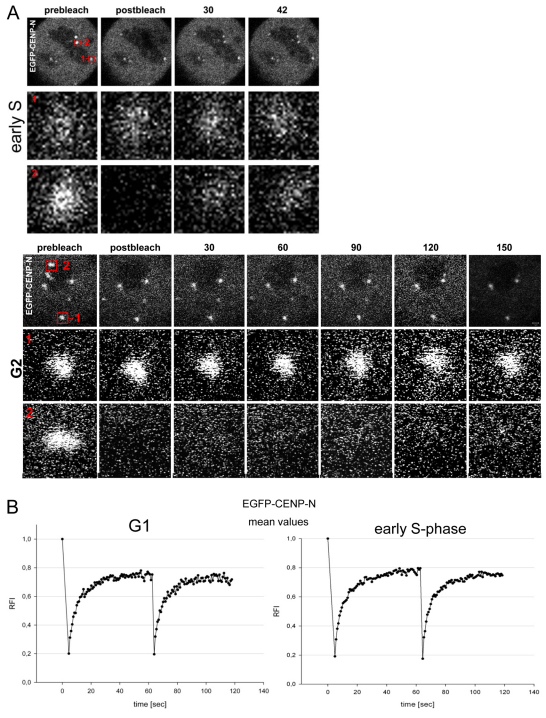
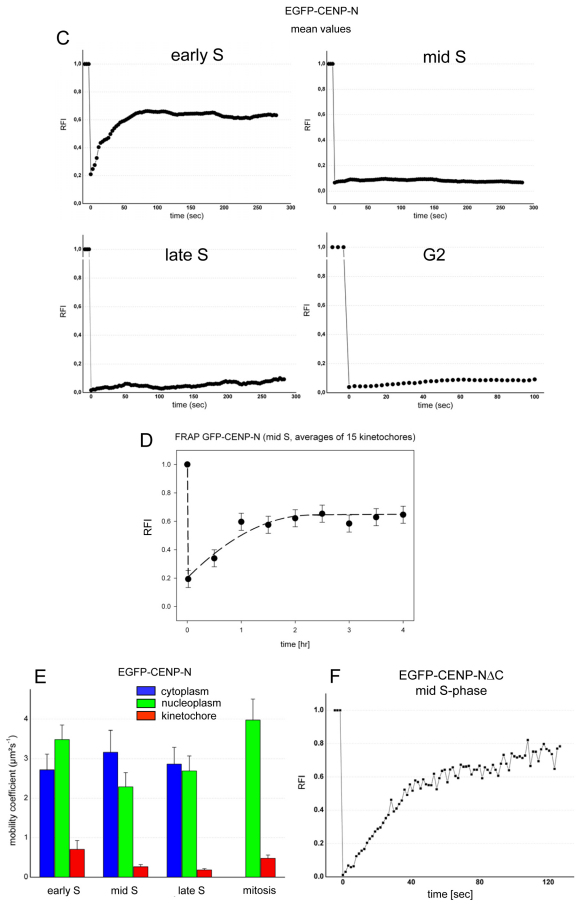
In order to refine our analysis, we carried out fluorescence recovery after photobleaching (FRAP) experiments to monitor kinetochore binding of EGFP–CENP-N in various cell cycle phases as well as measure its binding dynamics in living cells (see Fig. 5). In G1, we observed recovery of CENP-N at the kinetochores after photobleaching. In order to determine whether CENP-N is recruited to the kinetochore in G1 by a loading-only mechanism or by free exchange, the EGFP–CENP-N proteins were bleached a second time (double FRAP experiment), during recovery 60 seconds after the initial bleaching event (Hemmerich et al., 2008). This second recovery of EGFP–CENP-N had very similar kinetics of fluorescence recovery to that seen after the first bleach event (see Fig. 5B). During early S phase, we observed a partial recovery of CENP-N at the kinetochores after photobleaching (mean recovery of 50%) with half amplitude recovery times t1/2=20±2 seconds (Fig. 5C). Again, the EGFP–CENP-N proteins were bleached a second time. As after the first bleach, EGFP–CENP-N showed fluorescence recovery (Fig. 5A,B). This indicates that CENP-N, loaded in G1 or early S phase, is not irreversibly bound but can dissociate from the complex and rebind. In contrast to this behaviour of CENP-N, we previously observed irreversible ‘loading-only’ binding of CENP-A and CENP-I to the kinetochore (Hemmerich et al., 2008). In middle and late S phase, we observed a very low (0–7%) EGFP–CENP-N fluorescence recovery after photobleaching within 200 seconds (see Fig. 5C), as in G2 within 100 seconds (see Fig. 5A,C). Analysing EGFP–CENP-N fluorescence recovery over a much longer time period, now over 4 hours, in the second half of S phase, we found slow assembly of CENP-N at kinetochores with t1/2=38±7 minutes and a recovery fluorescence intensity increase of 45±6% (see Fig. 5D). These FRAP data confirm our SNAP-tag results.
Fig. 5.
EGFP–CENP-N binding dynamics to kinteochores. (A) EGFP–CENP-N binding to several kinetochores in HEp-2 cells in early S phase (upper three rows) and in G2 (lower three rows). In both cases, two kinetochore foci 1 and 2 were selected: spot 1 was not bleached and served as control, whereas spot 2 was photobleached. The changes in spots 1 and 2 over time are shown in the rows below at higher magnification. In early S phase, after photobleaching, fast recovery was observed in spot 2. In G2, after photobleaching, no FRAP was observed in spot 2. Quantification is presented in B and C. (B) Mean FRAP recovery curves of 29 kinetochores in 12 cells in late G1 (left) and 28 kinetochores in 12 cells in early S phase (right, see A). After 60 seconds during recovery, the spots were bleached a second time and again a similar recovery was measured, indicating exchange of CENP-N at the binding site. Experimental error was ±0.10. (C) Mean values of 5–20 experiments were blotted for early, middle and late S phase and G2. Partial recovery of the fluorescence intensity relative to the prebleach intensity (RFI) was observed for EGFP–CENP-N in early S phase, but there was hardly any recovery during 300 seconds in middle and late S phase and during 100 seconds in G2 (see A). Experimental error was ±0.02. (D) Very slow FRAP recovery (t1/2=38 minutes) of 45% EGFP–CENP-N was observed during 4 hours in the second half of S phase. Experimental error was ±0.06. (E) Diffusion of CENP-N measured by RICS. Mobility coefficient values of EGFP–CENP-N at early, middle and late S phase as well as in mitosis were measured in the cytoplasm (blue), nucleoplasm (green) and at kinetochores (red). CENP-N was strongly immobilised at kinetochores. (F) The CENP-N mutant EGFP–CENP-NΔC, lacking the C-terminal domain, showed fast exchange and strong recovery in middle S phase.
RICS
To verify CENP-N binding to kinetochores and its mobility in other cellular compartments, we used raster image correlation spectroscopy (RICS) (Digman et al., 2005) to measure the diffusion of EGFP–CENP-N in the cell cytoplasm, nucleoplasm and at kinetochores (Fig. 5E). Whereas in fluorescence correlation spectroscopy (FCS) experiments the protein mobility is measured in a single confocal volume in the cell, in RICS this FCS experiment is repeated many times with the confocal volume scanning a confocal plane in the cell. In S phase, CENP-N showed a mobility coefficient of about 3 μm2/second in the cytoplasm as well as in the nucleoplasm (Fig. 5E), which was comparable to the diffusion properties of CENP-H and CENP-I (Hemmerich et al., 2008). During mitosis, in the cellular plasma the mobility coefficient increases to about 4 μm2/second. At kinetochores, we observed a mobility of up to one order of magnitude slower (see Fig. 5E). At kinetochores during S phase, the CENP-N mobility coefficient decreases from 0.7±0.2 μm2/second (early S phase) to 0.2 μm2/second (late S phase) and increases again to 0.5 μm2/second in mitosis (Fig. 5E). Low residual mobility of 0.2 μm2/second was also observed for other CCAN proteins like CENP-T (unpublished observations). The dynamic RICS data support our FRAP results and confirm stable kinetochore attachment of CENP-N, in particular during late S phase.
CENP-NΔC mutant
Previous in vitro experiments have shown that the CENP-NΔC mutant still binds to the CENP-A nucleosomes, although binding is reduced in vivo (Carroll et al., 2009). This in vivo result was suggested to be due to additional interactions of wild-type CENP-N with CCAN subunits. We thus tested in vivo whether the C-terminal region contributes to the initial CENP-N binding in early S phase or to the stable retention in middle S phase. We analysed by FRAP the kinetochore binding of CENP-NΔC in middle S phase (when the wild-type protein shows no fast exchange). We found that CENP-NΔC exchanges fast (t1/2=23±2 seconds; within experimental error this is the same time constant as wild-type CENP-N in early S phase) with a high mobile fraction (70%; see Fig. 5F). These data show that in S phase the C-terminal region mediates stable CENP-N binding to kinetochores. This is presumably due to interactions with one or more other CCAN subunit(s). The best candidate is CENP-L, which interacts with CENP-N in a C-terminal region-dependent manner in vitro (Carroll et al., 2009) as well as in mitotic Xenopus egg extracts (unpublished observations). The highly mobile binding of CENP-NΔC to the CENP-A-containing centromeric chromatin can explain why CENP-NΔC binding to CENP-A nucleosomes could not be detected by co-immunoprecipitation (Carroll et al., 2009). The CENP-N mutants R11A and R196A displayed reduced CENP-A nucleosome binding and inefficiently localise to kinetochores (Carroll et al., 2009). We found their kinetochore binding too low for quantitative FRAP measurements.
In conclusion, our data show that CENP-N binds to kinetochores by rapid exchange during G1 and early S phase, but is stably associated with kinetochores during the remainder of S phase and G2. During the second half of S phase, CENP-N is loaded to kinetochores more than 100 times slower than the fast exchange in early S phase. CENP-N gradually dissociates from kinetochores during G2.
Discussion
The central CENP-A targeting domain (CATD) confers nucleosomal rigidity to CENP-A-containing nucleosomes at the centromere (Black et al., 2007b; Sekulic et al., 2010), providing a unique structure distinguishing it from H3-containing nucleosomes. This structural difference is recognised by the N-terminal region of CENP-N because CENP-N binds to CENP-A-containing nucleosomes but not to H3-containing nucleosomes in vitro (Carroll et al., 2009; Sekulic and Black, 2009). Here, we show by in vivo FRET that the N-terminal region of CENP-N is indeed in close proximity to the N-terminal region of CENP-A, confirming this interaction in living human cells. Our data further show that CENP-N binds to the kinetochore in a dynamic and complex cell cycle-dependent manner as, to a large extent, CENP-N is recruited to the kinetochore during S phase and then leaves the complex in G2. These data suggest a dynamic CCAN network, as opposed to a pure structural scaffold at the kinetochores. They are also consistent with recent data on the mitotic population of the CCAN subunit CENP-I, which rapidly switches between two sister-kinetochores during metaphase, as kinetochore–microtubules change from a growing to a shrinking phase (Amaro et al., 2010). The CENP-N properties described here might be unique, but alternatively might well be shared by other CCAN proteins.
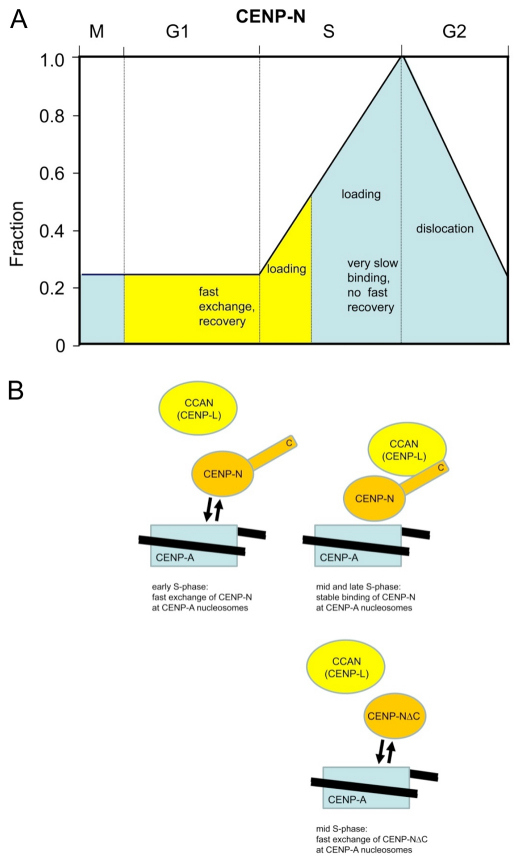
We observed two- to threefold amounts of CENP-N in the cell in S phase compared with G1 or mitosis, while CENP-N binding to kinetochores increases fivefold from G1 to late S phase. Then, CENP-N levels gradually decrease during G2 to reach low values at M phase (Fig. 6A), confirming previous initial results (McClelland et al., 2007). We postulate that CENP-N accumulation at kinetochores during S phase is driven by increasing CENP-N levels in the cell, potentially supported by post-translational modification. However, western-blot analysis showed no indications for a modification of CENP-N. When cells are arrested in mitosis by nocodazole treatment, CENP-N shows reduced electrophoretic mobility, indicating a potential phosphorylation event. This modification is, however, due to the nocodazole treatment because it is not detected when the cells are synchronised by a double-thymidine block.
Fig. 6.
Scheme of cell cycle-dependent CENP-N levels and its dynamic binding to kinetochores. (A) CENP-N is present at kinetochores at low levels (about 20%) during mitosis and G1. CENP-N exchanges fast at kinetochores during G1 and early S (yellow) and binds there stably during middle and late S phase with very slow loading (blue), probably due to CENP-L binding to the CENP-N C-terminus. CENP-N continuously dislocates from kinetochores during G2. (B) CENP-N binds to CENP-A-containing nucleosomes by fast exchange during early S phase (top left). In middle and late S phase (top right), CENP-N is stably bound due to an interaction of its C-terminus with a CCAN protein, probably CENP-L. Below: The C-terminal deletion mutant CENP-NΔC cannot be bound and fixed by CCAN (CENP-L). Accordingly, CENP-NΔC shows fast exchange also in middle S phase, with the same time constant as wild-type CENP-N in early S phase, and high recovery. A quantitative computer model is consistent with this interpretation (Bashar Ibrahim, Mathematics and Computer Science, FSU, Ernst-Abbe-Platz 1-3, 07743 Jena, Germany, personal communication; various assumptions of this model have to be verified experimentally).
Stable CENP-N kinetochore association during middle and late S phase seems to be regulated by binding of one or more CCAN protein(s), probably CENP-L, to the CENP-N C-terminal region. During S phase, one or more CCAN proteins might contribute to the changes in CENP-N dynamics by increasing their cellular levels or by post-translational modification. During G2, CENP-N dissociates from the kinetochores, potentially due to a dual mechanism involving both protein turnover and some other regulatory mechanism, to alter the dynamics of association. CENP-N is degraded during G2 (Fig. 3C), therefore if CENP-N falls off the CENP-A-containing nucleosome it will not be replaced from the cytosolic pool, followed by degradation of released CENP-N. Reduction of CENP-N during G2 is not due to decreasing levels of CENP-A because we found constant levels of CENP-A at kinetochores during S phase and G2 (Fig. 3D). It remains unclear what initiates CENP-N synthesis at the G1–S transition and its degradation at the end of S phase.
We detected CENP-N loading to kinetochores during G1 and S phase. We observed fast exchange during G1 and early S phase but slow loading in the second half of S phase. This slow loading dynamics of CENP-N corresponds quantitatively to the slow loading of CENP-T and CENP-W during second half of S phase (Prendergast et al., 2011). Our dynamics data clearly show that the CENP-N C-terminal region contributes considerably to the CENP-N binding stability in vivo, probably by binding to CENP-L (Fig. 6B) because both proteins interact in vitro (Carroll et al., 2009) and in Xenopus egg extracts (unpublished observations). The observed CENP-N binding dynamics differ from those of CENP-A, CENP-B, CENP-C, CENP-H and CENP-I (Hemmerich et al., 2008), suggesting a different functional role for CENP-N. Centromeric DNA is preferentially replicated during middle and late S phase with only few centromeres being replicated during early S phase (Shelby et al., 2000; Weidtkamp-Peters et al., 2006; Wu et al., 2006). Because major parts of CENP-N are loaded to kinetochores during S phase, we speculate that CENP-N, together with its binding partner of the CCAN complex, might have a role in securing functional centromeric chromatin structure through replication. As CENP-A nucleosomes are redistributed to both sister kinetochores, CENP-N as being part of the CCAN protein complex, might ensure that CENP-A remains associated with the centromeric DNA. Depletion of CENP-N led to kinetochore assembly defects and resulted in reduced assembly of nascent CENP-A into centromeric chromatin (Foltz et al., 2006; Carroll et al., 2009). In the absence of CENP-N, replicated centromeric chromatin might be unable to correctly assemble as required for mitosis, resulting in chromosome segregation defects and potentially also in incorrect loading of CENP-A in telophase and early G1. We thus speculate that CENP-N might be a fidelity factor for the kinetochore complex during centromeric replication. The Saccharomyces cerevisiae orthologue of CENP-N, Chl4p (Meraldi et al., 2006), seems to have a similar role as fidelity factor: although CHL4 is non-essential in budding yeast, CHL4 depletion becomes essential for kinetochore assembly under conditions in which the kinetochore has been disrupted by a transient strong transcriptional activity (Mythreye and Bloom, 2003). This could suggest an evolutionary conserved role of CENP-NChl4 as kinetochore fidelity factor.
Materials and Methods
Plasmids
For the expression of human CENP-A fused to the C-terminus of mCherry, the required plasmid was constructed from the vector pCerulean-C1–CENP-A (Orthaus et al., 2008). This vector was digested with AgeI-BsrGI. Into the resulting purified 4.433 bp fragment, we ligated a 713 bp AgeI-BsrGI fragment resulting from the vector pmCherry-C1 (Orthaus et al., 2009). For expressing human CENP-N fused to the C-terminus of EGFP, we used vector pH-G–CENP-N (Hellwig et al., 2009). All clones were verified by sequencing (MWG Biotech, Ebersberg, Germany). Full-length protein expression of the fusion constructs was confirmed by western blots. Under the control of the SV-40 promoter, CENPA was introduced at the 3′-end (yielding the fusion SNAP-tag–CENP-A) and CENPN at the 5′-end of the SNAP-tag gene (yielding the fusion CENP-N–SNAP-tag). Complementary DNAs were amplified from already existing constructs by PCR. Correct cloning of the obtained plasmids was verified by restriction analysis.
Cell culture, transfection, siRNA treatments and western blots
HeLa E1 and HEp-2 cells were cultured (McAinsh et al., 2006) and western blots were carried out as described recently (Orthaus et al., 2008). In order to determine cell cycle-dependent CENP-N levels, double-thymidine block synchronised HEp-2 cells were treated as above. Aliquots of equal cell numbers were taken after 3, 7, 9 and 12 hours after release and lysed. The aliquots were analysed by western blotting. In the blot, CENP-N was identified by the CENP-N-specific antibody, and CENP-N amounts quantified by the ODYSSEY Infrared Imaging System (LiCor, Lincoln, NE) following the protocol of the manufacturer.
Immunofluorescence
For indirect immunofluorescence detection, HEp-2 cells (from the DSMZ, Braunschweig, Germany) were fixed with 3% paraformaldehyde in PBS for 15 minutes, permeabilised with 0.5% Triton X-100 in PBS for 10 minutes and blocked with 3% bovine serum albumin (BSA) in PBS. Primary antibodies were diluted as follows in PBS and 3% BSA: anti-CENP-N (McClelland et al., 2007) 1:1000; anti-CENP-A antibodies (Abcam, Cambridge, UK) 1:500. Anti-CENP-N antibodies were detected with fluorescently labelled goat anti-rabbit antibodies (Cy3 anti-rabbit, Dianova, Hamburg, Germany) at a dilution of 1:400. For live cell measurements, HEp-2 cells were co-transfected with EGFP–CENP-N and mRFP–CENP-A or mRFP–PCNA. Cell cycle phases were identified via the localisation pattern of mRFP–PCNA (Hemmerich et al., 2008), CENP-F localisation or DAPI staining identifying morphological characteristics of the cells. Fluorophores were stimulated with 561 nm argon laser line with low intensity and signals were detected via a 575 nm long path filter. 3D image stacks were acquired in 0.2 μm steps using a 63× oil NA 1.4 objective on a Zeiss LSM 510 microscope (Carl Zeiss, Jena, Germany). The 3D image stacks were quantitatively analysed using Metamorph (Molecular Devices, Sunnyvale, CA).
SNAP-tag analysis
Cells were grown in DMEM supplemented with 10% FCS and PS (100 units/ml penicillin G, 100 μg/ml strepdavidin). One day before transfecting, 100–200 μl of the cells from the confluent 10 cm dish were added to a coverslip in a six-well plate containing MEM (10% FCS and PS). For transfections, 3 μl Fugene6 (Invitrogen, Carlsbad, CA) was added to 100 μl OPTIMEM and incubated for 5 minutes. The mixture was then added dropwise to 1 μg DNA and incubated for 30 minutes. Then, it was added to one well of the six-well plate containing 2 ml of MEM (10% FCS and PS). The coverslips with the transfected cells were transferred to parafilm in a 10 cm dish and 150 μl of MEM (10% FCS and PS) containing 2 μM TMR-star was added. After 30 minutes incubation at 37°C, the coverslips were washed twice in prewarmed MEM (10% FCS and PS) and incubated in fresh media for 30 minutes at 37°C. For quenching the SNAP-tag with BTP, the same protocol was performed but the media was supplemented with 10 μM BTP instead of TMR-star.
The media in the six-well dishes was removed, 2 ml PTEMF buffer (0.2% Triton, 20 mM PIPES pH 6.8, 1 mM MgCl2, 10 mM EGTA, 4% formaldehyde) was added and incubated for 10 minutes. After washing three times with 1× PBS, 2 ml blocking buffer (500 ml 1× PBS, 15 g BSA, 0.02% w/v NaN3) was added and incubated for 30 minutes. After washing three times with 1× PBS, the coverslips were placed on parafilm and antibodies were added in the appropriate dilutions (1:2000 for anti-CENP-A and anti-CENP-N) in 200 μl blocking buffer and incubated for 1 hour. After washing three times with 1× PBS, secondary antibodies (anti-mouse or anti-rabbit Alexa Fluor 488- or Alexa Fluor 647-conjugated) were added in 1:400 dilutions in 210 μl blocking buffer and incubated for 30 minutes. The coverslips were then placed on a microscope slide containing a drop of Vectashield (Vector Laboratories, Servion, Switzerland) supplemented with DAPI and fixed with nail polish.
3D image stacks were acquired in 0.5 μm steps on a Leica DMI 6000B microinjection microscope (Leica, Wetzlar, Germany) equipped with a Leica EL6000 metal halide light source, a 63× oil NA 1.4–0.6 objective, a DAPI-EGFP-Rhod-Cy5 filter set and a Hamamatsu high resolution digital camera (ORCA-HR). All images shown are maximum intensity projections of deconvolved 3D image stacks using the software Slidebook (Intelligent Imaging Innovations, Denver, CO). Maximum intensity projections were processed using the software Adobe Photoshop.
Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching (FRAP) experiments were carried out on a Zeiss LSM 510Meta confocal microscope (Carl Zeiss, Jena, Germany) using a C-Apochromat infinity-corrected 1.2 NA 63× water objective and the 488 nm laser line for GFP. Five or ten images were taken before the bleach pulse and 50–200 images after bleaching of two to four kinetochores of a nucleus, with an image acquisition frequency of 0.5–1.0 frames per second at 1% laser transmission to avoid additional bleaching. In long-term FRAP experiments, the pinhole was adjusted to 1 airy unit and image stacks were taken every 30 minutes. Relative fluorescence intensities were quantified according to previously described methods (Chen and Huang, 2001; Schmiedeberg et al., 2004) using Microsoft Excel and Origin (OriginLab, Northampton, MA) software.
Cell cycle synchronisation
For cell cycle-dependent analysis, HEp-2 cells were reversibly arrested via double-thymidine treatment at the initiation of early S phase. In some experiments, HeLa and U2OS cells were synchronised by double-thymidine and aphidicolin block, in other cases by nocodazole treatment. The cells were seeded with a density of about 50–60%. After attachment to reaction vessel, cells were blocked with a final concentration of 5 mM thymidine for 16 hours. Cells were released and grown in fresh media for 10 hours, followed by a second cell cycle block with 5 mM thymidine. After 16 hours, cells were washed and cultured in fresh media. To investigate the cell cycle-dependent behaviour of CENP-N, every 90 minutes for 9 hours cells were harvested and used for further steps of different procedures. Cells were fixed, labelled with antibodies, and the fluorescence intensity at kinetochores measured until mitotic cells were observed.
Raster image correlation spectroscopy
Raster image correlation spectroscopy (RICS) (Digman et al., 2005) experiments were accomplished on a Zeiss LSM 710 confocal microscope (Carl Zeiss, Jena, Germany) using a C-apochromat infinity-corrected 1.2 W 40× water objective and the 488 nm laser line for EGFP–CENP-N. Time series of 50 pictures were recorded with an image acquisition frequency of 1 frame per 3.9 seconds at low laser intensity. During the measurement, the pinhole was adjusted to a diameter of 40 μm. The emission of EGFP fluorophores was detected through a 505–550 nm band path filter. For evaluation of the data, the LSM-RICS software (Zeiss, Jena, Germany) was used, yielding the diffusion coefficients of the moving fluorescent particles (Digman et al., 2005).
Acceptor photobleaching-based FRET measurements
When FRET occurs, both the intensity and lifetime of the donor fluorescence decrease whereas the intensity of the acceptor emission increases. Here, the FRET pair EGFP–mCherry was used. FRET experiments were conducted as described (Orthaus et al. 2008; Orthaus et al., 2009). If the orientation of the fluorophore dipole moment of the acceptor relative to that of the donor were known, or if at least one of them rotated freely faster than nanoseconds, a more detailed distance between donor and acceptor could be deduced from the measured FRET efficiency (EFRET) values. In our live cell experiments, however, this information was not available to us. We therefore could not deduce defined distance values but interpreted the appearance of FRET as an indication that donor and acceptor chromophores are close to one another within 10 nm.
FLIM-based FRET measurements
Experiments were carried out as described (Orthaus et al., 2009). In short, the donor fluorescence lifetime was determined by time-correlated single photon counting (TCSPC) in living human HEp-2 cells. To calculate the fluorescence lifetime, the SymPhoTime software package (v4.7, Picoquant, Berlin, Germany) was used. Selected areas of the images corresponding to single kinetochores were fitted by a maximum likelihood estimation (MLE). Depending on the quality of a fit indicated by the value of χ2, a mono- or bi-exponential fitting model including background was applied. In this way, the presence of scattered light in a few measurements could be identified and separated. However, due to low photon numbers and similar time constants, the simultaneous presence of two different donor fluorescence lifetimes for complexes with donor-only and donor-plus-acceptor in one kinetochore could not be separated by a bi-exponential fit.
RNA preparation and RT-PCR
Human HEp-2 cells were synchronised by a double-thymidine block, harvested in time intervals of 90 minutes, centrifuged and frozen in liquid nitrogen. Pellets were resuspended in 1 ml RNApure (Peqlab, Göttingen, Germany), then 200 ml chloroform was added and the mixture was vortexed. The homogenous solution was incubated at 4°C for 5 minutes, then centrifuged at 22°C for 15 minutes at 13,400 g. The upper phase was separated, the same volume of 2-propanol and 2 μl glycogen added, the solution mixed and incubated at 4°C for 10 minutes followed by centrifugation at 22°C for 10 minutes at 13,400 g. The supernatant was removed, 1 ml of 70% ethanol added and the solution centrifuged at 22°C for 5 minutes at 5200 g. The supernatant was removed, the pellet air dried at 22°C and resuspended in 100 μl nuclease-free water. RNA content and purity was determined using the Nanodrop (Peqlab, Göttingen, Germany). The solution was stored at −80°C. For RNA amplification, the following primers were used: β-actin forward 5′-CATCATGAAGTGTGAACGTGGAC-3′, reverse 5′-AGCATTTGCGGTGGACGATG-3′; CENP-N forward 5′-CAACTCTACGACACCTCTACAG-3′, reverse 5′-TGCTAAGGATTTCAATGCTTCCAG-3′. These primers amplify fragments of about 300 bp at the 3′ end of the genes. Quantitative RT-PCR was performed with a SYBR Green Kit (Eurogentec, Liege, Belgium) according to the protocols of the manufacturer. For each reaction, 100 ng template RNA was used. Data were acquired using the Applied Biosystems 7300 Real Time PCR Light Cycler (Applied Biosystems, Foster City, CA) with temperature cycles as suggested by the manufacturer, and analysed with the 7300 Real Time PCR System Sequence Detection software, version 1.2.3 (Applied Biosystems).
Acknowledgement
We thank Nikolaj Klocker (University of Freiburg, Freiburg, Germany), Daniel Foltz (University of Virginia, Charlottesville, VA) and Iain Cheeseman (Whitehead Institute for Biomedical Research, Cambridge, MA) for the kind gift of plasmids. We acknowledge the expert technical support by S. Pfeiffer, M. Koch and S. Ohndorf.
Footnotes
Funding
This work was funded by the Marie Curie Cancer Care program; the Deutsche Forschungsgemeinschaft (DFG) [grant numbers SPP1128, SPP 1395/2]; the National Institutes of Health (NIH) [grant number GM074728 to A.F.S.]; and the Thueringer Aufbaubank [grant number 2007 FE 9011]. We thank the Swiss SNF for a Foerderungs-Professur [for P.M.]. Deposited in PMC for release after 12 months.
References
- Amano M., Suzuki A., Hori T., Backer C., Okawa K., Cheeseman I. M., Fukagawa T. (2009). The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 186, 173-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro A. C., Samora C. P., Holtackers R., Wang E., Kingston I. J., Alonso M., Lampson M., McAinsh A. D., Meraldi P. (2010). Molecular control of kinetochore-mircotuble dynamics and chromosome oscillations. Nat. Cell Biol. 12, 319-329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B. E., Jansen L. E. T., Maddox P. S., Foltz D. R., Desai A. B., Shah J. V., Cleveland D. W. (2007a). Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell 25, 309-322 [DOI] [PubMed] [Google Scholar]
- Black B. E., Brock M. A., Bedard S., Woods V. L., Cleveland D. W. (2007b). An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 104, 5008-5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C. W., Silva M. C. C., Godek K. M., Jansen L. E. T., Straight A. F. (2009). Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11, 896-902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C. W., Milks K. J., Straight A. F. (2010). Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189, 1143-1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Desai A. (2008). Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33-46 [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Niessen S., Anderson S., Hyndman F., Yates J. R., 3rd, Oegema K., Desai A. (2004). A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18, 2255-2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. (2006). The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983-997 [DOI] [PubMed] [Google Scholar]
- Chen D., Huang S. (2001). Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153, 169-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho U. S., Harrison S. C. (2011). Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc. Natl. Acad. Sci. USA 108, 9367-9371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y., Furuyama T., Vermaak D., Henikoff S. (2007). Structure, dynamics and evolution of centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 104, 15974-15981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa M. L., Wyns K., Li M., Hall M. A., Wang M. D., Luger K. (2011). Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat. Commun. 2, 313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A. D., Sorger P. K. (2003). Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17, 2902-2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman M. A., Brown C. M., Sengupta P., Wiseman P. W., Horwitz A. R., Gratton E. (2005). Measuring fast dynamics in solutions and cells with a laser scanning microscope 89, 1317-1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis E. K., Weber C., Gill R. K., Diekmann S., Dalal Y. (2010). Tetrameric organization of vertebrate centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 107, 20317-20322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L. E. T., Black B. E., Bailey A. O., Yates J. R., 3rd, Cleveland D. W. (2006). The human CENP-A centromeric complex. Nat. Cell Biol. 8, 458-469 [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., Ikemura T. (2001). CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertrebrate cells. EMBO J. 20, 4603-4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T., Henikoff S. (2009). Centromeric nucleosomes induce positive DNA supercoils. Cell 138, 104-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig D., Hoischen C., Ulbricht T., Diekmann S. (2009). Acceptor-photobleaching FRET analysis of core kinetochore and NAC proteins in living human cells. Eur. Biophys. J. 38, 781-791 [DOI] [PubMed] [Google Scholar]
- Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., Diekmann S. (2008). Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180, 1101-1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C. B., Welburn J. P., Dong Y., McEwan B. F., Shang W.-H., Suzuki E., Okawa K., et al. (2008a). CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135, 1039-1052 [DOI] [PubMed] [Google Scholar]
- Hori T., Okada M., Maenaka K., Fukagawa T. (2008b). CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol. Biol. Cell 19, 843-854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Liu Y., Wang M., Fang J., Huang H., Yang N., Li Y., Wang J., Yao X., Shi Y., Li G., Xu R. M. (2011). Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25, 901-906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L. E. T., Black B. E., Foltz D. R., Cleveland D. W. (2007). Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795-805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler A., Gendreizig S., Gronemeyer T., Pick H., Vogel H., Johansson K. (2003). A general method for the covalent labelling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86-89 [DOI] [PubMed] [Google Scholar]
- Liao H., Winkfein R. J., Mack G., Rattner J. B., Yen T. J. (1995). CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell Biol. 130, 507-518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., McLeod I., Anderson S., Yates J. R., 3rd, He X. (2005). Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24, 2919-2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen P., Celis J. E. (1985). S-phase patterns of cyclin (PCNA) antigen staining resemble topographical patterns of DNA synthesis. A role for cyclin in DNA replication? FEBS Lett. 193, 5-11 [DOI] [PubMed] [Google Scholar]
- McAinsh A. D., Meraldi P., Draviam V. M., Toso A., Sorger P. K. (2006). The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J. 25, 4033-4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S. E., Borusu S., Amaro A. C., Winter J. R., Belwal M., McAinsh A. D., Meraldi P. (2007). The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 26, 5033-5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., McAinsh A. D., Rheinbay E., Sorger P. K. (2006). Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7, R23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mythreye K., Bloom K. S. (2003). Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J. Cell Biol. 160, 833-843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V. S., Smith M. A., Peak-Chew S., Kilmartin J. V. (2003). Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 4931-4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., Yoda K. (2004). Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localised with the centromeric region in interphase. Genes Cells 9, 105-120 [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R., 3rd, Desai A., Fukagawa T. (2006). The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8, 446-457 [DOI] [PubMed] [Google Scholar]
- Orthaus S., Biskup C., Hoffmann B., Hoischen C., Ohndorf S., Benndorf K., Diekmann S. (2008). Assembly of the inner kinetochore proteins CENP-A and CENP-B in living human cells. Chembiochem 9, 77-92 [DOI] [PubMed] [Google Scholar]
- Orthaus S., Klement K., Happel N., Hoischen C., Diekmann S. (2009). Linker Histone H1 is present in centromeric chromatin of living human cells next to inner kinetochore proteins. Nucleic Acids Res. 37, 3391-3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast L., van Vuuren C., Kaczmarczyk A., Doering V., Hellwig D., Quinn N., Hoischen C., Diekmann S., Sullivan K. F. (2011). Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state. PLoS Biol. 9, e1001082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka M. R., Venkei Z., Bolanos-Garcia V. M., Debski J., Dadlez M., Glover D. M. (2011). CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21, 399-405 [DOI] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. (2009). The life and miracles of kinetochores. EMBO J. 28, 2511-2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedeberg L., Weisshart K., Diekmann S., Meyer Zu Hoerste G., Hemmerich P. (2004). High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol. Biol. Cell 15, 2819-2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G. M., Petrovic A., Melis T., Nogales E., Musacchio A. (2011). Direct binding of CENP-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21, 391-398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N., Black B. E. (2009). A reader for centrometric chromatin. Nat. Cell Biol. 11, 793-795 [DOI] [PubMed] [Google Scholar]
- Sekulic N., Bassett E. A., Rogers D. J., Black B. E. (2010). The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature 467, 347-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby R. D., Monier K., Sullivan K. F. (2000). Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151, 1113-1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanathan S., Suchyna T. M., Siegel A. J., Berezney R. (2001). Targeting of PCNA to sites of DNA replication in the mammalian cell nucleus. J. Cell Biochem. 81, 56-67 [DOI] [PubMed] [Google Scholar]
- Sporbert A., Domaing P., Leonhardt H., Cardoso M. C. (2005). PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res. 33, 3521-3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. U., Desai A. (2008). Kinetochore-microtubule interactions: the means to the end. Curr. Opin. Cell Biol. 20, 53-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso A., Winter J. R., Garrod A. J., Amaro A. C., Meraldi P., McAinsh A. D. (2009). Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol. 184, 365-372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidtkamp-Peters S., Rahn H. P., Cardoso M. C., Hemmerich P. (2006). Replication of centromeric heterochromatin in mouse fibroblasts takes place in early, middle, and late S phase. Histochem. Cell Biol. 125, 91-102 [DOI] [PubMed] [Google Scholar]
- Wu R., Singh P. B., Gilbert D. M. (2006). Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J. Cell Biol. 174, 185-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Feng H., Zhou B. R., Ghirlando R., Hu K., Zwolak A., Miller Jenkins L. M., Xiao H., Tjandra N., Wu C., et al. (2011). Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472, 234-237 [DOI] [PMC free article] [PubMed] [Google Scholar]