Abstract
Ethylene signaling pathway leads to rapid gene activation by two hierarchies of transcription factors with EIN3/EIL proteins as primary ones and ERF proteins as secondary ones. The role of chromatin modifications during the rapid gene activation is not known. In this work we studied trimethylated histone H3 lysine 4 (H3K4me3) and lysine 27 (H3K27me3), two opposite histone methylation marks for gene activity, during the induction course of three ethylene-responsive genes (ERF1, AtERF14 and ChiB). We found that the three genes displayed different histone modification profiles before induction. After induction, H3K4me3 was increased in the 5′ region and the gene body of ERF1, while H3K27me3 was decreased in the promoter of AtERF14. But the modification changes were later than the gene activation. Analysis of other rapidly inducible ERF genes confirmed the observation. In addition, histone H2A.Z occupancy on the three genes and the association of the H3K27me3-binding protein LHP1 with AtERF14 and ChiB were not affected by the inductive signal. However, the mutation of genes encoding H2A.Z and LHP1 attenuated and enhanced respectively the induction of target genes and altered H3K4me3. These results indicate that the induction of ethylene-responsive genes does not require immediate modulation of H3K4me3 and H3K27me3 and dissociation of LHP1 and H2A.Z from the targets, and suggest that the chromatin structure of the genes before induction is committed for transcriptional activation and that H3K4me3 is not required for ethylene-responsive gene activation, but may serve as a mark for gene activity.
Introduction
In addition to transcription factors chromatin structure plays an important role in the regulation of gene expression. The basic unit of chromatin is nucleosome that is formed by histone octamer containing two copies of H3, H4, H2A and H2B wrapped around by 147 base pairs of DNA. Chromatin structure change includes histone modifications and DNA methylation, histone variant deposition and chromatin remodeling. Histone modifications, especially H3K4 trimethylation and H3K27 trimethylation, have been largely reported to be tightly associated with gene transcription activity [1], [2]. H3K4me3 is associated with highly expressed and/or housekeeping genes whereas H3K27me3 marks under-expressed and/or repressed tissue-specific genes [1], [2]. Both modification marks could be recognized by different chromatin factors through specific protein domains. For example, the Plant Homeodomain (PHD) of ING2 (Inhibitor of Growth 2) can bind to H3K4me3 and the chromodomain of Polycomb proteins in animal cells and LIKE HETEROCHROMATIN PROTEIN1 (LHP1) in Arabidopsis can bind to H3K27me3 [3], [4]. The recognitions may serve as a mechanism by which histone modifications regulate gene expression. Histone variant H2A.Z is another important regulator of gene expression which is deposited into nucleosome by SWR complex. Recent analysis in various species has revealed that activation of H2A.Z-regulated genes was accompanied by eviction of H2A.Z or replacement of H2A.Z with H2A by INO80 complex [5], [6], [7]. Other studies have suggested that H2A.Z may act as an epigenetic mark to promote gene reactivation [8], [9].
In plants, H3K4me3 and the H3K27me3/LHP1 module have been shown to mediate developmental genes expression such as FLC (FLOWERING LOCUS C), AG (AGAMOUS), FUS3 (FUSCA 3) and FT (FLOWERING LOCUS T) [10], [11], [12], [13]. However, how these modifications affect rapidly induced gene activation was not clear. Ethylene is a plant hormone participating in different processes including germination, flower and leaf senescence, fruit ripening, leaf abscission, root nodulation, programmed cell death, and response to stress and pathogen attack. Genetic and molecular analyses have revealed a response pathway from perception to a series of MAP kinase and finally transduced to two hierarchies of transcription regulation [14]. The primary transcription regulation is that transcription factors EIN3 (ETHYLENE-INSENSITIVE3)/EIL1 (ETHYLENE-INSENSITIVE3-LIKE 1) directly bind to EREBP (ethylene-responsive element binding protein) genes such as ERF1 (ETHYLENE RESPONSE FACTOR 1) to activate their expression. Subsequently EREBP proteins activate downstream effecter genes (e.g. ChiB, basic chitinase and PDF1.2, Plant Defensin 1.2). However, it was not known whether the rapid activation of ethylene-responsive genes involves change of chromatin structure. Here, we chose ERF1 and AtERF14 (Arabidopsis thaliana Ethylene-responsive element binding factor 14) as well as 5 other ERF genes as primary and ChiB as secondary regulation targets to analyze whether chromatin structures of these target genes changed during rapid induction by ethylene. We used 1-aminocyclopropane-1-carboxylic acid (ACC) which is converted to ethylene by 1-aminocyclopropane-1-carboxylic acid oxidase (ACO) in plants to treat 12 day-old seedlings. Increase of H3K4me3 and decrease of H3K27me3 were observed during the treatment, but the changes of both marks were much later than the gene activation. H2A.Z occupancy and LHP1 binding did not respond to the treatment indicating that the gene induction by ethylene signaling did not require immediate change of the cognate chromatin structure. However, mutation of genes encoding H2A.Z and LHP1 affected the induction of ethylene-responsive genes, suggesting that the committed chromatin structure of these genes before induction is important for the transcriptional activation.
Results
Histone methylation profile and H2A.Z deposition over ethylene-responsive genes before induction
To assess the chromatin structure of ethylene-responsive genes before induction we tested H3K4me3, H3K27me3 and H2A.Z deposition in the promoter, the 5′ region and the gene body of ERF1, AtERF14 and ChiB (Fig. 1A). RBCS-1A (RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN 1A), AG and HSP70 (heat shock protein 70) were used as positive controls respectively for H3K4me3, H3K27me3 and H2A.Z deposition [1], [5], [13]. The At4g07700 locus was used as negative control for H2A.Z deposition [5]. Moderate levels of H3K4me3 were detected in the 5′ region and the gene body, but not the promoter, of ERF1 and ChiB compared to that of RBCS-1A (Fig. 1B). In contrast, H3K4me3 was not detected over AtERF14 (Fig. 1B). H3K27me3 was enriched in the 5′ region and the gene body of both AtERF14 and ChiB but not in ERF1 (Fig. 1C). Analysis of five additional ERF genes (ORA59, TDR1, AtERF1, AtERF2 and AtERF11) revealed that TDR1 displayed a high level of H3K27me3 but a low level of H3K4me3, while the other four genes showed a high level of H3K4me3 but a low level of H3K27me3 (Fig. S2). Similar to what found in ERF1 and ChiB, H3K4me3 levels on the promoter of these genes were relatively low (Fig. S2). This analysis revealed that the ethylene-inducible genes displayed different histone modification profiles before induction.
Figure 1. Chromatin status of ERF1, AtERF14 and ChiB before ACC induction.
(A) Diagrams of the gene structure of ERF1, AtERF14, ChiB, AG, and RBCS1A. The solid boxes indicate the coding regions, and the open boxes indicate untranslated regions (UTRs). The solid bars indicate the regions in which primers were designed for ChIP tests. (B) H3K4me3 on the three genes before induction. The relative enrichments were calculated by compared to input. RBCS was used as a positive control. The regions (P1, P2 and P3) shown in A represent promoter, 5′ region and gene body respectively. (C) H3K27me3 on the three genes before induction. AG was used as a positive control. (D) H2A.Z abundance on the three genes before induction. HSP70 was used as a positive control and At4g07700 was used as a negative control. Bars represent mean values +/−SD from three repeats.
To test whether H2A.Z was present in the chromatin of these genes, chromatin fragments isolated from H2A.Z-GFP transgenic plants were precipitated with GFP antibody. We found that H2A.Z was incorporated into chromatin over the three genes with highest levels in the gene bodies and lowest levels in the promoters (Fig. 1D). The presence of H2A.Z was detected also over the five additional ERF genes (Fig. S2).
Histone methylation dynamics during induction of ethylene responsive genes
In order to study histone modification dynamics during gene activation, we chose five time points to monitor ACC induction time course of ethylene-responsive genes by quantitative RT-PCR. The induction of ERF1 by ACC was early, which was 4 folds after 1 hour and elevated to 40 folds after 8 hours (Fig. 2A). For AtERF14, the expression began to increase after 2 hours and reached to 12 folds after 8 hours (Fig. 2A). The induction of ChiB was moderate, only 2 to 3 folds after 8 hours (Fig. 2A). ORA59, AtERF1, AtERF2 and AtERF11were induced as early as ERF1, while the induction of TDR1 was delayed (Fig. S2).
Figure 2. Expression and histone modification changes of three ethylene responsive genes during ACC induction.
(A) Induction time course of the three genes by ACC. Twelve day-old seedlings were treated with 50 µM ACC and harvested at the indicated time points. Relative fold changes were determined by normalization with ACTIN2 transcript levels. (B) H3K4me3 detected on the three genes during ACC induction. The relative enrichments were calculated by first comparing to input and then to the reference gene RBCS-1A. The three regions were analyzed for ChIP. (C) H3K27me3 detected on the three genes during ACC induction. The relative enrichments were calculated by first comparing to input and then to the reference gene AGAMOUS (AG). The three regions were analyzed for ChIP. Bars represent mean values +/−SD from three repeats. Significance of H3K4me3 and H3K27me3 differences between after ACC treatment (at different time points) and before treatment (0) was tested by two-tailed Student's t-test, * p<0.05, **p<0.005.
H3K4me3 and H3K27me3 are two opposite histone modification marks associated with gene transcription activity. However, it is still not clear whether the two modifications are involved in gene activation process. Therefore, we tested the levels of these modifications over ethylene responsive genes during the ACC induction process. For ERF1, H3K4me3 in the 5′ region and the gene body began to increase only after 4 hours, which was later than the initial increase of gene expression (Fig. 2B). Similarly, the increase of H3K4me3 over ORA59, AtERF1, AtERF2 and AtERF11 was also later than gene activation (Fig. S2). This suggested that the induction of ERF1 did not require a concurrent increase of H3K4me3. The late increase might indicate that H3K4me3 served as a mark of elevated transcription activity of the genes. H3K4me3 remained undetectable over AtERF14 and TDR1and did not change over ChiB during the induction (Fig. 2B) (Fig. S2). These results indicated that H3K4me3 was not necessary for the induction of AtERF14 and TDR1. However, it was not clear whether the basal levels of H3K4me3 over ERF1 and ChiB before induction was required for the induction of the genes.
Due to the low level of H3K27me3 over ERF1 before induction we did not expect that there would be any change during the induction. So we tested H3K27me3 over AtERF14 and ChiB. H3K27me3 was not much changed in the gene body of both genes during induction, but was decreased in the promoter of the genes, especially AtERF14 (Fig. 2C). However the decrease of H3K27me3 was delayed compared to the gene induction, suggesting that rapid gene activation did not require or lead to immediate demethylation of H3K27me3 and that the presence of H3K27me3 did not prevent the induction process. Analysis of H3K27me3 over ORA59, TDR1, AtERF1, AtERF2 and AtERF11 confirmed the results (Fig. S2).
Negative function of LHP1 on the induction of AtERF14 and ChiB
H3K27me3 is recognized and bound by LHP1 that is suggested to be an H3K27me3 effector. To explore the role of LHP1 in rapid gene activation, we analyzed the induction of ethylene responsive genes in the lhp1 mutant. For AtERF14 and ChiB that displayed high levels of H3K27me3 the induction by ACC was clearly enhanced in the mutant (Fig. 3A), indicating that LHP1 had a repressive function on induction of the two genes. However, we also detected an elevated expression of ERF1 in lhp1 (Fig. 3A). Considering that there was a low level of H3K27me3 over ERF1 we speculated that this might be an indirect effect of increased expression of AtERF14, as it has been reported that overexpression of AtERF14 could lead to increased expression of ERF1 [15]. In addition, we tested H3K4me3 levels over the target genes in lhp1 in comparison with the wild type. We found that in lhp1 H3K4me3 was increased in the 5′ region and the gene body, but not the promoter, of ERF1 and ChiB. The increased H3K4me3 levels may be also a consequence of increased transcription activity of the genes as mentioned before. However, H3K4me3 remained undetectable over AtERF14 despite the increased expression of this gene in lhp1 (Fig. 3B). The early induction of 4 of the 5 additional ERF genes was found to be enhanced in the lhp1 mutant. Except TDR1 that had no H3K4me3, the other three genes displayed increased H3K4me3 (Fig. S3).
Figure 3. The role of LHP1 on the induction of ethylene responsive genes.
(A) Transcript levels of the three genes at different time points during ACC induction in wild type (WT) and lhp plants. Relative fold changes were determined by normalization with ACTIN2 transcripts. (B) H3K4me3 levels on ChiB, ERF1 and AtERF14 in wild type (WT) and lhp1 mutants during treatment. The relative enrichments (to RBCS-1A) on three regions of the three genes are presented. Bars represent mean values +/−SD from three repeats. Significance of H3K4me3 differences between WT and lhp1at different time points was determined by two-tailed Student's t-test, * p<0.05, **p<0.005.
To study whether LHP1 was bound to ChiB and AtERF14, ChIP analysis of the lhp1 mutant complemented by LHP1::LHP1-MYC was performed by using anti-MYC antibodies[16]. The analysis revealed that LHP1 was associated with ChiB and AtERF14 as well as with AG, but not with ERF1 (Fig. 4). Importantly, ACC treatment did not lead to dissociation of LHP1 from these genes (Fig. 4). Therefore, although the lhp1 mutation had an effect on the induction of ethylene-induced genes, the presence of the H3K27me3 /LHP1 module on the genes was irresponsive to the inductive signal.
Figure 4. LHP1 binding to different genes before and after ACC treatment.
WT and lhp1 mutants complemented by LHP1::LHP1-MYC were used for ChIP analysis with anti-MYC antibodies. The relative enrichments were normalized with input. Bars represent mean values +/−SD from three repeats.
Requirement of H2A.Z for the induction of ERF1 and ChiB
In Arabidopsis, H2A.Z has been shown to be involved in multiple responses such as temperature and phosphate starvation [5], [7]. It either activates or represses target genes expression by eviction from nucleosomes occupying around the transcription start site. The presence of H2A.Z was detected over the three ethylene-inducible genes before ACC induction (Fig. 1D). During ACC treatment, no immediate decrease of H2A.Z abundance over ethylene-responsive genes was detected, albeit a slight decrease was observed after induction (Fig. 5; Fig. S2), suggesting that a clear H2A.Z eviction was not required for the initial induction of the genes.
Figure 5. H2A.Z deposition over ethylene responsive genes during ACC induction.
H2A.Z-GFP transgenic plants were used for ChIP analysis with GFP antibody. The enrichments on three regions are shown. Bars represent mean values +/−SD from three repeats. Significance of H2A.Z level difference between induced (at different time points) and non induced (0) was determined by two-tailed Student's t-test, * p<0.05, **p<0.005.
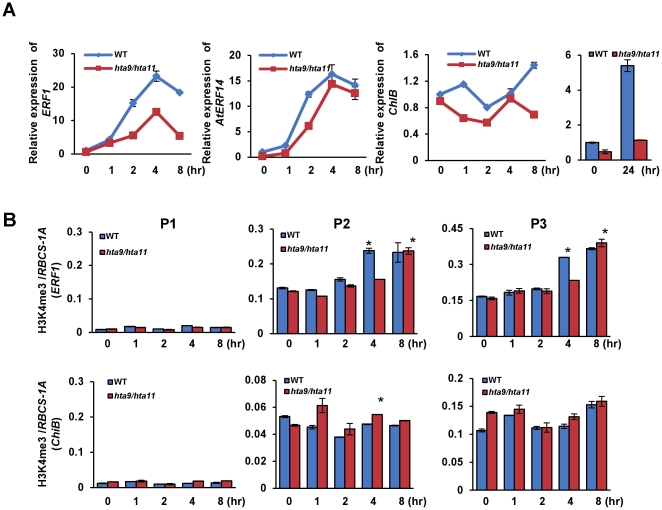
In order to study whether H2A.Z was required for the induction, we tested the expression of these genes in hta9/hta11 double mutants that have a reduced level of H2A.Z [17]. In the mutant the induction of ERF1 was reduced, while that of AtERF14 was not clearly affected (Fig. 6A). The effect of the mutations on the induction of ChiB was detected after 8 hours, but was more severe after 24 hours (Fig. 6A). These results indicated that H2A.Z was involved in the induction of ERF1 and ChiB. Then we tested H3K4me3 over ERF1 and ChiB during ACC induction in the hta9/hta11 mutants. Increase of H3K4me3 over ERF1 observed in wild type was delayed in the mutants, but the basal levels of H3K4me3 over both ERF1 and ChiB were not affected (Fig. 6B). We speculated that H2A.Z had no effect on H3K4me3 and the delayed increase of H3K4me3 over ERF1 might be a result of decreased transcription activity of the gene in the mutants.
Figure 6. Function of H2A.Z in the ACC induction of ERF1, AtERF14 and ChiB.
(A) Transcript levels of the three genes at different time points during ACC induction in wild type (WT) and hta9/hta1 mutants. Relative fold changes were determined by normalization with ACTIN2 transcripts. (B) H3K4me3 levels on ERF1 and ChiB during ACC induction in WT and hta9/hta11. The relative enrichments (to RBCS-1A) on three regions are shown. Bars represent mean values +/−SD from three repeats. Significance of H3K4me3 difference between WT and hta9/hta11at different time points was determined by two-tailed Student's t-test, * p<0.05, **p<0.005.
Discussion
H3K4me3 serves as a mark of gene transcription activity
Genome-wide analysis in plants indicates that H3K4me3 is associated with actively transcribed genes. But how it affects gene expression remains unclear [1], [18]. Some researchers have proposed that this modification may be recognized and bound by specific proteins which act as effectors to control transcription [3], [19], [20]. But other studies have suggested that H3K4me3 could serve as a memory or a mark of active genes [21]. In this study we tried to find out the role of H3K4me3 during activation of ethylene-inducible genes. Our results indicated that elevated H3K4me3 was not necessary for the ethylene-induced gene activation but may serve as a mark of transcription activity of the genes. First, H3K4me3 was not detected over AtERF14 and TDR1 before induction and was not increased after ACC treatment (Fig. 1B, Fig. 2B; Fig. S2). Second, although some increase of H3K4me3 was detected in the 5′ region and/or the gene body of ERF1, ORA59, AtERF1, AtERF2 and AtERF11 after induction, the increase of H3K4me3 lagged behind that of gene activation (Fig. 2B; Fig. S2). Finally, the level of H3K4me3 was associated with that of gene expression in lhp1 and hta9/hta11 mutant and wild type plants. For instance, H3K4me3 was increased when the induction of ERF1 and ChiB was enhanced in lhp1 over these genes (Fig. 3). Conversely, the increase of H3K4me3 was delayed when the induction of the genes was attenuated by the mutation of H2A.Z genes (Fig. 6). Although there was no concomitant increase of H3K4me3 with activation of gene expression, it is not excluded the possibility that the basal level of H3K4me3 over ERF1 and ChiB might contribute to the chromatin commitment of these genes for the induction.
Repressive function of the H3K27me3/LHP1 module on inducible genes
The observations that H3K27me3 did not change over AtERF14, ChiB and TDR1 during ACC induction are consistent with recent results showing that although H3K27me3 on the floral time repressor FLC is inversely correlated with transcriptional activity, the abundance of this mark is not diminished in the first 12h following activation of transcription [22]. Analysis of cold-inducible genes has detected H3K27me3 to decline only one to several days after application of the inductive signals [23]. These observations suggest that transcription activation may not involve immediate demethylation of H3K27me3, or the presence of H3K27me3 is not sufficient to impair transcriptional activation during induction. This is supported by a recent result showing that vernalization-mediated induction of VERNALISATION INSENSITIVE 3 (VIN3) does not lead to any decrease of H3K27me3 on the locus 40 days after exposure to cold temperature [24], [25].
The observations that the binding of LHP1 to AtERF14 and ChiB was not affected by ACC induction and that the mutation of LHP1 led to increased induction of the genes suggest that the H3K27me3 /LHP1 module is required for the repression of the genes and the repressive effect could be lifted by additional events during the induction. The constitutive association of LHP1 with these targets is reminiscent of the data showing that LHP1 remains to be associated with VIN3 chromatin many days after induction by vernalization [25], indicating that the LHP1 binding in that conditions does not lead to gene silencing. These observations collectively suggest that additional elements associated with the H3K27me3/LHP1 module, which can be inactivated by inductive signals, might be involved in H3K27me3/LHP1–mediated gene silencing.
Involvement of H2A.Z in the induction of gene expression
Our data showing that deposition of histone variant H2A.Z over the eight ethylene-responsive genes was not evicted after ACC induction are in agreement with the findings that H2A.Z is present in both silent and active FLC chromatin (Fig. 5; Fig. S2) [8]. Probably, the presence of H2A.Z may mark these genes for induction, supporting the notion that H2A.Z serves to mark active gene and poise silent genes for reactivation [8], [9]. Recent results have shown that H2A.Z is required for both gene activation and repression in responding to warmer temperature [5]. In contrast to the observations on the ethylene-responsive genes, H2A.Z-containing nucleosomes are found to be lost from both up-regulated and down-regulated genes after an increase of temperature. Therefore, the role of H2A.Z in chromatin structure and in gene activity is complex, which may be dependent on the chromatin context of the gene. In addition, our results showed that mutation of H2A.Z genes had an obvious negative effect on the induction of ERF1 and ChiB but not AtERF14. Considering the undetectable level of H3K4me3 over AtERF14 and moderate levels over ERF1 and ChiB we speculate that H2A.Z may have a coordinated effect with H3K4me3 on the activation of these genes during ACC induction.
Chromatin structure is considered as an important regulator of transcription in addition to transcription factors especially for the developmental genes. In this work we tried to figure out whether chromatin modifications take place during activation of rapidly inducible genes. Our work revealed that histone modifications including H3K4me3 and H3K27me3 and presence of chromatin proteins such as LHP1 and H2A.Z did not display any immediate change upon ACC treatment. However, mutation of LHP1 and H2A.Z genes had an effect on the induction suggesting that basal chromatin structure before induction is important for the induction.
Materials and Methods
Plant material and Exogenous ACC treatment
The mutants used in this study are lhp1 [26], hta9/hta11 [17], lhp1 complemented by LHP1::MYC [16] and H2A.Z-GFP transgenic plants [5]. Arabidopsis seeds were surface-sterilized and growth at 22°C with a 16 h light/8 h dark (long day) cycle. Twelve days after germination 50 µM ACC solution was added. The samples were harvested at indicated time points.
RNA extraction and reverse transcription
Total RNA was extracted from twelve day-old seedling using Trizol (Invitrogen). Four µg total RNA were treated first with 1 unit of DNase I (Promega) and then reverse transcribed in a total volume of 20 µL with 0.5 µg oligo(dT)15, 0.75 mM dNTPs, 2.5 mM MgCl2, 1 µl ImProm-II reverse transcriptase (Promega). The resulting products were tested by Real-Time PCR with gene specific primers (Table S1).
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) experiment was performed most as described in [27]. One gram of 12 day-old seedlings before and after ACC treatment were harvested and crosslinked in 1% formaldehyde under vacuum. Nuclei were then extracted with extraction buffers. Chromatin was fragmented to 200–2000 bp by sonication and ChIP was performed using antibodies: c-Myc (Sigma, M4439), H3K4me3 (Cell Signaling, 9751S) H3K27me3 (Millipore, 07–449) and GFP antibody (Abcam, ab290). The precipitated and input DNAs were then analyzed by real-time PCR with gene specific primer sets (Fig. S1, Table S1). At least three biological repeats were performed for the ChIP experiments.
Real-Time PCR
Real-time PCR was performed in a total volume of 20 µL with 1.0 µl of the reverse transcription or ChIP products, 0.25 µM primers, and 10 µl SYBR Green Master mix (Roche) on a LightCycler 480 real-time PCR machine (Roche) according to the manufacturer's instructions. All primers were annealed at 60°C and run 45 cycles. The ChIP enrichment for GFP, H3K27me3 and H3K4me3 was quantified by comparing the thresholdcycle (Ct) of the ChIP samples with that of the input and then normalized with the levels of control genes: 2(Ct of input-Ct of sample ChIP) /2(Ct of input-Ct of control ChIP). The expression level of target genes was normalized with that of ACTIN: 2(Ct of actin- Ct of target).
Supporting Information
Genes used as controls in this study were not affected by ACC treatment. The expression of ACTIN2, AGAMOUS (AG) and RBCS1A was not affected by ACC treatment. For ACTIN2, three biological replication of ACC induction were performed. Data represent average means and the expression before ACC induction was set as 1. The expressions of AG and RBCS-1A were normalized with that of ACTIN2.
(TIF)
Expression, histone methylation and H2A.Z deposition of five additional ethylene responsive factor (ERF) genes during ACC induction. RNA levels (A), H3K4me3 (B, C), H3K27me3 (D) and H2A.Z (E) of ORA59 (At1g066160), TDR1 (At3g23230), AtERF1(At4g17500), ATERF2(At4g47220) and ATERF11(At1g28370) were measured at the different time points during ACC treatment as indicated. Bars represent mean values +/− SD from three repeats. For ChIP experiments, primers corresponding to the promoter (B) and gene bodies (C-E) were used. Significance of H3K4me3, H3K27me3 and H2A.Z induction compared to that before treatment (0) was determined by two-tailed Student's t-test, * p<0.05, **p<0.005.
(TIF)
Expression and H3K4me3 of additional ethylene responsive factor (ERF) genes between WT and lhp1 during ACC induction. RNA levels (upper) and H3K4me3 (lower) were measured during ACC treatment. Bars represent mean values +/− SD from three repeats. For ChIP experiments, primers corresponding to the gene bodies were used. Significance of H3K4me3 levels between WT and lhp1 before and after ACC treatment was determined by two-tailed Student's t-test, *p<0.05, **p<0.005.
(TIF)
Sequences of primers used in this study.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant from the French Agence Nationale de la Recherche. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, et al. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol. 2007;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- 5.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith AP, Jain A, Deal RB, Nagarajan VK, Poling MD, et al. Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 2010;152:217–225. doi: 10.1104/pp.109.145532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deal RB, Topp CN, McKinney EC, Meagher RB. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cel. 2007;19:74–83. doi: 10.1105/tpc.106.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly TK, Miranda TB, Liang G, Berman BP, Lin JC, et al. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol Cell. 2010;39:901–911. doi: 10.1016/j.molcel.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, et al. cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell. 2010;22:1425–1440. doi: 10.1105/tpc.110.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, et al. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 2006;7:947–952. doi: 10.1038/sj.embor.7400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, et al. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci U S A. 2006;103:5012–5017. doi: 10.1073/pnas.0507427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, et al. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. Embo J. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo H, Ecker JR. The ethylene signaling pathway: new insights. Curr Opin Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Onate-Sanchez L, Anderson JP, Young J, Singh KB. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 2007;143:400–409. doi: 10.1104/pp.106.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latrasse D, Germann S, Houba-Herin N, Dubois E, Bui-Prodhomme D, et al. Control of flowering and cell fate by LIF2, an RNA binding partner of the polycomb complex component LHP1. PLoS One. 2011;6:e16592. doi: 10.1371/journal.pone.0016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.March-Diaz R, Garcia-Dominguez M, Lozano-Juste J, Leon J, Florencio FJ, et al. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 2008;53:475–487. doi: 10.1111/j.1365-313X.2007.03361.x. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Wang X, He K, Ma Y, Su N, et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell. 2008;20:259–276. doi: 10.1105/tpc.107.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 20.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 21.Muramoto T, Muller I, Thomas G, Melvin A, Chubb JR. Methylation of H3K4 Is required for inheritance of active transcriptional states. Curr Biol. 2010;20:397–406. doi: 10.1016/j.cub.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 22.Buzas DM, Robertson M, Finnegan EJ, Helliwell CA. Plant J; 2010. Transcription-dependence of histone H3 lysine 27 trimethylation at the Arabidopsis polycomb target gene FLC. [DOI] [PubMed] [Google Scholar]
- 23.Kwon CS, Lee D, Choi G, Chung WI. Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 2009;60:112–121. doi: 10.1111/j.1365-313X.2009.03938.x. [DOI] [PubMed] [Google Scholar]
- 24.Jean Finnegan E, Bond DM, Buzas DM, Goodrich J, Helliwell CA, et al. Polycomb proteins regulate the quantitative induction of VERNALIZATION INSENSITIVE 3 in response to low temperatures. Plant J. 2011;65:382–391. doi: 10.1111/j.1365-313X.2010.04428.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim DH, Zografos BR, Sung S. Vernalization-mediated VIN3 Induction Overcomes the LIKE-HETEROCHROMATIN PROTEIN1/POLYCOMB REPRESSION COMPLEX2-mediated epigenetic repression. Plant Physiol. 2010;154:949–957. doi: 10.1104/pp.110.161083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudin V, Libault M, Pouteau S, Juul T, Zhao G, et al. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development. 2001;128:4847–4858. doi: 10.1242/dev.128.23.4847. [DOI] [PubMed] [Google Scholar]
- 27.Huang L, Sun Q, Qin F, Li C, Zhao Y, et al. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007;144:1508–1519. doi: 10.1104/pp.107.099473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes used as controls in this study were not affected by ACC treatment. The expression of ACTIN2, AGAMOUS (AG) and RBCS1A was not affected by ACC treatment. For ACTIN2, three biological replication of ACC induction were performed. Data represent average means and the expression before ACC induction was set as 1. The expressions of AG and RBCS-1A were normalized with that of ACTIN2.
(TIF)
Expression, histone methylation and H2A.Z deposition of five additional ethylene responsive factor (ERF) genes during ACC induction. RNA levels (A), H3K4me3 (B, C), H3K27me3 (D) and H2A.Z (E) of ORA59 (At1g066160), TDR1 (At3g23230), AtERF1(At4g17500), ATERF2(At4g47220) and ATERF11(At1g28370) were measured at the different time points during ACC treatment as indicated. Bars represent mean values +/− SD from three repeats. For ChIP experiments, primers corresponding to the promoter (B) and gene bodies (C-E) were used. Significance of H3K4me3, H3K27me3 and H2A.Z induction compared to that before treatment (0) was determined by two-tailed Student's t-test, * p<0.05, **p<0.005.
(TIF)
Expression and H3K4me3 of additional ethylene responsive factor (ERF) genes between WT and lhp1 during ACC induction. RNA levels (upper) and H3K4me3 (lower) were measured during ACC treatment. Bars represent mean values +/− SD from three repeats. For ChIP experiments, primers corresponding to the gene bodies were used. Significance of H3K4me3 levels between WT and lhp1 before and after ACC treatment was determined by two-tailed Student's t-test, *p<0.05, **p<0.005.
(TIF)
Sequences of primers used in this study.
(DOC)