Abstract
Background
The penetrance of CDKN2A mutations is subject to geographic and latitudinal variation and is presumably dictated by UVR exposure and possibly other co-inherited genetic factors. The frequency of mutations increases with the number of family members affected and the number of primary tumors and also fluctuates with geography. Up to date, little is known about the prevalence of CDKN2A mutations in melanoma patients from Greece.
Objective
To characterize the frequency of CDKN2A and CDK4 mutations in a hospital-based population of Greek patients with melanoma.
Methods
Three-hundred and four consecutive single primary melanoma (SPM), 9 familial melanomas (FM) and 7 multiple primary melanoma cases (MPM) were assessed for sequence variants in exons 1α, 1β and 2 of CDKN2A and exon 2 of CDK4.
Results
Germline CDKN2A mutations were detected in 10 of 304 SPM (3.29%), in 4 of 7 MPM (57.0%) and in 2 of 9 FM (22.2%) cases. The most common mutation was a Northern European allele (p16 p.R24P) detected in 8 individuals. Five previously unreported CDKN2A variants were also identified: −34G>C, c.41_43delins20bp, c.301G>C(p.G101R), c.301G>A(p.G101E) and c.296_297insGACC. We also describe the first report of a Cdk4 p.R24H substitution in a Greek family.
Conclusions
The Greek population appears to harbor a higher prevalence of CDKN2A mutation than other reported cohorts. This supports the notion that genetic susceptibility may play a stronger influence in a country with a relatively low incidence of melanoma. Furthermore, the identification of Northern European alleles suggests that gene migration may be responsible, in part, for the observed cases in Greece.
Keywords: melanoma, Greek, CDKN2A, CDK4, mutations
Introduction
Greece is a Mediterranean country with a relatively low incidence of melanoma (4/100,000 inhabitants) compared to other European countries. Although the prevalence of germline CDKN2A and CDK4 variants among Northern European populations have been described, 1, 2 little is known about the mutational status at these loci among melanoma patients from Greece.3, 4, 5 In a previous study, we reported a 15% of CDKN2A mutations among early onset and multiple primary melanoma cases suggesting a more widespread genetic susceptibility to melanoma within this population.5 We thus performed a comprehensive CDKN2A and CDK4 mutation analysis of all melanoma patients evaluated at a single hospital in Athens, Greece.
Methods
The Study was approved by the Ethics Committee of A.Sygros Hospital and the Institutional Review Board at the Massachusetts General Hospital (legacy #94-138). Between January 2004 and December 2008, 319 single primary melanoma (SPM) cases along with 16 genetically-enriched cases (7 multiple primary melanoma (MPM) and 9 familial melanoma (FM) patients) were evaluated at the Melanoma Clinic of the A.Sygros Hospital in Athens, Greece and were enrolled in the study. All participants donated 3ml of peripheral blood for mutation analysis.
Genomic DNA was extracted from peripheral blood leucocytes cells using commercially available kits (QIAGEN miniblood kit, Hilden, Germany). We performed direct sequencing of CDKN2A exons 1α, 1β and 2 and of CDK4 gene using primers and conditions that have been previously published.6 We also performed direct sequencing for exons1β and for CDK4 gene, using the following sets of primers: for exon1β: p14ARF-96F(5′GCTCAGGGAAGGCGGGTGC 3′), and P14ARF-644R (5′AGGGCTGTGTGAAGGGAGGT-3′), for CDK4 exon2: CDK4-2AF(5′GCTGCAGGTCATACCAATCCT-3′), CDK4-2AR (5′CTCTCACACTCTTGAGGGCC3′).
The MGH familial melanoma cohort has been previously described in detail. 7, 8, 9 DNA was extracted from peripheral blood leucocytes and subjected to the same mutational analysis as listed above. Statistical analyses were performed with SigmaStat.
Results
In our sample there was a relatively even representation between men and women (49.2% men vs 50.8% women). The median age at diagnosis was 55 years. The most common histologic type was superficial spreading melanoma, which corresponded to 57.3% of the melanoma cases. The median Breslow thickness was 1.75 mm. Regarding the genetically-enriched Greek population, there were 9 probands from distinct familial melanoma pedigrees-4 individuals had one 1st degree affected relative, 2 had a second degree affected relative, 2 patients had a 1st and a 2nd degree affected relative and 1 patient with both FM and MPM came from a very loaded family with at least 6 family member affected by melanoma.
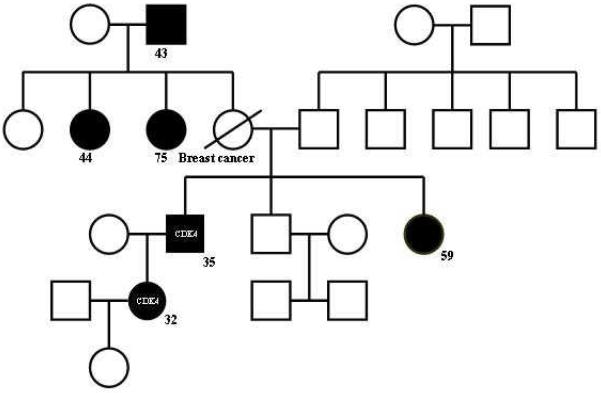
Three hundred and four of 319 SPM cases and all 16 FH and MPM cases had amplifiable DNA for analysis. Despite multiple attempts, 13 samples failed CDK4 amplification and 36 samples failed exon1β amplification. The mutations detected in the Greek sample set are summarized in Table 1. Overall, we detected 9 CDKN2A variants in 16 melanoma patients and 1 CDK4 alteration but no ARF-specific changes. Ten of 304 sporadic cases (3.29%), 4 of 7 multiple melanoma cases (57.2%) and 2 of 9 familial cases (22.2%) harbored CDKN2A mutations, which included 5 previously unreported variants: −34G>C, c.41_43delins20bp, c.301G>C(p.G101R), c.301G>A(p.G101E) and c.296_297insGACC(Table 1). We also detected a CDK4 p.R24H substitution in one patient with familial melanoma and multiple primary tumors, the first of which occurred at age of 54. The patient carried the p.R24H mutation. Five additional members of his family had also developed melanoma (Figure 1). Finally, the c.442 G>A (p.A148T) polymorphism was identified in 22/304 sporadic cases (7.2%) and in 3/9 familial cases (33.3%).
TABLE 1. CDKN2A/CDK4 mutations detected in 17 Greek melanoma patients.
| Gene | Site | Nucleotide position | Effect on p16/ARF |
CDKN2A/CDK4 Mutations (n=17) |
|---|---|---|---|---|
| CDKN2A | Exon 1α |
−34G>C* |
Not known/ Presumably none |
1 SPM |
| CDKN2A | Exon 1α |
−25C>T |
Not known/ Presumably none |
1 SPM |
| CDKN2A | Exon 1α |
c.71G>C | p.R24P/none | 5 SPM 3 MPM |
| CDKN2A | Exon 1α |
c.41- 43delinsCCGTGGCTGGCCACGGCCAC* |
Frameshift | 1 MPM+FM |
| CDKN2A | Exon 2 | c.259C>T | p.R87W/p.P142L | 1 SPM |
| CDKN2A | Exon 2 | c.301G>C* | p.G101R/p.R156P | 1 FM |
| CDKN2A | Exon 2 | c.302G>A* | p.G101E/none | 1 SPM |
| CDKN2A | Exon 2 | c.330G>A | p.W110X/p.G125R | 1 MPM |
| CDKN2A | Exon 2 | c.296_297insGACC* | Frameshift | 1 SPM |
| CDK4 | Exon2 | c.71G->A | p.R24H (Cdk4) | 1 FM |
Novel mutations
Figure 1.
Pedigree of family with CDK4 p.R24H mutation
In terms of prevalence, the frequency of CDKN2A/CDK4 mutations among Greek patients with a SPM was 3.29% (10/304; Table 2). Although not directly comparable due to the differences in the study design and the targeted population (hospital based study vs population based study) our study suggests a higher rate of CDKN2A mutations on sporadic cases compared with a SPM cohort from the Genes,Environment and Melanoma (GEM) consortium (3.29% vs 1.24%, p=0.011 chi square test). Individuals with a single primary lesion accounted for the vast majority of patients since familial and MPM cases account for only 3.7% and 2.8% of the patients evaluated in the A. Sygros melanoma clinic, respectively (data not shown). Strikingly, the observed rate of CDKN2A/CDK4 mutations reached 22.2% (3/9) for those patients with FM and 57.0% (4/7) for those with MPMs (total for high-risk, or genetically-enriched, cases: 7/16, 43.8%). For comparison, we analyzed 236 unrelated familial melanoma patients from a Boston-area melanoma clinic (a largely Northern European cohort) and found 14 probands from unrelated families (5.93%; 14/236) who harbored germline CDKN2A mutations (some of these families were published earlier) 8- a rate that is significantly lower than the Greek mutation rate for high-risk individuals (43.8% vs. 5.93%, p<0.001, chi square test). Likewise, the rate of CDKN2A/CDK4 mutations in the Greek high risk population was also much higher than that reported for an MPM cohort from the GEM study (43.8% vs. 2.94%, p<0.001, chi square test), which carried out the largest MPM analysis to date.10
TABLE 2.
Comparison of CDKN2A/CDK4 mutations in various cohorts
| Greek SPM (this study) |
GEM SPM |
Greek FH/MPM (this study) |
MGH FH1 |
GEM MPM |
|
|---|---|---|---|---|---|
| N | 304 | 2424 | 16 | 236 | 1189 |
|
CDKN2A mutations |
10 | 30 | 6 | 14 | 35 |
|
CDK4 mutations |
0 | ? | 1 | 0 | ND |
| CDKN2A-/CDK4- | 294 | 2394 | 9 | 222 | 1154 |
| % | 3.29 | 1.24 | 43.8 | 5.93 | 2.94 |
MPM: Multliple Primary Melanomas ; FH: Family History; SPM: Sporadic Primary Melanoma, ND: Not Done
Fourteen mutations from distinct families detected in Boston cohort including c.32_33ins9-32 (N=3), p.W15X (N=2), p.M53I (N=4), p.M53V (N=1), p.G101W (N=1), c.240_253del14 (N=2) and p.V126D (N=1); details of cohort can be found in Ref. 3 (Greek); Ref 7-9 (MGH) and Ref. 2, 10 (GEM).
Discussion
Unlike the Northern European populations, there have been sparse data about the genetic landscape of Greek melanoma patients. Overall, we detected 16 CDKN2A variants and 1 CDK4 alteration. It is noteworthy that several Greek CDKN2A mutations have been previously described in other European populations, including the p.R24P mutation, which is the most common mutation in our Greek population (N=8). This differs from another Mediterranean cohort- the Ligurian melanoma kindreds in Italy- where a single founder p.G101W alteration is frequently observed.11 The presence of a Northern European allele suggests that migration from Northern Europe may have introduced the CDKN2A mutations into the Greek population as opposed to a locally pervasive founder alteration that arose around Greece. Nevertheless, there were CDKN2A changes that appear unique to the Greek population. A previously described non-coding variant was also found (−25C>T). In various reporter assays, this 5-UTR substitution had a weak-moderate effect on transcript levels. At best, this change would be a low-risk. 12
Five CDKN2A variants have not been previously described to the best of our knowledge. The two frameshift mutations (c.41-43delinsCCGTGGCTGGCCACGGCCAC and c.296_297insGACC) are likely deleterious. In addition, two distinct p.G101 mutations- p.G101R and p.G101E-were identified. Together with the aforementioned p.G101W founder change, codon 101 appears to be a hotspot for mutagenesis. The transcriptional effect of the novel exon1α promoter (−34G>C) is not known though a −34G>T transition creates an alternative start codon and is associated with melanoma risk.13 Given their unique nature, these novel CDKN2A alterations may represent private mutations.
We also detected the CDK4 p.R24H substitution in one male with MPM who belongs in family with 6 affected members; his daughter is also affected and a carrier. Alterations in the CDK4 gene render the protein kinase resistant to p16 inhibition and represent a rare cause of familial melanoma. To date, two mutations (ie. p.R24H and p.R24C) and only a handful of families bearing a CDK4 mutation have been described. 14, 15 To the best of our knowledge, this is the first report of a CDK4 mutation in a Greek family.
Taken together, our study supports a role for CDKN2A and CDK4 mutations in susceptibility to melanoma among Greek individuals. We hypothesized that genetic input may have broader influence in low ambient risk regions and indeed found a higher prevalence of CDKN2A/CDK4 mutations in both sporadic and genetically-enriched cases from the Hellenic region. Although the role of genetic screening in the management of melanoma is unclear, genetic testing could be considered for Greek patients at hereditary risk for melanoma, especially in those patients who harbor a family history or those individuals with MPMs.
What’s already known about this topic? What does this study add?
The penetrance of CDKN2A mutations is subject to geographic variation and is presumably dictated by UVR exposure and possibly other co-inherited genetic factors.
Our study provides information on the contribution of genetic factors in melanoma risk in a large clinic population of Greek patients with melanoma
A comparison with reported cohorts revealed a higher mutation rate in both single primary melanoma patients and genetically enriched cases
The results support the hypothesis that genetic risk factors may have a stronger influence in melanoma development in populations residing in low prevalence areas
ACKNOWLEDGEMENTS
This activity was supported in part by the NIH (P50 CA93683, CA083115-06, K24 CA149202-01), the American Cancer Society (RSG-07-085-01-MGO) and the generous donors to the MGH Millennium Melanoma Fund.
Funding Sources: This activity was supported in part by the NIH (K24 CA149202-01 to HT), the American Cancer Society (RSG-07-085-01-MGO to HT) and the generous donors to the MGH Millennium Melanoma Fund.
Footnotes
Conflicts of interest: None declared
References
- 1.Goldstein AM, Chan M, Harland M, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet. 2007;44:99–106. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berwick M, Orlow I, Hummer AJ, et al. The prevalence of CDKN2A germ-line mutations and relative risk for cutaneous malignant melanoma: an international population-based study. Cancer Epidemiol Biomarkers Prev. 2006;15:1520–1525. doi: 10.1158/1055-9965.EPI-06-0270. [DOI] [PubMed] [Google Scholar]
- 3.Stefanaki I, Stratigos AJ, Dimisianos G, et al. p53 codon 72 Pro homozygosity increases the risk of cutaneous melanoma in individuals with dark skin complexion and among noncarriers of melanocortin 1 receptor red hair variants. Br J Dermatol. 2007;156:357–362. doi: 10.1111/j.1365-2133.2006.07645.x. [DOI] [PubMed] [Google Scholar]
- 4.Stratigos AJ, Dimisianos G, Nikolaou V, et al. Melanocortin receptor-1 gene polymorphisms and the risk of cutaneous melanoma in a low-risk southern European population. J Invest Dermatol. 2006;126:1842–1849. doi: 10.1038/sj.jid.5700292. [DOI] [PubMed] [Google Scholar]
- 5.Stratigos AJ, Yang G, Dimisianos R, et al. Germline CDKN2A mutations among Greek patients with early-onset and multiple primary cutaneous melanoma. J Invest Dermatol. 2006;126:399–401. doi: 10.1038/sj.jid.5700078. [DOI] [PubMed] [Google Scholar]
- 6.Hogg D, Liu L, Lassam N. Genetic testing in familial melanoma. In: Nickoloff BJ, editor. Melanoma: Methods and Protocols. Vol. 61. Humana Press Inc.; Totowa, N.J.: 2001. [DOI] [PubMed] [Google Scholar]
- 7.Lang JM, Shennan M, Njauw JC, et al. A flexible multiplex bead-based assay for detecting germline CDKN2A and CDK4 variants in melanoma-prone kindreds. J Invest Dermatol. 2011;131:480–486. doi: 10.1038/jid.2010.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niendorf KB, Goggins W, Yang G, et al. MELPREDICT: a logistic regression model to estimate CDKN2A carrier probability. J Med Genet. 2006;43:501–506. doi: 10.1136/jmg.2005.032441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Niendorf KB, Patel D, et al. Estimating CDKN2A carrier probability and personalizing cancer risk assessments in hereditary melanoma using MelaPRO. Cancer Res. 2010;70:552–559. doi: 10.1158/0008-5472.CAN-09-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orlow I, Begg CB, Cotignola J, et al. CDKN2A germline mutations in individuals with cutaneous malignant melanoma. J Invest Dermatol. 2007;127:1234–1243. doi: 10.1038/sj.jid.5700689. [DOI] [PubMed] [Google Scholar]
- 11.Ghiorzo P, Ciotti P, Mantelli M, et al. Characterization of ligurian melanoma families and risk of occurrence of other neoplasia. Int J Cancer. 1999;83:441–448. doi: 10.1002/(sici)1097-0215(19991112)83:4<441::aid-ijc2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 12.Bisio A, Nasti S, Jordan JJ, et al. Functional analysis of CDKN2A/p16INK4a 5′-UTR variants predisposing to melanoma. Hum Mol Genet. 2010;19:1479–91. doi: 10.1093/hmg/ddq022. [DOI] [PubMed] [Google Scholar]
- 13.Liu L, Dilworth D, Gao L, et al. Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat Genet. 1999;21:128–132. doi: 10.1038/5082. [DOI] [PubMed] [Google Scholar]
- 14.Soufir N, Avril MF, Chompret A, et al. Prevalence of p16 and CDK4 germline mutations in 48 melanoma-prone families in France. Human Molecular Genetic. 1998;7:209–216. doi: 10.1093/hmg/7.2.209. [DOI] [PubMed] [Google Scholar]
- 15.Zuo L, Weger J, Yang Q, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nature Genetics. 1996;12:97–99. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]