Abstract
In bacterial communities, ‘tight economic times’ are the norm. Of the many challenges bacteria face in making a living, perhaps none are more important than generating energy, maintaining redox balance, and acquiring carbon and nitrogen to synthesize primary metabolites. The ability of bacteria to meet these challenges depends heavily on the rest of their community. Indeed, the most fundamental way in which bacteria communicate is by importing the substrates for metabolism and exporting metabolic end products. As an illustration of this principle, we will travel down a carbohydrate catabolic pathway common to many species of Bacteroides, highlighting the interspecies interactions established (often inevitably) at its key steps. We also discuss the metabolic considerations in maintaining the stability of host-associated microbial communities.
Introduction
Tight economic times remind us of two important lessons: our biggest challenge is often just finding a way to make a living, and our ability to make a living depends heavily on the people around us.
These lessons are especially pertinent in bacterial communities, where ‘tight economic times’ are the norm. Of the many challenges bacteria face in making a living, perhaps none are more important than generating ATP, maintaining redox balance, and acquiring carbon and nitrogen to synthesize primary metabolites. So persistent and fundamental are these challenges that bacteria devote extraordinary effort to them, as illustrated here.
The ability of bacteria to make a living depends on the rest of their community. While much more is known about certain forms of bacterial signaling such as quorum sensing, the process by which a microbial species detects the levels of a specific metabolite to monitor its own abundance (Bassler and Losick, 2006), it is easy to forget that the most fundamental way in which bacteria communicate is by importing the substrates for metabolism and exporting its end products. If your neighbor eats starch and produces 5 mM succinate, you will probably take notice. If you can turn the succinate into butyrate and generate ATP in the process, your chances of thriving are even better. Of course, your neighbor will likely sense the decreased succinate and increased butyrate, and respond accordingly. A pair of bacterial species carrying out housekeeping metabolism is thus an important example of systems biology at work, with the proviso that the system does not end at the cell membrane.
In this review, we will travel down a carbohydrate catabolic pathway common to many species of Bacteroides, highlighting the interspecies interactions established (often inevitably) at its key steps. Primary fermenters, like the Bacteroides, are the gateway through which carbohydrates enter the network of syntrophic links (inter-species metabolic interactions) within a gut microbiota. (Fig. 1A). Carbohydrates consumed by gut microbiota are typically oligo- or polysaccharides derived from diet, host mucosal secretion, or other resident (or dietary) microbes. In the simplest manifestation, Bacteroides and other glycophagic (carbohydrate-eating) species, degrade the complex carbohydrates to their component monosaccharides, which in turn are metabolized through the sequential action of (i) one of three prototypic glycolytic pathways to yield phosphenolpyruvate (PEP), followed by (ii) conversion of PEP to fermentation end-products such as an organic acid, solvent, or alcohol. Below we discuss factors that influence the operation of these pathways and the resulting spectrum of secreted by-products and end-products which serve as the currency of syntrophy within the gut. By focusing on a these pathways, our goal is to offer illustrative examples and highlight existing questions rather than provide an exhaustive account of what is known, and we apologize in advance to our colleagues whose work has been omitted inadvertently and due to space constraints.
Figure 1. Simplified illustrative schematic of some trophic networks within the intestinal microbiota.
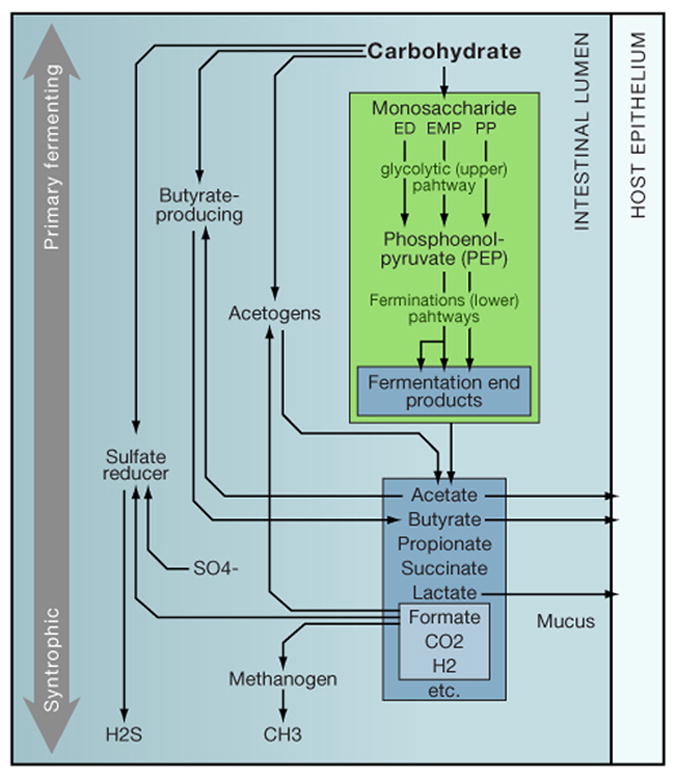
Dietary or host-derived substrates may be metabolized by different microbial groups, which are divided in this schematic by major metabolic function (e.g., acetogen). The sequential action of glycolytic and fermentation pathways (shown within the green box representing a Bacteroides cell; ED= Entner–Doudoroff pathway, EMP= Embden-Meyerhof-Parnas pathway, and PP= pentose phosphate pathway) result in fermentation end-products that become the metabolic inputs for other syntrophic microbes, such as acetogens, sulfate-reducers and butyrate-producers, or the host.
Setting the Table: Polysaccharides as fuel for syntrophy and community dynamics
Multiple bacterial taxa compete for a limited quantity of carbohydrate in the distal gut, and changes in host diet are a major driver of change in community composition and function (Table 1) (Faith et al., 2011; Martinez et al., 2010; Sonnenburg et al., 2010; Sonnenburg et al., 2005; Walker et al., 2011; Wu et al., 2011). Given the variety of fermentation end products generated by different members of the microbiota, changes in the relative level of each species -- and their success in acquiring polysaccharides -- have an important effect on the final mixture of fermentation end products. Whether the inulin from an onion is converted into butyrate, propionate, lactate, or acetate can greatly influence subsequent metabolic transformations and help determine which syntrophs are likely to flourish. Differences in the ratio of fermentation products can also modulate diverse host pathways and steer caloric benefit toward specific host tissues.
Table 1. Nutrient sources and requirements for Bacteroides and Clostridium.
Typical nutrient sources and requirements for Bacteroides and host-associated Clostridium species are shown. Note that nutrient sources and requirements can vary considerably among species, so the ones shown here should be interpreted as a general guideline.
| Bacteroides | Clostridium | |
|---|---|---|
| Carbon | saccharides1 | amino acids6, saccharides2,3 SCFAs4 |
| Nitrogen | NH31 | amino acids2,3 |
| Other Requirements | CO2, B12, hemin, cysteine or sulfide, menaquinone1 | pantothenate, pyridoxine, (biotin)2,3 |
| Major Excreted Products | acetate, propionate, H2, succinate1 | acetate, butyrate, H2, alcohols5 |
Palette breadth and community stature
Not all primary fermenters are equally capable of consuming a given carbohydrate substrate. Even closely related Bacteroides species exhibit significant differences in their ability to use glycans common to the gut such as mucin, xylan, and fructans (Salyers et al., 1977a; Sonnenburg et al., 2010). Differences in substrate utilization are meaningful in vivo, and can result in predictable shifts in population abundance due to the presence of a given substrate within the diet (Kolida et al., 2007; Ramirez-Farias et al., 2008; Sonnenburg et al., 2010). The intentional enrichment of an oligosaccharide (‘prebiotic’) within the diet of humans has been used as a strategy to selectively expand subsets of the resident microbiota; for example, the expansion of Bifidobacterium species has been attempted through the dietary supplementation of inulin, a fructan (Kolida et al., 2007). However, prebiotic supplementation can lead to unexpected changes in the microbiota composition—perhaps not surprising considering the variability in microbiota among individuals, a dearth of functional information about the relative efficiencies with which gut taxa catabolize specific prebiotics, and the tendency of single-node perturbations to ripple through the network of a community.
Since differences in community composition can alter the function of a resident species, the apparent uniqueness of an individual’s microbiota poses an interesting challenge for understanding species (and ultimately community) function. Despite the conservation in broad functional categories represented in human intestinal metagenomes (Turnbaugh et al., 2009), three factors will make inter-individual differences important: 1) The levels of key genes and pathways can have a profound impact on the host and microbiota; for example, a single toxin-encoding gene can elicit a potent response from the host. 2) While broad functional categories may be present at similar levels across individuals, these functions will partition in distinct ways among taxa, which will in turn influence how the levels of individual species respond to a perturbation. 3) At a higher level of resolution, broad categories splinter into functions that are polymorphic among individuals. The presence of seaweed-oligosaccharide-degrading porphyranases in the microbiota of Japanese individuals illustrates how averaging over a single category (in this case, ‘carbohydrate transport and metabolism’) obscures functionally meaningful differences among individuals (Hehemann et al., 2010).
What you eat might depend on your company
Even in cases where two communities harbor the same bacterial strain, the functions the bacteria carry out in individual settings may differ greatly, depending on the presence or absence of other community members. In other words, the same set of genes may carry out distinct functions depending upon community context. Much insight has been provided into functional shifts induced by microbiota membership alteration by studying model microbial communities that isolate and compare one- and two-species communities in vivo and in vitro. The study of two primary fermenters of different phyla, Bacteroides thetaiotaomicron and Bifidobacterium longum, living alone or together within the gnotobiotic mouse intestine has demonstrated that these two prodigious consumers of polysaccharides undergo niche partitioning. Both strains upregulate putative xylanases while reciprocally regulating putative mannosidases, suggesting that molecular correlates of synergism and competition, respectively, occur simultaneously in vivo between these two species (Sonnenburg et al., 2006).
Another study in which B. thetaiotaomicron was paired with a prominent representative of the Firmicutes phylum, Eubacterium rectale, similarly demonstrates niche adjustments by the two species (Mahowald et al., 2009). In the presence of B. thetaiotaomicron, E. rectale shifts away from complex glycan use and increases import of simple carbohydrates and amino acids. However, E. rectale impacts the strategy of B. thetaiotaomicron in a manner similar to the impact exerted by B. longum described above --B. thetaiotaomicron expands the repertoire of glycoside hydrolases expressed when bi-associated with either microbe indicating that it exploits new carbohydrate niches in the presence of other primary fermenters. The predicted classes of carbohydrates targeted during B. thetaiotaomicron niche adjustments were distinct when paired with B. longum or E. rectale, but in both cases B. thetaiotaomicron exhibits an expansion in the number and type of glycoside hydrolases that are highly expressed (Mahowald et al., 2009), suggesting that each of these two co-habitating species increases the diversity of glycan substrates that B. thetaiotaomicron consumes.
In contrast, Marvinbryantia formatexigens, an acetogen with extensive primary fermentation capabilities including >100 glycoside hydrolases, only marginally impacts the regulation of B. thetaiotaomicron’s extensive polysaccharide utilization machinery (Rey et al., 2010). The presence of Methanobrevibacter smithii, the prominent gut archeal methanogen, which is poorly equipped to degrade carbohydrates but consumes fermentation end products, decreases the diversity of carbohydrate substrates that B. thetaiotaomicron accesses—this methanogen-induced niche contraction contrasts sharply to B. thetaiotaomicron’s bi-associations with B. longum and E. rectale (Samuel and Gordon, 2006). Together, these studies reveal how the specifics of community membership, the represented functionalities, the carbohydrate inputs, and the nature of metabolic links between species are critical in dictating the functional output of a given set of genes (in this case, the genes encoded within the B. thetaiotaomicron genome).
Polysaccharides to phosphoenolpyruvate (PEP) and beyond: Which pathways to use?
The chief metabolic feat of Bacteroides, and many other primary fermenting members of the gut microbiota, is the conversion of carbohydrates into a variety of organic acids and alcohols. Once a monosaccharide is imported into a bacterial cell, it may flow through one of multiple glycolytic pathways including the Embden-Meyerhof-Parnas pathway (EMP), the pentose phosphate pathway (PP) or the Entner-Doudoroff pathway (ED). Multiple factors govern which route of glycolytic transformation is chosen by the bacterial cell: (i) the carbohydrate substrate being metabolized, e.g., consumption of pentoses may bias a bacterium toward PP due to the ease of shunting a 5-carbon sugar into this pathway; (ii) production of metabolic precursors for biosynthetic reactions, e.g., PP produces ribose-5-phosphate and erythrose-4-phosphate, which are necessary for nucleic acid and aromatic amino acid biosynthesis, respectively; (iii) quantity of ATP produced via substrate-level phosphorylation; and (iv) NAD+ consumed by the pathway and the accompanying consideration of redox balance. While most of these factors have been determined or inferred by experiments based on cells grown in pure culture, pathway choice is undoubtedly tied to the community in which a microbe finds itself, and how neighbors directly or indirectly influence these factors. Importantly, all three glycolytic pathways generate PEP, which serves as the common precursor for production of organic acids and alcohols that result from fermentation and anaerobic respiration. Since the “lower” pathways (fermentation and anaerobic respiration) differentially influence multiple physiological aspects of the cell, such as energy production and redox balance, bacterial choice/regulation of upper (glycolytic) and lower pathways are tightly linked.
Phosphoenolpyruvate (PEP) to oxaloacetate: CO2 fixation in the gut
With plenty of mucus to adhere to, a well-regulated temperature, and a warm meal every few hours, the gut would seem like a great place to live. But there is a catch: the gut has no O2 to support aerobic respiration, nor are there appreciable levels of other terminal electron acceptors such as nitrate and sulfate (although low levels of these compounds do not necessarily imply their absence; they might be rapidly reduced by anaerobic respiration and therefore difficult to detect) (Levine et al., 1998; Saul et al., 1981). Since respiration is generally much more efficient than fermentation, bacteria often innovate to enable respiration. Bacteroides is no exception; it takes advantage of the high levels of CO2 -- a peculiarity of the gut over other anaerobic environments -- to set up a primitive electron transport chain based on the reduction of fumarate to succinate (detailed in the next section) (Table 1).
If fumarate can be respired, how would a bacterium with a glut of PEP (three carbons) convert it to something that resembles fumarate (four carbons)? The ideal approach would be to pull the fourth carbon right out of the air. Bacteroides does just that, employing exactly the same carbon-fixing reaction used by C4 plants: the fusion of CO2 and PEP to form oxaloacetate (Fig. 2C). In addition to capturing CO2, this reaction allows Bacteroides to generate one equivalent of ATP or GTP from the high-energy PEP phosphate (Macy et al., 1978; Macy and Probst, 1979).
Figure 2. A carbohydrate catabolic pathway from Bacteroides.
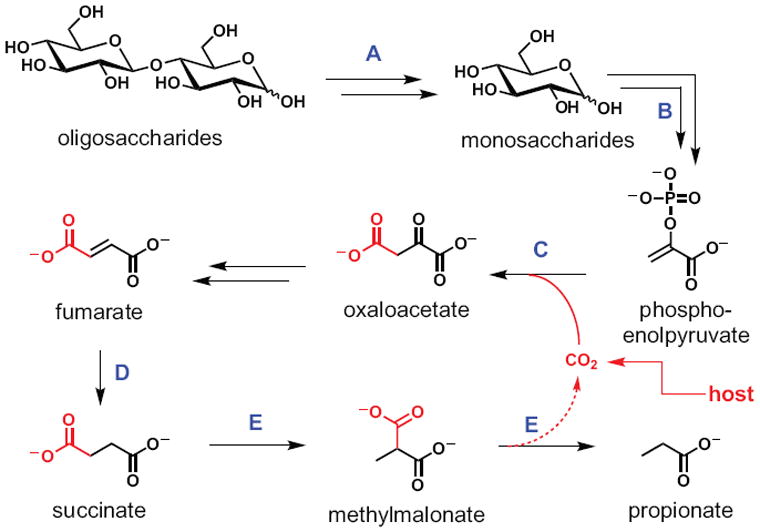
This pathway serves as a template for our review. (A) Oligosaccharide foraging and hydrolysis to monosaccharides. (B) The Embden-Meyerhof-Parnas and Entner Doudoroff pathways convert process monosaccharides to PEP, generating ATP and NADH. (C) PEP carboxykinase fixes CO2 by appending it to PEP, generating oxaloacetate in a reaction that generates one equivalent of ATP or GTP. (D) Fumarate serves as the terminal electron acceptor in a primitive Bacteroides electron transport chain, enabling anaerobic respiration. (E) Methylmalonyl-CoA mutase isomerizes succinate to methylmalonate, which is then decarboxylated to regenerate CO2 and produce propionate as an end product. Double arrows represent multiple steps in a pathway.
Competition for CO2?
Bacteroides is not the only gut species that utilizes CO2; several of its counterparts have a similar requirement, suggesting that CO2 is likely to be in demand in the gut lumen (Dehority, 1971). For instance, Eubacterium rectale upregulates PEP carboxykinase in the presence of B. thetaiotaomicron, potentially reflecting a competition for CO2 (Mahowald et al., 2009). In addition, the level of propionate in a E. rectale/B. thetaiotaomicron biassociated mouse is lower than in a B. thetaiotaomicron monoassociated mouse, supporting a model in which E. rectale depletes CO2, nudging B. thetaiotaomicron toward producing acetate instead of propionate. Interestingly, this might provide further benefit to E. rectale, which can generate ATP by converting acetate to butyrate. Acetogens that employ the Wood-Ljungdahl pathway which consumes CO2, such as Blautia hydrogenotrophica, may be similarly engaged in competition with Bacteroides species. Bi-association of gnotobiotic mice with Bacteroides thetaiotaomicron and B. hydrogenotrophica revealed decreased succinate generation consistent with less CO2 availability for Bacteroides anaerobic respiration (Rey et al., 2010).
Where does the CO2 come from? Much of it likely comes from the host; an average human expels 0.68 kg of CO2 per day (el-Khoury et al., 1994). Most of the CO2 in host circulation is in the form of bicarbonate (HCO3-) and host transporters secrete bicarbonate into the gut lumen (Hopfer and Liedtke, 1987). An important challenge will be to understand how the flow of CO2 is regulated. On the host side, how is the CO2 spigot controlled, and do CO2 gradients exist in the lumen that help determine community geography? On the bacterial side, how is CO2 imported and sequestered, and how does the competition for CO2 modulate community dynamics?
Fumarate to succinate: A primitive electron transport chain
Electron transport chains give organisms a great bang for their buck. Depending on their configuration, these systems -- in which electrons from a donor such as NADH are made to do the work of generating a proton gradient by passing through a bucket brigade of electron carriers -- can yield far more ATP per mole glucose than substrate-level fermentation. An electron transport chain works something like a battery; the anode is the electron source (e.g., NADH) and the cathode is the electron sink (e.g., O2) -- also known as the terminal electron acceptor. The ‘voltage’ of an electron transport chain can be calculated by subtracting the reduction potential of the source reaction (NADH to NAD+, E0’ = -0.32 V) from that of the sink reaction (O2 to H2O, E0’ = +0.82 V). The larger the difference, the more work each electron can do, which translates into more moles of ATP synthesized per mole glucose consumed.
O2 is a better terminal electron acceptor than anything else commonly found in biology; the unbeatable reduction potential for its conversion to water maximizes the proton-gradient-generating work performed by each electron. In the absence of O2, some microbes can use NO3-, SO42-, or Fe3+ as alternative terminal electron acceptors.
A DIY approach to a terminal electron acceptor
What if the organism does not have access to a good terminal electron acceptor? Bacteroides uses what it has in abundance -- sugars and CO2 -- to fashion its own electron acceptor. By reductively carboxylating PEP (see previous section), Bacteroides generates the C4 metabolite oxaloacetate. Using a pathway that runs in the reverse direction of the widely known citric acid cycle, oxaloacetate is then reduced to malate and dehydrated to fumarate (Macy et al., 1978) (Fig. 2D).
Fumarate is the key here; its reduction to succinate is the final step in a primitive electron transport chain that uses NADH as its major electron donor (Fig. 3). While the Bacteroides pathway is abbreviated, it has the trappings of a typical electron transport chain: a series of electron handoffs that involve membrane-bound cytochromes and quinones, which helps explain the ability of hemin and menaquinone to influence Bacteroides growth and fermentation end-product profile (Chen and Wolin, 1981; Robins et al., 1973). The resulting proton gradient helps drive the synthesis of ATP from ADP by ATP synthase. Indeed, fumarate is the most common terminal electron acceptor for anaerobic respiration (Kroger et al., 1992).
Figure 3. Bacteroides’ primitive electron transport chain.
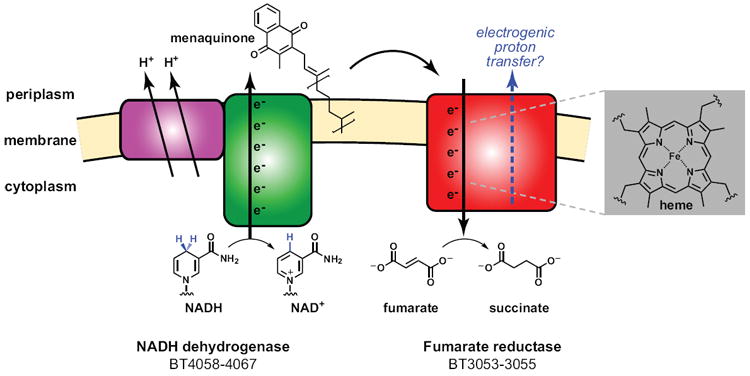
A proposed model for the anaerobic electron transport chain common to many Bacteroides species is shown; this model is based on crystal structures of NADH dehydrogenase and fumarate reductase from other bacteria (Efremov et al., 2010; Lancaster et al., 1999). The chain would consist of two enzyme complexes: a proton-translocating NADH-quinone oxidoreductase (NADH dehydrogenase, left) and a quinol:fumarate reductase (fumarate reductase, right). The action of NADH dehydrogenase would shuttle electrons from the oxidation of NADH to NAD+ down a pathway involving a flavin cofactor and multiple iron-sulfur clusters, ultimately reducing a membrane-bound menaquinone. This menaquinone would carry the electrons to fumarate reductase, which would shuttle them down a pathway involving one or two heme cofactors, possibly a second quinone, multiple iron-sulfur clusters, and a flavin cofactor that would reduce the terminal electron acceptor fumarate to succinate. ‘BT-numerical’ labels are the locus tags for genes within the B. thetatiotaomicron (VPI-5482) genome.
Why go to the trouble of fixing CO2 to make fumarate? Instead, why not build an electron transport chain in which the abundant intermediate pyruvate is the terminal electron acceptor? One reason might be that fumarate/succinate (E0’ = +0.03 V) is a better cathode than pyruvate/lactate (E0’ = -0.19 V). An electron transport chain based on fumarate therefore runs more efficiently, synthesizing a greater quantity of ATP per mole glucose consumed. While it is not nearly as efficient as aerobic respiration, it is still quite impressive for an anaerobe.
Succinate to propionate: Waste not, want not
When CO2 is plentiful, Bacteroides does not have much use for the succinate generated by its electron transport chain, so this reduced C4 diacid is excreted as a major metabolic end product (and, in turn, it becomes the input metabolite for secondary fermenters -- see below). When CO2 is limiting, however, Bacteroides has a clever way of regenerating CO2 from succinate (Fig. 2E).
Some bacteria harbor a pathway for degrading branched-chain amino acids that converges on succinate; the last step in this pathway is catalyzed by the vitamin-B12-dependent enzyme methylmalonyl-CoA mutase, which converts the branched C4 diacid methylmalonate to its linear isomer succinate. Taking advantage of the fact that this reaction is reversible, Bacteroides runs it in the opposite direction, using methylmalonyl-CoA mutase to convert succinate to methylmalonate (Fig. 2E). Although methylmalonate is a structural isomer of succinate, the two compounds differ in an important way: unlike succinate, methylmalonate is easy to decarboxylate. Eager to increase the size of its CO2 pool, Bacteroides decarboxylates methylmalonate, regenerating CO2 and yielding propionate as the new end product. Illustrating this point, above 25.5 mM CO2, the growth rate of B. fragilis does not increase by adding additional CO2, but below 10 mM, a marked decrease in growth rate was observed. At limiting CO2 concentrations, the ratio of propionate to succinate increases, as does the specific activity of the CO2-fixing enzyme PEP carboxykinase, indicating its upregulation to increase the capture of CO2 (Caspari and Macy, 1983).
Thus, the ambient CO2 concentration appears to modulate a metabolic switch between the production of succinate and propionate. Availability of vitamin B12, the cofactor for methylmalonyl-CoA mutase, to the auxotrophic Bacteroides, who appear to derive B12 or B12-precursors from other prototrophic community members such as certain Firmicutes, may represent another important factor in modulating the Bacteroides conversion of succinate to propionate (Goodman et al., 2009). Major questions: How is the regenerated CO2 sequestered and ‘delivered’ to PEP carboxylase? And, as a bridge to the next section: How does Bacteroides sense low CO2 levels and switch to one of its fermentation pathways?
How microbes choose which end products to produce…
The choice between succinate and propionate is one example of a larger question: Given that many gut microbes can choose from a menu of possible metabolic end products, how do they make a decision?
One way to think about it is that every end product has advantages and disadvantages that can be summarized with a metabolic ‘scorecard’ (Fig. 4). Although there are other important considerations, for the sake of simplicity we will look at four: redox balance (how many equivalents of NADH are converted to NAD+); energy production (how many equivalents of ATP are synthesized); acid production; and H2 production.
Figure 4. A metabolic scorecard.
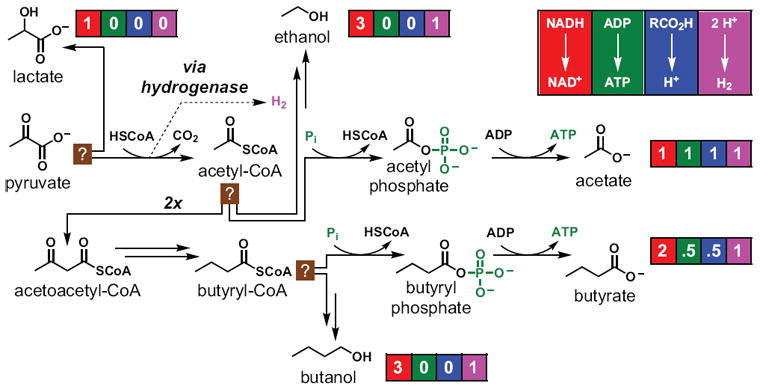
The choice of a lower pathway for the metabolism of pyruvate by Clostridium pasteuranium is shown. Lactate, ethanol, acetate, butyrate, and butanol are the major metabolic end products; next to each one is a ‘scorecard’ showing, from left to right: equivalents of NADH reduced to NAD+; equivalents of ADP converted to ATP by substrate-level phosphorylation; equivalents of acid produced; and equivalents of H2 produced (all figures are in units of per mole pyruvate). Double arrows represent multiple steps in a pathway; see text for details.
Solving an optimization problem
A study on the metabolism of Clostridium pasteurianum provides an instructive example of how microbes sense their environment and adjust their metabolic end product accordingly (Fig. 4) (Dabrock et al., 1992). C. pasteurianum ferments glucose to a mixture of acetate and butyrate under normal growth conditions. Starting from pyruvate, the production of acetate results in the conversion of 1 NADH to NAD+, 1 ADP to ATP, and 1 equivalent of acid. Butyrate production has a slightly different outcome: the bad news is ATP production is only half as efficient, but the good news is that only half the acid is produced and an additional equivalent of NADH is oxidized to NAD+. By modulating the ratio of acetate to butyrate, C. pasteurianum is presumably optimizing the number of ATP equivalents it can synthesize, consistent with maintaining redox balance (complete conversion of NADH to NAD+) and avoiding too low a pH (too much acid).
Importantly, a strain that specializes in gut metabolism will often have a better answer to the optimization problem than a generalist. For example, under anaerobic conditions, Bacteroides fragilis produces 4.5 moles of ATP per mole glucose, compared to 2.5 for E. coli (Baughn and Malamy, 2003). This 80% advantage in efficiency would give B. fragilis a strong edge in a competitive setting.
As noted earlier, a bacterium’s choice in the upper pathway is intimately linked with its choice in the lower pathway. When C. pasteruanium’s carbon source changes from glucose to glycerol, C. pasteuranium responds by producing a mixture of ethanol and butanol (Fig. 4) (Dabrock et al., 1992). Conversion of pyruvate to ethanol or butanol has the distinct disadvantage of yielding zero ATP (but since 2 equivalents of ATP are generated by converting glycerol to pyruvate in the new upper pathway, all is not lost). The advantage of butanol and ethanol is the opportunity to restore 3 equivalents of NADH to NAD+, which is important because an additional NAD+ is consumed during the first step of the new upper pathway of glycerol fermentation. Indeed, C. pasteuranium is willing to forgo ATP altogether to maintain redox balance: it produces the end product 1,3-propanediol directly from glycerol using an alternative upper pathway (not shown), yielding zero ATP but reducing one NADH to NAD+ to help maintain redox balance (Dabrock et al., 1992). Of course, neighbors of C. pasteuranium would experience a radically different environment following its switch to glycerol fermentation since acid levels would drop and solvent levels would rise.
The environment can influence decision making and the resulting metabolic end-products
Environmental influences other than a change in carbon source can have an equally important effect. When the hydrogenase inhibitor carbon monoxide is introduced, pyruvate is reduced directly to lactate (Fig. 4). This yields zero ATP and regenerates only one NADH, but importantly it circumvents the requirement for hydrogenase. Similar results are seen under iron limitation, likely due to a blockade of pyruvate-ferredoxin oxidoreductase, which harbors an Fe-S cluster (Dabrock et al., 1992). Importantly, similar conditions can arise through interspecies interactions: for example, competition for iron often leads to a limiting concentration, which could impact hydrogenase activity and elicit a change in fermentation end product.
…and how those end products influence interspecies interactions
Molecular hydrogen (H2) is excreted by many primary fermenters and serves as a sink for electrons in an anaerobic environment that lacks many external electron acceptors (the same is true for other electron-rich metabolic end products such as formate). If H2 accumulates within the system, the ability of primary fermenters to regenerate NAD+ from NADH may be inhibited, slowing fermentation and growth and resulting in reduced bacterial density (Miller and Wolin, 1982). H2-consuming microbes such as acetogens, methanogens, and sulfate-reducing bacteria therefore play an important role in maintaining the metabolic efficiency of primary fermenters. Indeed, H2 is an important mediator of interspecies interactions. In addition to the interspecies dynamics established by H2 commerce, consumption of H2 is accompanied by the depletion and production of organic acids and other metabolites (e.g., H2S). Depending upon which taxa are responsible for H2 consumption, diverse influences on host biology, from calorie harvest in the case of acetate production (acetogenesis) to genotoxic effects in the case of H2S, may result (Huycke and Gaskins, 2004).
Methanogens consume H2, providing a valuable service to primary fermenters. Similarly, hydrogenotrophic acetogens like Blautia hydrogenotrophica deplete H2, but convert it to acetate instead of the comparatively inert methane. Acetate can further potentiate the primary fermenting (and acetate consuming) butyrate producers such as Roseburia intestinalis (Barcenilla et al., 2000; Chassard and Bernalier-Donadille, 2006; Duncan et al., 2004). So while methanogens serve as a faithful trash collection service for primary fermenters, acetogens will pick up the trash, convert it to compost, and return a valuable metabolic commodity.
Hydrogen-fueled interactions with B. hydrogenotrophica
B. hydrogenotrophica has been studied in detail in a series of in vivo experiments that illustrate its ability to transform the metabolic milleu within the intestine (Rey et al., 2010). B. hydrogenotrophica encodes all of the enzymes required for the Wood-Ljungdahl reductive acetogenesis cycle, which converts CO2 and H2 into acetate. The paucity of glycoside hydrolases within its genome suggests that its niche within the microbiota is a syntrophy with primary fermenting bacteria.
Indeed, when paired with the glycan degrading and fermenting B. thetaiotaomicron in bi-association within the intestines of gnotobiotic mice, B. hydrogenotrophica density increases 1000-fold compared to colonization in the absence of the primary fermenter (Rey et al., 2010). In the presence of B. thetaiotaomicron, B. hydrogenotrophica expresses the H2-consuming pathway and is able to increase the NAD+/NADH ratio within the intestinal contents, suggesting a transformation of the ecosystem that is favorable for fermentation. Consistent with B. hydrogenotrophica-potentiated fermentation is the increased depletion of glucans from the intestinal contents and increased concentration of acetate in the lumen and host serum (Rey et al., 2010). These changes, combined with the reduced levels of succinate, provides a nice demonstration of how a syntrophic bacterium can promote the conversion of substrates that cannot be harvested by the host into a lipogenic short-chain fatty acid (indirectly: glucans via B. thetaiotaomicron fermentation; directly: H2 and CO2). Notably, the nature of acetogenesis is important since a comparison with Marvinbryantia formatexigens, an acetogen that does not consume H2 but is adept at primary fermentation, did not alter the NAD+/NADH ratio or acetate level, but increased the levels of succinate, and decreased the density of B. thetaiotaomicron.
Impact on an outsider’s odds of ‘invading’ the gut microbiota
Several lines of evidence support the importance of the microbiota’s production of fermentation end products, such as short chain fatty acids, in dictating the favorability of the environment for other species. Two independent human trials conducted in Bangladesh and Venezuela, respectively, have recently found that feeding children a diet enriched in cooked green bananas can significantly hasten recovery from shigellosis, a diarrheal disease caused by the bacterium Shigella (Alvarez-Acosta et al., 2009; Rabbani et al., 2009). Although the exact mechanistic basis for this effect has not been established, the observed increase in short-chain fatty acid production by the distal microbiota that occurs upon enrichment of dietary starch has been proposed to result in an intestinal environment unfavorable to Shigella. These studies suggest the tantalizing prospect of altering diet to influence microbiota production of a metabolite that, in turn, can eradicate a pathogen.
The ability of exotic species to ‘invade’ a microbiota can also be influenced by the identity and functionality that exists within a microbiota. Stetcher and colleagues have used a gnotobiotic mouse model to illustrate that the ability of either a Lactobacillus species or a Salmonella species to hold a niche within a microbiota depends upon the residents they encounter (Stecher et al., 2010). Both invading species were more likely to take up residence in the microbiota if species closely related to the invading organism were already well represented. These data suggest that an invading species’ odds within a microbiota may depend upon whether the necessary metabolic links are already in place. A corollary to this idea is that microbes capable of flexibility in syntrophic relationships (e.g., generalists) may have a better chance of invading a foreign environment. Further exploration of how syntrophic networks (and metabolites) are established within a microbiota may contribute to understanding why probiotic bacteria, which often have a short residence time in the gut microbiota, sometimes persist within a small fraction of study participants.
How about nitrogen? Acrobatics with amino acids
Bacteroides clearly specializes in carbohydrate utilization. Its appetite for sugar explains from where it gets the carbon it needs to synthesize primary metabolites, but it leaves an interesting question: where does Bacteroides get its nitrogen?
Bacteroides’ nitrogen source: The host, and possibly other bacteria
Acquisition of nitrogen by intestinal bacteria remains an important and open question. Bacteroides and many other gut taxa are capable of fixing NH3, primarily via glutamate dehydrogenase (Yamamoto et al., 1984), which suggests that NH3 may be the most common source of nitrogen in the intestine (Table 1). Although this view is speculative and likely will not apply for all species in the genus, it is supported by the inability of many Bacteroides species to substitute free amino acids, peptides, nitrate, or urea for ammonia as a nitrogen source (Varel and Bryant, 1974). The source of ammonia has been debated, but host secretion of urea into the intestine followed by bacterial-urease-mediated breakdown is well supported: (i) treating rodents or humans with antibiotics results in the accumulation of urea in the feces (Dintzis and Hastings, 1953; Wilson et al., 1968); (ii) germ-free rats exhibit a 100-fold decrease in the degradation of isotopically labeled urea, whether it is administered intragastrically or subcutaneously, compared to conventionally reared controls (Levenson et al., 1959). Human studies suggest that 25-45% of the daily urea pool of humans (~7 g) is degraded in the intestine, where bacteria expressing ureases are well represented (Walser and Bodenlos, 1959; Wrong, 1978).
A major question is how Bacteroides species manage to mono-associate germ-free mice so well in the absence of urease-expressing strains. Four other sources might contribute to Bacteroides’ nitrogen supply. First, the host could supply nitrogen through amino sugars and proteins that are present in secreted mucus and epithelial cells, as B. fragilis is capable of using porcine gastric mucin as its sole carbon and nitrogen source (Macfarlane and Gibson, 1991); however, the quantity of liberated nitrogen would have to be quite large to satisfy the demand. Second, amino acids derived from diet are also present in the colon, although Bacteroides cannot use them as its sole nitrogen source (see above) and the bulk of dietary amino acids are absorbed in the small intestine, independent of microbiota presence (Whitt and Demoss, 1975). Third, NH3 is excreted by other gut taxa including amino-acid-metabolizing Firmicutes (see below). While this might establish an important syntrophy under normal circumstances, it does not explain how Bacteroides acquires nitrogen during mono-association. Fourth, the host could conceivably transport NH3 (or NH4+) directly into the gut lumen; this would represent a new way to regulate the growth of NH3-dependent strains in the microbiota. Overall, this should be a rich area to explore, and previously generated transcriptional profiling data from monoassociation experiments might yield clues about candidate nitrogen assimilation genes.
Turning CO2 and NH3 into amino acids
If the major source of nitrogen for Bacteroides is free ammonia, then where do its amino acids come from? Some might come directly from peptides that reach the colon, although Firmicutes adapted to ferment peptides might present fierce competition. Surprisingly, many come from a two-step process in which Bacteroides transforms a group of end products it makes in abundance -- short-chain fatty acids -- into amino acids (Allison, 1969; Allison et al., 1984) (Fig. 5A). The first step is to reductively carboxylate the short-chain fatty acid using a thiamine-pyrophosphate-dependent enzyme in the pyruvate ferredoxin oxidoreductase family, creating an alpha-ketoacid. Second, this alpha-ketoacid is transaminated by NH3, generating an amino acid. The net conversion yields an amino acid in which the ‘amino’ and the ‘acid’ come from two important host waste products: NH3 and CO2.
Figure 5. Amino acid acrobatics.
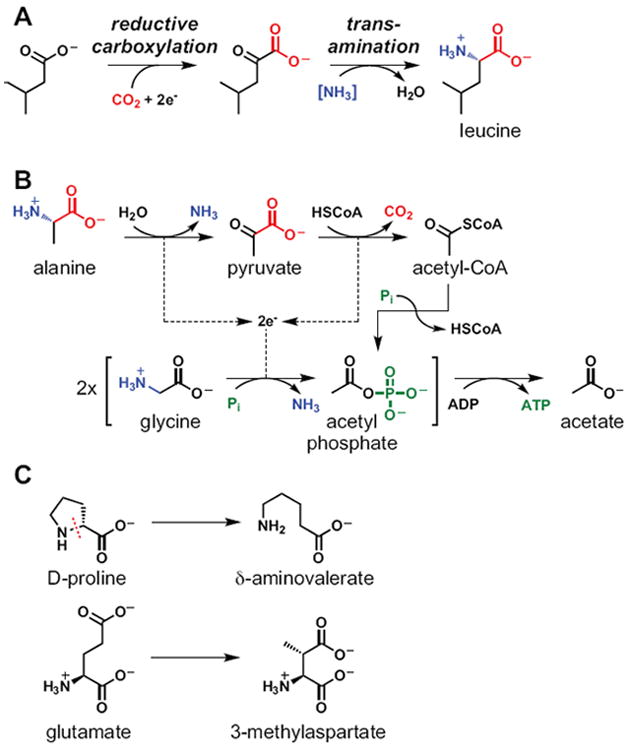
(A) Bacteroides species can synthesize certain amino acids from short-chain fatty acids by a two step process: an initial reductive carboxylation to generate an alpha-ketoacid extended by one carbon, and its subsequent transamination to an amino acid. The amino group of the resulting amino acid derives originally from host NH3 (possibly via urea), while the carboxylic acid comes from host CO2. The bracketed NH3 indicates that the proximal nitrogen donor for transamination is unknown. (B) The Stickland reaction, in which two amino acids are co-fermented, with one serving as electron donor and the other as electron acceptor. (C) Unusual fragmentations and rearrangements of amino acids, which allow certain Clostridium species to use amino acids as electron donors and acceptors.
Firmicutes: 101 ways to use amino acids
In contrast to the Bacteroidetes, a group that has been well documented for its ability to use peptides and amino acids is the Firmicutes, including species of Clostridium. Clostridium species harbor a staggering array of enzymes that catalyze exotic reactions in the service of fermenting peptide-derived amino acids. While a comprehensive review of these enzymes is beyond the scope of this perspective, we will highlight two representative examples: the Stickland reaction (Fig. 5B), in which two amino acids are co-fermented, with one used as an electron donor and the other as an electron acceptor; and fragmentations or rearrangements of amino acids (Fig. 5C), which allow Clostridium species to use amino acids as an electron source or sink (Barker, 1981; Buckel and Golding, 2006; Lovitt et al., 1986; Mead, 1971; Whitehead et al., 2008).
A toy model for the flow of carbon and nitrogen in the gut
Species of Bacteroidetes and Firmicutes, the two dominant gut phyla, seem to have distinct metabolic specialties: the former are glycan foragers while the latter harvest amino acids from peptides (Table 1). Based on this dichotomy, we propose a simplistic model for the flow of carbon and nitrogen in the gut between these two species (Fig. 6A). In our model, dietary oligosaccharides are harvested by Bacteroides, while dietary peptides are taken up by Clostridium (representing the Firmicutes). Amino acid fermentation by Clostridium generates NH4+ for Bacteroides, while the hydrolysis of oligosaccharide-coated food particles liberates additional peptides for Clostridium to ferment. Both Bacteroides and Clostridium generate short-chain fatty acids that are taken up by host enterocytes, and the host provides CO2 to support Bacteroides anaerobic respiration. Needless to say, this model is overly simplistic and potentially flawed—many species of Firmicutes are adept at polysaccharide use (Salyers et al., 1977b; Scott et al., 2011), and abundance of Bacteroides species in humans has recently been correlated with a high fat, high protein diet (Wu et al., 2011). Nevertheless, models such as this one will be an important starting point for experimentation; they generate hypotheses about syntrophies, competitions, and feedback loops that can be tested using mock communities of two or more bacterial species in germ-free mice.
Figure 6. Interspecies interactions in metabolic networks.
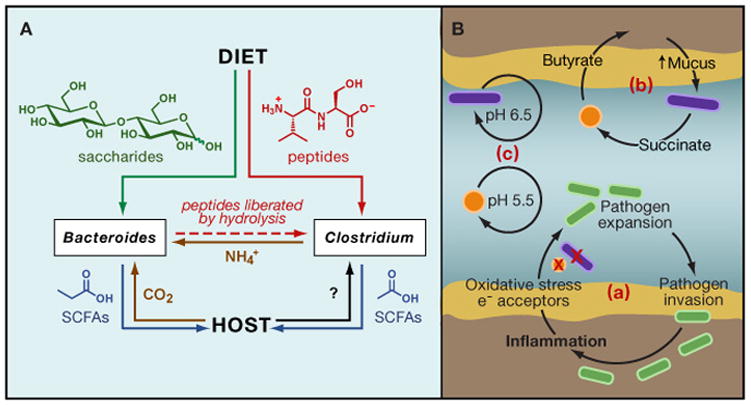
(A) A toy model for carbon and nitrogen flows in the gut. Bacteroides’ ability to access protein-encapsulated plant carbohydrates via proteases may provide a source of amino acids for other genera such as Clostridium, which, in turn, provide a pool of free nitrogen (NH4+) derived from amino acid deamination. The provision of short-chain fatty acids to the host is reciprocated by CO2 excretion among other, to be defined metabolites. (B) Positive feedback loops in the gut (lumen in blue and mucus in light brown). Although the intestine is a dynamic environment, continually being perfused with new dietary components and microbial species, the relative temporal stability of microbiota composition suggests that communities may tend toward self-reinforcing configurations. These may be simple feedback loops in which a species’ metabolism creates a favorable local chemical milieu (e.g., optimal pH) for that species (c). Alternatively, multiple species may act syntrophically to reinforce advantageous metabolism of one another, and in some cases this may involve host responses. For example, the fermentation of host mucus to succinate by one species (represented in purple) may be accompanied by another microbe (represented in orange) converting succinate to butyrate, which in turn can be taken up by the host and lead to increased mucus production (b). Alternatively, a pathogen (green rods) may induce inflammation, which leads to host production of oxidative effectors that have the dual effect of killing mutualistic, pathogen-excluding anaerobes (represented as orange circles and purple rods) and providing terminal electron acceptors to the facultative pathogen. Both effects lead to additional pathogen expansion, pathogen invasion and more inflammation (a).
Maintaining Community Stability: Many solutions to the same problem
Many recent studies that define the bacterial taxa within human microbiota demonstrate significant variability between individuals. One of the most impressive differences yet reported in a human microbiota comes from a study of children living in rural Western Africa, whose Bacteroidetes dominated fecal microbiota is populated by Xylanibacter and Prevotella spp., rather than the typical Bacteroides spp. seen in most Westerners (De Filippo et al., 2010). Additional analysis of the microbiota of Europeans and Americans suggests that while differences are not as profound as the comparison with Africa, many differences are evident. Strikingly, these differences do not amount to a continuum of variability, but rather separate into three distinct “enterotypes”, each of which harbors a distinct composition signature (Arumugam et al., 2011). These compositions likely reflect relatively stable alternative “solutions” to formation of an intestinal ecosystem.
Two properties that are used to characterize the intestinal microbiota are resistance and resilience, the ability to defy change and the ability to return to an initial state after a perturbation, respectively. The remarkable stability of the human microbiota in adult healthy humans over time suggests the intestinal ecosystem is well endowed with both properties. However, there are clear limits to the microbiota’s stability, as oral antibiotic treatment in humans and in mice results in a sudden and marked alteration in community composition and density (Dethlefsen et al., 2008; Dethlefsen and Relman, 2010; Stecher et al., 2007). Tracking the microbiota’s recovery after cessation of two sequential treatments of ciprofloxacin separated by six months in three humans revealed individual variation in the rate and extent of microbiota recovery. The fecal communities of all three individuals stabilized after treatment, but were compositionally distinct from pre-antibiotic treatment (Dethlefsen and Relman, 2010). These results demonstrate that in the absence of a harsh perturbation like antibiotic treatment, an intestinal microbial community fluctuates around a stable ecological state. An extreme perturbation appears to be required to shift the community to a new, distinct stable state over a short time period. Similarly, short-term (10-day) dietary change rapidly impacts microbiota composition, but the change is not sufficient to shift an individual’s microbiota to a new enterotype (Wu et al., 2011). An intriguing, unproven alternative would be that milder perturbations, if focused on key nodes in the community or the community’s metabolic network, could effect a ‘surgical’ shift in community composition.
Positive feedback loops and gut community stability
The molecular underpinnings of gut ecosystem stability remain poorly defined, but the specific syntrophic links within a community -- and the resulting positive feedback loops -- are likely to help enforce resistance and resilience. One of the best understood examples of a microbe establishing a positive feedback loop to enhance its own stability within the ecosystem comes from studying intestinal pathogenesis. Orally acquired bacterial pathogens are often unable to colonize at high densities or invade host tissue when the host harbors an unperturbed microbiota (Freter, 1962; van der Waaij et al., 1971). However, upon antibiotic treatment, pathogens such as Salmonella typhimurium, can expand to high densities and induces inflammation (Lawley et al., 2008; Stecher et al., 2007). The host immune response that Salmonella elicits appears to be deleterious to competing species and potentiates Salmonella expansion via production of oxygen radicals that generate tetrathionate, which Salmonella can use as an electron acceptor for anaerobic respiration (Winter et al., 2010). An avirulent Salmonella mutant that cannot induce host inflammation expands ephemerally after antibiotic treatment, and then contracts as the normal microbiota recovers, illustrating the necessity of host inflammation for S. typhimurium to establish a positive feedback loop that is self-sustaining (Stecher et al., 2007) (Fig. 6B).
Other similar circuits may reinforce the stabilitiy of a microbiota. Butyrate, produced by members of the Firmicutes (or the presence of Bacteroides and butyrate-producing Firmicute species) has been suggested to enhance host mucus production (Barcelo et al., 2000; Burger-van Paassen et al., 2009; Finnie et al., 1995; Shimotoyodome et al., 2000). In turn, many Bacteroides species convert mucin into fermentation end products such as succinate and acetate, which can be used to drive butyrate production by Firmicutes (Fig. 6B) (Louis et al., 2007; Mahowald et al., 2009; Martens et al., 2008). Such feedback loops are likely to range in complexity from one to multiple species, sometimes including the host. The influence of pH on growth differs among species within the microbiota, providing an example of a simple feedback loop in which a single species’ metabolism can potentiate its own growth (Duncan et al., 2009). Species of Bacteriodes and Firmicutes may be continually dueling over slightly higher vs. slightly lower pH, controlled by their own metabolism.
Interactions among feedback loops
These examples illustrate that multiple feedback loops exist simultaneously, and the degree to which they cooperate or conflict varies. Such feedback loop interactions may contribute to community stability or instability. The properties of each circuit are context dependent, and range from rare and unstable to pervasive and highly stable; how each circuit responds to perturbation is surely impacted by competing circuits that may establish new stable states. For example, a Bacteroides-Firmicute syntrophy may be largely disrupted upon antibiotic treatment, and its reestablishment may be dictated by other factors, e.g., whether Salmonella is lying in wait to induce inflammation and establish a new stable state.
Considering the susceptibility of a perturbed ecosystem to invasion and accompanying changes in composition, a mother would do well to attract community members to her infant’s intestine that can persist and help foster a smooth transition from suckling to solid food. Two recent studies have demonstrated that genes dedicated to plant polysaccharide degradation are embedded within the microbiome of breast-fed infants, despite the absence of the corresponding carbohydrate substrates (Koening et al., 2010; Vaishampayan et al., 2010). These data suggest that the infant microbiota contains species that, instead of being lost upon switching to solid food, can merely undergo a shift in function and continue to thrive. Recently, Marcobal and colleagues have established that Bacteroides species, known for their ability to consume plant polysaccharides, are indeed adept at consuming the oligosaccharides supplied to the infant microbiota in mothers’ milk, and employ the same polysaccharide acquisition pathways they use for mucin glycan consumption. The similarity between the structures of human milk oligosaccharides (HMO) and intestinal glycans suggests that HMOs mimic mucus carbohydrates to facilitate the recruitment of mucin-adapted bacteria to the infants’ intestine (Marcobal et al., In press at Cell Host & Microbe). Such a strategy could be highly adaptive in recruiting mucus-adapted mutualists that are also well endowed to consume the plant polysaccharides in the post-weaning diet. While the importance of a stable microbiota is just starting to be understood, it is likely that the host facilitates and participates in many feedback loops to promote a stable microbiota harboring species with defined functions in the intestine.
Prospects
Here we have focused on one metabolic pathway of gut-resident Bacteroides to highlight how basic catabolic processes feed into inter-species metabolic interactions. While tremendous strides are currently being made in describing changes in the composition of taxa and genes within the microbiota due to numerous factors (e.g., diet, disease, genotype, geographic location), much work remains to truly understand the biology that drives and results from community changes. Our attempt to revisit classic literature has revealed many gaps in our knowledge that can now be examined in the light of modern systems approaches. At the same time, descriptive sequence-based studies are serving an important complementary role in defining priorities for pursuing mechanism-level understanding of this otherwise exceedingly complex interplay between the microbial species that inhabit humans and the resulting interactions that occur between these species and the host.
Acknowledgments
We thank members of the Fischbach and Sonnenburg labs for helpful discussions. Research in the authors’ laboratories is supported by grants from the NIH (DP2 OD006515 to J.L.S. and DP2 OD007290 to M.A.F.); NIDDK (R01 DK085025 to J.L.S.); the Burroughs Wellcome Foundation (J.L.S.); the W.M. Keck Foundation (M.A.F.); the Program for Breakthrough Biomedical Research (M.A.F.); and a Young Investigator Grant from the Global Probiotics Council (M.A.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez-Acosta T, Leon C, Acosta-Gonzalez S, Parra-Soto H, Cluet-Rodriguez I, Rossell MR, Colina-Chourio JA. Beneficial role of green plantain [Musa paradisiaca] in the management of persistent diarrhea: a prospective randomized trial. Journal of the American College of Nutrition. 2009;28:169–176. doi: 10.1080/07315724.2009.10719768. [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Applied and environmental microbiology. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker HA. Amino acid degradation by anaerobic bacteria. Annu Rev Biochem. 1981;50:23–40. doi: 10.1146/annurev.bi.50.070181.000323. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Baughn AD, Malamy MH. The essential role of fumarate reductase in haem-dependent growth stimulation of Bacteroides fragilis. Microbiology. 2003;149:1551–1558. doi: 10.1099/mic.0.26247-0. [DOI] [PubMed] [Google Scholar]
- Caspari D, Macy JM. The role of carbon dioxide in glucose metabolism of Bacteroides fragilis. Arch Microbiol. 1983;135:16–24. doi: 10.1007/BF00419476. [DOI] [PubMed] [Google Scholar]
- Chassard C, Bernalier-Donadille A. H2 and acetate transfers during xylan fermentation between a butyrate-producing xylanolytic species and hydrogenotrophic microorganisms from the human gut. FEMS microbiology letters. 2006;254:116–122. doi: 10.1111/j.1574-6968.2005.00016.x. [DOI] [PubMed] [Google Scholar]
- Chen M, Wolin MJ. Influence of heme and vitamin B12 on growth and fermentations of Bacteroides species. Journal of bacteriology. 1981;145:466–471. doi: 10.1128/jb.145.1.466-471.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrock B, Bahl H, Gottschalk G. Parameters Affecting Solvent Production by Clostridium pasteurianum. Appl Environ Microbiol. 1992;58:1233–1239. doi: 10.1128/aem.58.4.1233-1239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehority BA. Carbon dioxide requirement of various species of rumen bacteria. J Bacteriol. 1971;105:70–76. doi: 10.1128/jb.105.1.70-76.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Microbes and Health Sackler Colloquium: Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences of the United States of America. 2010 doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dintzis RZ, Hastings AB. The Effect of Antibiotics on Urea Breakdown in Mice. Proceedings of the National Academy of Sciences of the United States of America. 1953;39:571–578. doi: 10.1073/pnas.39.7.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Applied and environmental microbiology. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Thomson JM, Flint HJ. The role of pH in determining the species composition of the human colonic microbiota. Environmental microbiology. 2009;11:2112–2122. doi: 10.1111/j.1462-2920.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- el-Khoury AE, Sanchez M, Fukagawa NK, Gleason RE, Young VR. Similar 24-h pattern and rate of carbon dioxide production, by indirect calorimetry vs. stable isotope dilution, in healthy adults under standardized metabolic conditions. The Journal of nutrition. 1994;124:1615–1627. doi: 10.1093/jn/124.9.1615. [DOI] [PubMed] [Google Scholar]
- Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science (New York, NY) 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter R. In vivo and in vitro antagonism of intestinal bacteria against Shigellaflexneri. II. The inhibitory mechanism. The Journal of infectious diseases. 1962;110:38–46. doi: 10.1093/infdis/110.1.38. [DOI] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell host & microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- Hopfer U, Liedtke CM. Proton and bicarbonate transport mechanisms in the intestine. Annu Rev Physiol. 1987;49:51–67. doi: 10.1146/annurev.ph.49.030187.000411. [DOI] [PubMed] [Google Scholar]
- Huycke MM, Gaskins HR. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Experimental biology and medicine (Maywood, NJ) 2004;229:586–597. doi: 10.1177/153537020422900702. [DOI] [PubMed] [Google Scholar]
- Karasawa T, Ikoma S, Yamakawa K, Nakamura S. A defined growth medium for Clostridium difficile. Microbiology. 1995;141(Pt 2):371–375. doi: 10.1099/13500872-141-2-371. [DOI] [PubMed] [Google Scholar]
- Koening JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proceeding of the Natural Academy of Science of the United States of America. 2010 doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolida S, Meyer D, Gibson GR. A double-blind placebo-controlled study to establish the bifidogenic dose of inulin in healthy humans. European journal of clinical nutrition. 2007;61:1189–1195. doi: 10.1038/sj.ejcn.1602636. [DOI] [PubMed] [Google Scholar]
- Kroger A, Geisler V, Lemma E, Theis F, Lenger R. Bacterial fumarate respiration. Arch Microbiol. 1992;158:311–314. [Google Scholar]
- Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infection and immunity. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson SM, Crowley LV, Horowitz RE, Malm OJ. The metabolism of carbon-labeled urea in the germ free rat. J Biol Chem. 1959;234:2061–2062. [PubMed] [Google Scholar]
- Louis P, Scott KP, Duncan SH, Flint HJ. Understanding the effects of diet on bacterial metabolism in the large intestine. Journal of applied microbiology. 2007;102:1197–1208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Gibson GR. Formation of glycoprotein degrading enzymes by Bacteroides fragilis. FEMS Microbiol Lett. 1991;61:289–293. doi: 10.1016/0378-1097(91)90567-t. [DOI] [PubMed] [Google Scholar]
- Macy JM, Ljungdahl LG, Gottschalk G. Pathway of succinate and propionate formation in Bacteroides fragilis. J Bacteriol. 1978;134:84–91. doi: 10.1128/jb.134.1.84-91.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell host & microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I, Kim J, Duffy PR, Schlegel VL, Walter J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS ONE. 2010;5:e15046. doi: 10.1371/journal.pone.0015046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TL, Wolin MJ. Enumeration of Methanobrevibacter smithii in human feces. Arch Microbiol. 1982;131:14–18. doi: 10.1007/BF00451492. [DOI] [PubMed] [Google Scholar]
- Rabbani GH, Ahmed S, Hossain I, Islam R, Marni F, Akhtar M, Majid N. Green banana reduces clinical severity of childhood shigellosis: a double-blind, randomized, controlled clinical trial. The Pediatric infectious disease journal. 2009;28:420–425. doi: 10.1097/INF.0b013e31819510b5. [DOI] [PubMed] [Google Scholar]
- Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. The British journal of nutrition. 2008:1–10. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
- Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, Gordon JI. Dissecting the in vivo metabolic potential of two human gut acetogens. The Journal of biological chemistry. 2010;285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins DJ, Yee RB, Bentley R. Biosynthetic precursors of vitamin K as growth promoters for Bacteroides melaninogenicus. Journal of bacteriology. 1973;116:965–971. doi: 10.1128/jb.116.2.965-971.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Vercellotti JR, West SE, Wilkins TD. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Applied and environmental microbiology. 1977a;33:319–322. doi: 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, West SE, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Applied and environmental microbiology. 1977b;34:529–533. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KP, Martin JC, Chassard C, Clerget M, Potrykus J, Campbell G, Mayer CD, Young P, Rucklidge G, Ramsay AG, et al. Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4672–4679. doi: 10.1073/pnas.1000091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebald M, Costilow RN. Minimal growth requirements for Clostridium perfringens and isolation of auxotrophic mutants. Appl Microbiol. 1975;29:1–6. doi: 10.1128/am.29.1.1-6.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohling B, Gottschalk G. Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J Bacteriol. 1996;178:871–880. doi: 10.1128/jb.178.3.871-880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell. 2010 doi: 10.1016/j.cell.2010.05.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Chen CT, Gordon JI. Genomic and Metabolic Studies of the Impact of Probiotics on a Model Gut Symbiont and Host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science (New York, NY) 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Stecher B, Chaffron S, Kappeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, McCoy KD, von Mering C, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS pathogens. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biology and Evolution. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waaij D, Berghuis-de Vries JM, Lekkerkerk L-v. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. The Journal of hygiene. 1971;69:405–411. doi: 10.1017/s0022172400021653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varel VH, Bryant MP. Nutritional features of Bacteroides fragilis subsp. fragilis Applied microbiology. 1974;28:251–257. doi: 10.1128/am.28.2.251-257.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. The ISME journal. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt DD, Demoss RD. Effect of microflora on the free amino acid distribution in various regions of the mouse gastrointestinal tract. Applied microbiology. 1975;30:609–615. doi: 10.1128/am.30.4.609-615.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DR, Ing TS, Metcalfe-Gibson A, Wrong OM. In vivo dialysis of faeces as a method of stool analysis. 3. The effect of intestinal antibiotics. Clinical science. 1968;34:211–221. [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science (New York, NY) 2011 doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto I, Abe A, Saito H, Ishimoto M. The pathway of ammonia assimilation in Bacteroides fragilis. J Gen Appl Microbiol. 1984;30:499–508. [Google Scholar]


