Abstract
Carbonyl reductase activity catalyzes the two electron reduction of several endogenous and exogenous carbonyl substrates. Recent data indicate that the expression of human carbonyl reductase 3 (CBR3) is regulated by the master redox switch Nrf2. Nrf2 binds to conserved antioxidant response elements (AREs) in the promoters of target genes. The presence of functional AREs in the CBR3 promoter has not yet been reported. In this study, experiments with reporter constructs showed that the prototypical Nrf2 activator tert-butyl hydroquinone (t-BHQ) induces CBR3 promoter activity in cultures of HepG2 (2.7-fold; p<0.05) and MCF-7 cells (22-fold; p<0.01). Computational searches identified a conserved ARE in the distal CBR3 promoter region (−2698ARE). Deletion of this ARE from a 4212-bp CBR3 promoter construct impacted basal promoter activity and induction of promoter activity in response to treatment with t-BHQ. Deletion of −2698ARE also impacted the induction of CBR3 promoter activity in cells overexpressing Nrf2. Electrophoretic mobility shift assays (EMSA) demonstrated increased binding of specific protein complexes to −2698ARE in nuclear extracts from t-BHQ treated cells. The presence of Nrf2 in the specific nuclear protein-−2698ARE complexes was evidenced in EMSA experiments with anti-Nrf2 antibodies. These data suggest that the distal −2698ARE mediates the induction of human CBR3 in response to prototypical activators of Nrf2.
1. INTRODUCTION
Carbonyl reductase activity plays a crucial role during the reduction of several endogenous and exogenous carbonyl compounds [1]. In humans, there are three carbonyl reductases, CBR1, CBR3 and CBR4. CBR1 and CBR3 are cytosolic monomeric enzymes, whereas CBR4 forms a mitochondrial heterotetramer with 17β-hydroxysteroid dehydrogenase type 8 (17β-HSD8) [2, 3]. The CBR1 and CBR3 genes are located in chromosome 21 (CBR1, 21q22.13 and CBR3, 21q22.22), where they are separated by a relatively short distance of 62 kb. CBR3 catalyzes the reduction of 1,2-naphthoquinone, isatin, oracin, coniferyl aldehyde and acetohexamide [4, 5]. The anticancer anthracyclines doxorubicin and daunorubicin are also reduced by CBR3 into their corresponding C-13 alcohol metabolites doxorubicinol and daunorubicinol. Recent studies suggest that genetic polymorphisms in CBR3 contribute to the variable toxicodynamics of anthracycline drugs in cancer patients [6-9].
CBR3 is expressed in various tissues including heart, liver, kidney, spleen, lung and brain [5]. Variable CBR3 expression at the mRNA and protein level has been documented in liver [10, 11]. Absolute quantification of CBR3 with a new liquid chromatography/mass spectrometry assay showed that protein expression varied by 14-fold (range: 1.3 -17.9 ppm) [11]. Recent research has provided the first insights into the molecular mechanisms that dictate variable CBR3 expression. Several lines of evidence indicate that CBR3 expression is modulated by the transcription factor Nrf2 (nuclear factor [erythroid-derived 2]-like 2, official symbol: NFE2L2) [12-17]. For example, Hu et al. examined gene expression profiles in livers from nrf2 wild type and nfr2 knockout mice after treatment with the Nrf2 activator sulforaphane. The authors documented significant Cbr3 mRNA induction in livers of nfr2 proficient animals relative to the levels of transcript in nrf2 deficient mice (e.g., 4.0-fold after 3h treatment) [12]. More recently, Ebert et al. have shown that Nrf2 also mediates the induction of CBR3 in human cancer cell lines after treatment with prototypical Nrf2 activators (e.g., sulphorane and diethylmaleate) [14]. It is known that Nrf2 coordinates the induction of a battery of stress-responsive genes including NAD(P)H:quinone oxidoreductase-1 (NQO1), superoxide dismutase (SOD), glutathione S-transferases (GSTs) and glutathione peroxidases (GPXs) [18]. Mutational analyses identified a conserved DNA sequence element known as an antioxidant response element (ARE) in the promoter of genes regulated by Nrf2. Under conditions of oxidative stress, Nrf2 translocates to the nucleus and forms heterodimers with other transcription factors such as small Maf proteins. The Nrf2 heterodimers bind to functional AREs and mediate the transcriptional activation of a wide range of target genes encoding stress-responsive proteins [18-21].
The promoter of human CBR3 has been recently identified [10]. To the best of our knowledge, the functional role of conserved AREs in the CBR3 promoter region has not yet been elucidated. This study documents the identification and functional characterization of a conserved ARE (−2698ARE) in the promoter region of human CBR3.
2. MATERIALS AND METHODS
2.1 Cell culture and reagents
HepG2 (human hepatoblastoma, HB-8065), and MCF-7 (human breast adenocarcinoma, HTB-22) cell lines were obtained from the American Type Culture Collection (Manassas, VA). RPMI 1640 and other cell culture reagents were purchased from Invitrogen (Carlsbad, CA). Cells were routinely cultured in 10 cm2 dishes or 48-well plates using RPMI 1640 medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Sigma-Aldrich, St. Louis, MO), 100 U/ml penicillin, and 100 μg/ml streptomycin. Cultures were grown in an incubator at 37°C, 5% CO2, and 95% relative humidity. Cultures were maintained at low passage numbers (n < 12) and were free of mycoplasma contamination. Tert-butyl hydroquinone (t-BHQ) was purchased from Sigma-Aldrich.
2.2 Quantification of CBR3 mRNA
Total RNA (100 ng) was reverse-transcribed and amplified using one-step QuantiTect SYBR Green RT-PCR kits (Qiagen, Valencia, CA) with the following primers: 5′-GCTTCCACCAACTGGACATC-3′ (forward) and 5′-GGGCATTGGATCATCACTCT-3′ (reverse). Melting curve analyses demonstrated a unique PCR amplification product. Amplification reactions were performed as described [10]. In brief, relative CBR3 mRNA levels were determined by the comparative quantitation method using individual β-actin mRNA levels as normalizers. Amplification efficiencies for CBR3 and β-actin mRNAs were similar. Standard curves for both mRNAs were run in parallel (20-fold dynamic range, r2 ≥ 0.96). Experimental samples and standards for calibration curves were analyzed in quadruplicate [22].
2.3 Detection of CBR3 protein by immunoblotting
HepG2 cells (80% confluence) were incubated with DMSO (0.1% v/v) or 50 μM t-BHQ for 18 h. HepG2 cell lysates were prepared with ice-cold Pierce RIPA buffer (Thermo Fisher Scientific, Rockford, IL) supplemented with Halt Protease and Halt Phosphatase inhibitor cocktails (Thermo Fisher Scientific). The concentration of total proteins was determined with the BCA Protein Assay Reagent (Thermo Fisher Scientific). HepG2 cell lysates (50 μg of protein/lane) were loaded into 12% precast polyacrylamide gels (Invitrogen, Carlsbad, CA) and separated by electrophoresis. Protein blots were probed with a specific polyclonal anti-human CBR3 antibody (1: 2,000; sc-70220, Santa Cruz Biotechnology, Santa Cruz, CA) and a secondary goat anti-rabbit IgG antibody conjugated with horseradish peroxidase (1: 10,000; Sigma-Aldrich). Membranes were also probed with anti-β-actin antibody (1: 10,000; Santa Cruz) to correct for differences in protein loading. Immunoreactive bands were visualized with the ECL Plus Western blotting detection system (GE Healthcare, Chalfont St. Giles, UK). CBR3 and β-actin band intensity values (pixels/mm2) were quantified with a Molecular Imager® Gel Doc™ XR+ System (Bio-Rad Laboratories, Hercules, CA).
2.4 Reporter gene studies
DNA sequence upstream (4212 bp) from the translation start codon of CBR3 (A+1TG) was amplified by PCR from human DNA sample HD17249 (Coriell Institute for Medical Research, Camden, NJ). The amplicon was cloned into a pGL3 basic firefly luciferase vector (Promega, Madison, WI). The ARE core sequence (5′-GTGACCCTGC-3′; Fig. 1) was deleted from the −4212CBR3 construct by site-directed mutagenesis (QuikChange, Stratagene, La Jolla, CA) with the following primers: 5′-GCTGGGTTTGTTGCCAAACTTATAGAGACCCTTCG-3′ (forward) and 5′-CGAAGGGTCTCTATAAGTTTGGCAACAAACCCAGC-3′ (reverse). The human Nrf2 expression plasmid pNrf2-CMV6-XL5 was purchased from OriGene (Rockville, MD). The empty CMV6-XL5 vector was generated by removal of the Nrf2 insert from pNrf2-CMV6-XL5 with the restriction enzyme NotI-HF™ (New England Biolabs, Ipswich, MA). The identity of all constructs and the absence of mutations were verified by direct sequencing.
Figure 1.
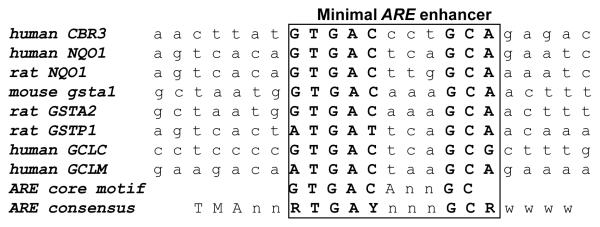
DNA sequence alignment for −2698ARE in the promoter of human CBR3 and AREs in the promoters of prototypical target genes. The conserved −2698ARE encompasses the −2704 to −2695 bp region upstream the translation start codon (A+1TG). Nucleotides in bold capitals indicate the consensus ARE sequence. ARE sequences are taken from Nioi and Hayes [20].
HepG2 and MCF-7 cells were plated 24 h before transfections in 48-well plates. Cell cultures at 60~80% confluence were co-transfected with CBR3 reporter constructs (250 ng) and the internal control plasmid pRL-TK (50 ng; Promega). In some experiments, cultures were co-transfected with various amounts of the Nrf2 expression plasmid (1, 10, and 100 ng), and the total amount of transfected DNA was adjusted up to 100 ng with the empty pCMV6-XL5 vector. The pGL3-basic empty vector was transfected into control cultures to correct for background luciferase activity. Firefly luciferase activities from each reporter construct and from the pGL3-basic empty vector were first normalized to their corresponding renilla reniformis luciferase activities. Normalized luciferase activities from each reporter construct were corrected by subtracting the mean luciferase activity from the pGL3-basic empty vector. Luciferase reporter gene activities were determined 24h after co-transfections with the Dual-Luciferase Reporter Assay System (Promega). Light intensities were measured in a Synergy HT luminometer (BioTek, Winooski, VT). In all cases, three independent experiments were performed in triplicate to evaluate reproducibility. Unpaired Student’s t tests were used to compare experimental means. Differences were considered to be significant at p<0.05. Computations were performed with Microsoft Excel 2007 (Redmond, WA) and GraphPad Prism, version 4.03 (GraphPad Software Inc., San Diego, CA).
2.5 Electrophoretic mobility shift assays (EMSA)
Nuclear extracts from HepG2 cells were prepared with the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific). The −2698ARE oligonucleotides: 5′-CAAACTTATGTGACCCTGCAGAGACC-3′ (−2698ARE-CBR3 forward) and 5′-GGTCTCTGCAGGGTCACATAAGTTTG-3′ (−2698ARE-CBR3 reverse) were synthesized and purified by Invitrogen. The following antibodies: anti-human Nrf2 C-20x (sc-722x, rabbit IgG), anti-human Nrf2 H-300x (sc-13032x, rabbit IgG), and anti-human aryl hydrocarbon receptor nuclear translocator 2 (Arnt 2; sc-5581x, M-165x, rabbit IgG) were purchased from Santa Cruz Biotechnology. Normal rabbit IgG suitable for EMSA (code number: 312-005-003) was obtained from Jackson Immunoresearch Laboratories (West Grove, PA) [23]. EMSA were performed with the LightShift Chemiluminescent Kit (Thermo Fisher Scientific) by following the manufacturer’s instructions. In brief, −2698ARE-CBR3 forward and −2698ARE-CBR3 reverse oligonucleotides were labeled with Biotin 3′ End DNA Labeling kits (Thermo Fisher Scientific). Equimolar amounts of biotin-labeled −2698ARE-CBR3 forward and −2698ARE-CBR3 reverse were annealed to generate double stranded oligonucleotides. Double stranded oligonucleotides (100 pmole) were incubated with nuclear extracts (10 μg) for 45 min at 4°C. Competition studies were performed by adding a 50-fold molar excess of unlabeled “wild type” or “mutant” −2698ARE-CBR3 oligonucleotides to the reaction mixtures.
Modified −2698ARE-CBR3 oligonucleotides:
−2698ARE-CBR3 mt-f (−2698ΔARE, forward)
5′-GCTGGGTTTGTTGCCAAACTTATAGAGACCCTTCG -3′
−2698ARE-CBR3 mt-r (−2698ΔARE, reverse)
5′- CGAAGGGTCTCTATAAGTTTGGCAACAAACCCAGC -3′
EMSAs were performed by first incubating double stranded −2698ARE-CBR3 oligonucleotides with nuclear extracts for 15 min followed by 30 min. incubation with the antibody (1 and 3 μg) at 4°C. In all cases, incubation mixtures (20 μl per binding reaction) were analyzed by electrophoresis on 5% non-denaturing polyacrylamide gels with 0.5X Tris/borate/EDTA buffer at 4°C. DNA-protein complexes were visualized with a Molecular Imager® Gel Doc™ XR+ System (Bio-Rad).
3. RESULTS
3.1 Identification of a conserved ARE in the human CBR3 promoter region
The presence of the ARE consensus (5′- TA/CAnnA/GTGAC/TnnnGCA/GA/TA/TA/TA/T-3′) and ARE core motifs (5′- TGACnnnGC-3′) in the promoter region of human CBR3 was investigated with the program FindPatterns. Four kilobases of DNA sequence upstream the translation initiation codon (A+1TG) of CBR3 were analyzed [19, 20]. A conserved ARE consensus sequence (100% match) was pinpointed at −2698 bp (−2698ARE). Comparisons revealed that −2698ARE was similar to prototypical AREs in the promoters of various Nrf2 target genes including NQO1, glutathione S-transferases (GSTs), and glutamate-cysteine ligase (GCLC) (Fig. 1) [18, 20].
3.2 Induction of CBR3 expression by the Nrf2 activator t-BHQ
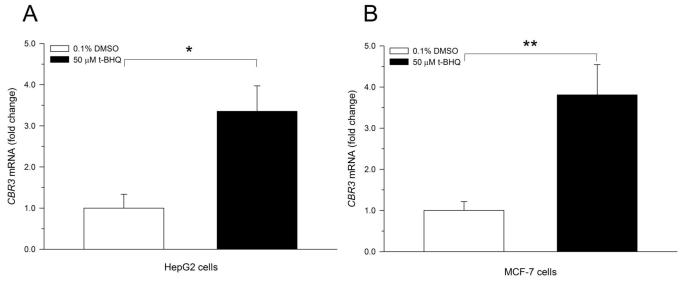
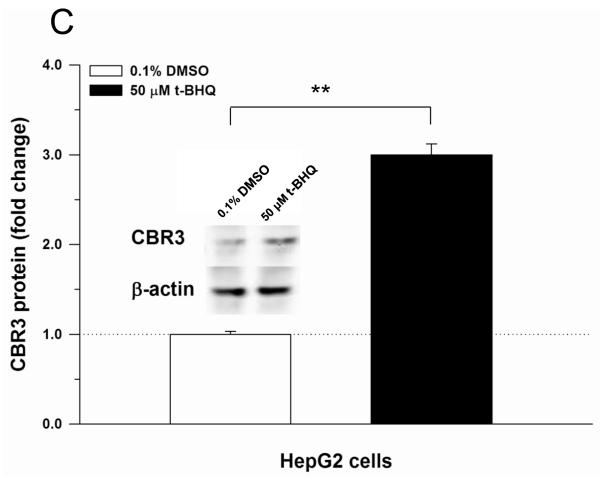
Recently, Ebert et al. described induction of CBR3 mRNA expression in cancer cell lines treated with the prototypical Nrf2 activator t-BHQ [14]. In this study, confirmatory experiments showed that t-BHQ (50 μM, 18 h) significantly induced CBR3 mRNA expression in cultures of HepG2 (3.4-fold, p<0.05) and MCF-7 cells (3.8-fold, p<0.01) (Fig. 2A and B). In HepG2 cells, immunoblotting analysis showed that t-BHQ treatment induced CBR3 protein levels by 3-fold (p<0.01) in comparison to vehicle treated controls (DMSO, 0.1% v/v. Fig. 2C).
Figure 2.
Induction of CBR3 mRNA by the Nrf2 activator t-BHQ in HepG2 (A) and MCF-7 cells (B). Induction of CBR3 protein expression in HepG2 cells by t-BHQ (C). Cells were grown in monolayer cultures to sub-confluence and treated with DMSO (0.1%, v/v) or 50 μM t-BHQ for 18 h. The expression of CBR3 mRNA and CBR3 protein was analyzed by quantitative real-time RT-PCR and immunoblotting as described under Materials and Methods. Each value represents the mean ± S.D. from 3 independent experiments analyzed in triplicates. Asterisks indicate significant differences from CBR3 mRNA (A and B) and CBR3 protein (C) levels from vehicle treated cells (*, p<0.05; **, p<0.01).
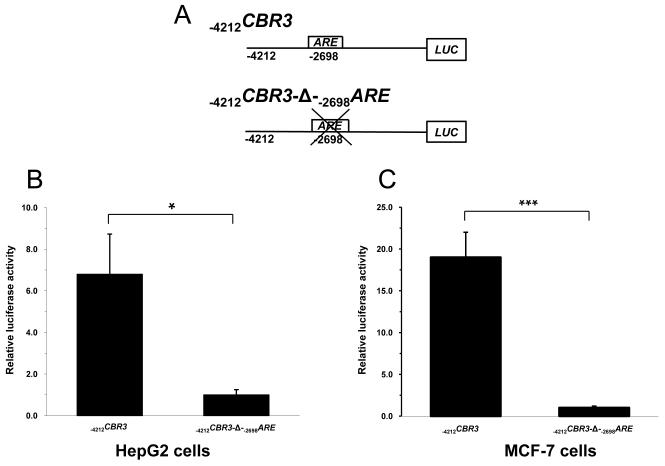
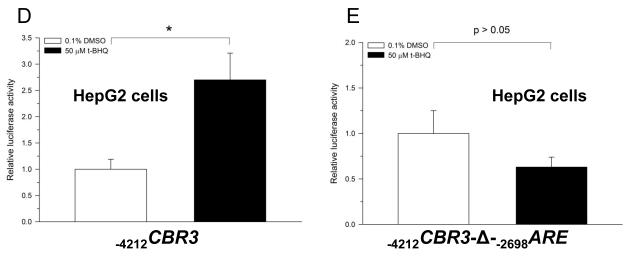
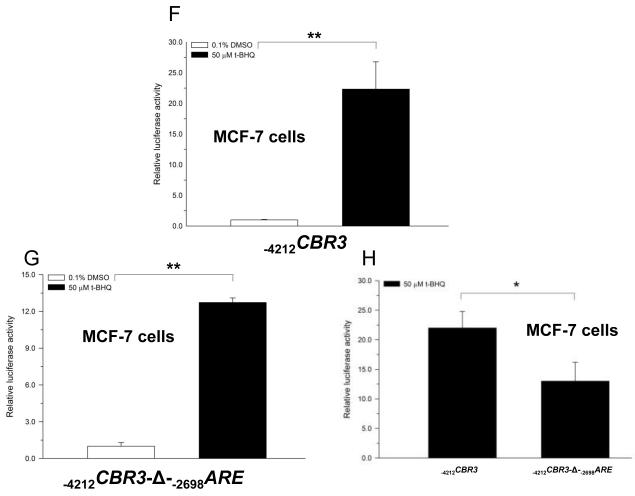
The functional impact of the distal −2698ARE in mediating the transcriptional regulation of CBR3 was investigated by performing gene reporter experiments with engineered promoter constructs. First, cultures of HepG2 cells and MCF-7 cells were transfected with an unmodified CBR3 promoter construct encompassing 4212 bp of the 5′ flanking region (−4212CBR3) or with a CBR3 promoter construct lacking the −2698ARE (−4212CBR3-Δ-−2698ARE) (Fig. 3A). Under basal conditions, the gene reporter activities of the unmodified CBR3 promoter construct were approximately 7- (HepG2; p<0.05) and 17-fold (MCF-7; p<0.001) higher than the reporter activities of the CBR3 construct without the distal −2698ARE (Fig. 3B and C). Next, cultures of cells transfected with −4212CBR3 or −4212CBR3-Δ-−2698ARE reporter constructs were incubated with t-BHQ (50 μM, 18 h). In HepG2 cells, t-BHQ treatment increased the reporter activity of −4212CBR3 by almost 3-fold in comparison to vehicle (DMSO, 0.1% v/v) treated controls (p<0.05) (Fig. 3D). T-BHQ treatment failed to induce the gene reporter activity of the modified −4212CBR3-Δ-−2698ARE construct (p>0.05) (Fig. 3E). In MCF-7 cells, t-BHQ treatment induced the reporter activity of the intact −4212CBR3 construct by 22-fold (p<0.01) (Fig. 3F). In contrast, t-BHQ treatment induced the reporter activity of the modified −4212CBR3-Δ-−2698ARE construct by 13-fold (p<0.01). Figure 3H shows that deletion of −2698ARE significantly decreased the induction of CBR3 reporter activity by t-BHQ in MCF-7 cells (p<0.05. Fig. 3H).
Figure 3.
Schematic representations of the CBR3 promoter constructs (A). Impact of the distal −2698ARE in mediating basal gene reporter activity of CBR3 promoter constructs in HepG2 (B) and MCF7 cells (C). Firefly luciferase activities from each reporter construct and from the pGL3-basic empty vector were first normalized to their corresponding renilla reniformis luciferase activities. Normalized luciferase activities from each reporter construct were corrected by subtracting the mean luciferase activity from the pGL3-basic empty vector. Luciferase activities were expressed as fold increases with respect to the values from transfections with the modified −4212CBR3-Δ-−2698ARE promoter construct, which was set arbitrarily at 1. Effect of t-BHQ on the luciferase activities of the intact −4212CBR3 and modified −4212CBR3-Δ-−2698ARE promoter constructs in HepG2 (D and E) and MCF-7 cells (F and G). For comparison, panel H shows luciferase activities for the intact and modified CBR3 promoter constructs after t-BHQ treatment in MCF-7 cells. Normalized luciferase activities were expressed as fold induction with respect to the values from vehicle incubations, which were set arbitrarily at 1. Each value represents the mean ± SD from 3 independent experiments performed in triplicates. The asterisks indicate significant differences between the luciferase activities exerted by both constructs (*, p<0.05; **, p<0.01; ***, p<0.001).
3.3 Transcriptional activation of CBR3 promoter constructs by Nrf2 over-expression
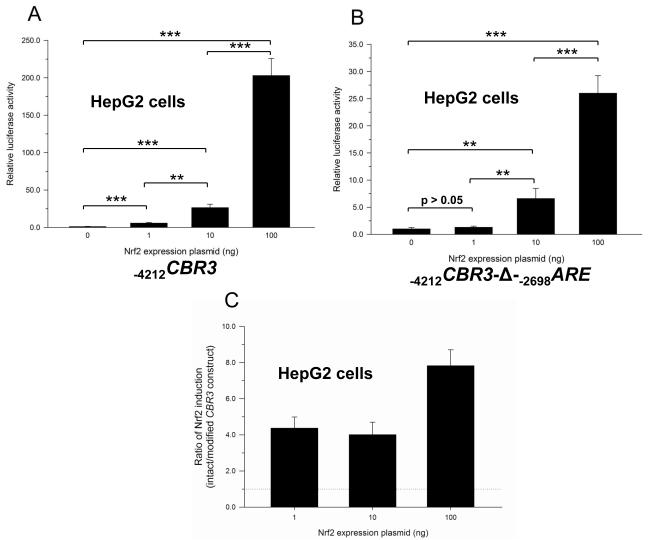
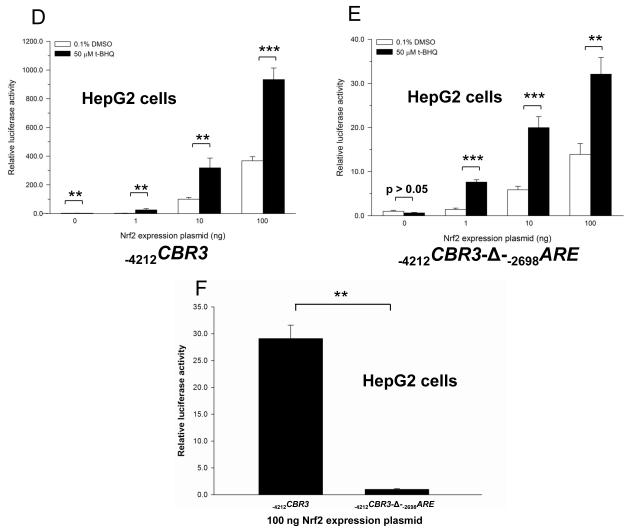
Wang and Jaiswall showed that over-expression of Nrf2 increases the reporter activity of the human NRH:quinone oxidoreductase 2 gene promoter (NQO2) [24]. Similarly, over-expression of Nrf2 in HepG2 cells increased the reporter activity of the intact −4212CBR3 promoter construct in a dose dependent manner (Fig. 4A). Nrf2 over-expression also induced the reporter activity of the modified −4212CBR3-Δ-−2698ARE promoter construct in a dose dependent manner (Fig. 4B). However, the lack of −2698ARE in the modified CBR3 promoter construct resulted in lower induction of reporter activity as a consequence of Nr2 over-expression. In this context, the modified CBR3 construct showed 4- (1 ng Nrf2 plasmid), 4- (10 ng Nrf2 plasmid) and 8-fold (100 ng Nrf2 plasmid) lower gene reporter activities than the intact construct (Fig. 4C). Similar trends were documented in experiments with MCF-7 cells (data not shown). Additional experiments with HepG2 cells showed that Nrf2 over-expression coupled to t-BHQ treatment further increased the reporter activity of the intact CBR3 construct in comparison to vehicle treated controls. For example, t-BHQ treatment induced the gene reporter activity of the intact −4212CBR3 promoter construct by over 900-fold in cells transfected with 100 ng of Nrf2 plasmid (Fig. 4D). In similar conditions, the magnitude of induction of reporter activity by t-BHQ in cells transfected with the modified −4212CBR3-Δ-−2698ARE promoter construct (no distal −2698ARE) was only 32-fold (Fig. 4E). Therefore, deletion of −2698ARE decreased the induction of promoter activity by approximately 29-fold in Nrf2 over-expressing cells (100 ng Nrf2 plasmid) treated with t-BHQ (Fig. 4F).
Figure 4.
Impact of Nrf2 overexpression on the on the luciferase activities of the intact −4212CBR3 (A) and modified −4212CBR3-Δ-−2698ARE promoter constructs (B) in HepG2 cells. For comparison, panel C shows the ratio in luciferase activities from the plots depicted in panels A and B, respectively. Firefly luciferase activities from each reporter construct and from the pGL3-basic empty vector were first normalized to their corresponding renilla reniformis luciferase activities. Normalized luciferase activities from each reporter construct were corrected by subtracting the mean luciferase activity from the pGL3-basic empty vector. Luciferase activities were expressed as fold induction with respect to the values from cells transfected with the empty pCMV6-XL5 vector, which were set arbitrarily at 1. Impact of t-BHQ treatment coupled to Nrf2 overexpression on the luciferase activities of the intact −4212CBR3 (D and F) and modified −4212CBR3-Δ-−2698ARE promoter constructs (E and F). Normalized luciferase activities were expressed as fold induction with respect to the values from vehicle incubations, which were set arbitrarily at 1. Each value represents the mean ± SD from 3 independent experiments performed in triplicates. Asterisks indicate significant differences between luciferase activities (*, p<0.05; **, p<0.01; ***, p<0.001).
3.4 Binding of nuclear proteins to CBR3 −2698ARE
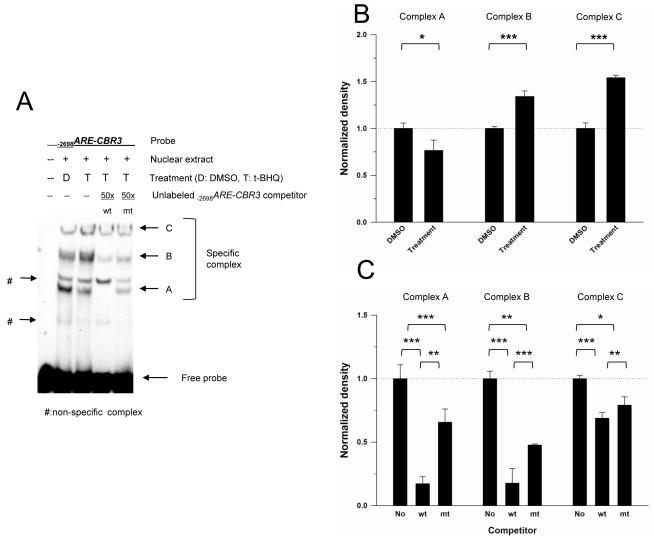
Nuclear extracts from HepG2 cells treated with vehicle or t-BHQ were incubated with biotin labeled −2698ARE-CBR3 oligonucleotides, and examined by EMSA followed by densitometric analysis. Five prominent DNA-nuclear protein complexes were detected (Fig. 5A). Densitometric analysis revealed that extracts from t-BHQ treated cells exhibited more intense bands for DNA-protein complexes B and C than extracts from vehicle treated cells (p<0.01 for both complexes), while nuclear extracts from vehicle treated cells showed a more intense band for DNA-protein complex A than extracts from t-BHQ treated cells (p<0.05. Figs. 5A and B). The specificity of the DNA-protein complexes was examined by performing competition experiments with unlabelled −2698ARE-CBR3 oligonucleotides. Incubations with a 50-fold molar excess of the unlabeled −2698ARE-CBR3 oligonucleotide (“wild type”, wt) diminished the formation of DNA-protein complexes A, B and C (p<0.001 for complexes A, B and C. Figs. 5A and C). Similar competition experiments with an oligonucleotide lacking the −2698ARE core motif (“mutant”, mt) showed relatively less impact on the formation of DNA protein complexes A, B and C than that observed for the “wild type” −2698ARE-CBR3 oligonucleotide. That is, competition with the “mutant” oligonucleotide decreased the intensity of the band for DNA-protein complex A by 34%, while competition with the “wild type” oligonucleotide decreased the intensity of the band for DNA-protein complex A by 83%, respectively (p<0.001. Figs. 5A and B). Also, competition with the “mutant” oligonucleotide decreased the intensity of the band for DNA-protein complex B by 52% whereas competition with the “wild type” oligonucleotide decreased the intensity of the band for complex B by 82% (p<0.01. Figs. 5A and B). Similar trends were observed for DNA-protein complex C (i.e., “mutant”: 21% vs. “wild type”: 31%, p<0.05. Figs. 5A and B). Together, these results suggest that protein(s) in complexes A, B and C bind specifically to −2698ARE-CBR3.
Figure 5.
Formation of specific DNA-nuclear protein complexes as evidenced by EMSA with the −2698ARE-CBR3 oligonucleotide (A). Nuclear extracts from vehicle- (DMSO, 0.1% v/v) and t-BHQ (50 μM) treated HepG2 cells were incubated with the labeled −2698ARE-CBR3 oligonucleotide. Competition experiments with a 50-fold molar excess of the unlabelled “wild type” (wt) and “mutant” (mt) −2698ARE-CBR3 oligonucleotides evidenced the formation of 3 specific complexes (A, B and C. See text for details). Panel B shows the densitometric analysis of complexes A, B, and C from incubations with vehicle and t-BHQ, respectively. Panel C shows the densitometric analysis of complexes A, B and C from incubations with the wt and mt oligonucleotides, respectively. EMSA experiments were repeated 3 times to evaluate reproducibility. Data represent the mean ±SD. Asterisks indicate significant differences between band intensity values (*, p<0.05; **, p<0.01; ***, p<0.001).
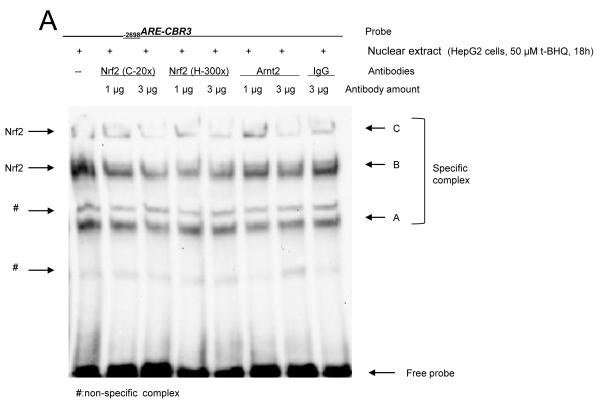
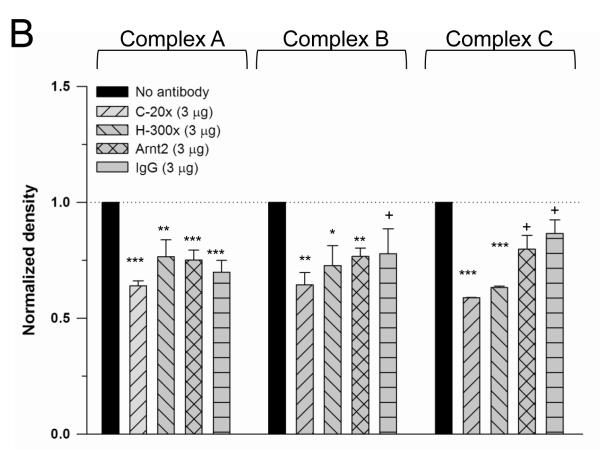
The presence of Nrf2 in the −2698ARE binding complexes was examined by performing EMSA experiments with the anti-Nrf2 antibodies C-20x and H-300x, respectively. The epitope recognized by the C-20x antibody is located at the C-terminus of Nrf2, whereas the H-300x antibody recognizes an epitope located at the N-terminus of the protein. Addition of the anti-Nrf2 antibodies to the binding reactions were expected to either yield larger DNA-protein complexes evident as “supershifted” EMSA bands or to diminish the formation of DNA-protein complexes containing Nrf2 [24, 25]. In line with the latter possibility, EMSA experiments with nuclear extracts from t-BHQ-treated cells showed that incubations with anti Nrf2 antibodies impact the binding of the specific protein complexes to −2698ARE (Fig. 6A). Densitometry analysis showed that incubations with 3 μg of anti-Nrf2 antibodies (C-20x and H-300x) decreased the formation of DNA-protein complex A in comparison to incubations with no antibody (p<0.01 for H-300x and p<0.001 for C-20x. Figs. 6A and B). Similarly, incubations with the anti-Arnt2 antibody M-165x and normal rabbit IgG (isotype control) diminished the formation of DNA-protein A in comparison to incubations with no antibody (p<0.001 for anti-Arnt2 and rabbit IgG, respectively). Therefore, Nrf2 may not be present in DNA-protein complex A (Fig. 6A and B). The formation of DNA–protein complexes B and C decreased significantly in nuclear extracts from t-BHQ treated cells incubated with anti-Nrf2 antibodies in comparison to incubations with no antibodies (complex B: p<0.01 for C-20x, p<0.05 for H-300H; complex C: p<0.001 for C-20x, p<0.001 for H-300). The decrease in band intensity for complexes B and C was more evident in incubations with the C-20x antibody (Figs. 6A and B). Incubations with the rabbit IgG isotype control did not significantly impact the formation of DNA-protein complexes B and C (Figs. 6A and B). These findings suggest that Nrf2 may be present in specific complexes B and C.
Figure 6.
Analysis of DNA-nuclear protein complexes by EMSA with the anti-Nrf2 antibodies C-20x and H-300x (A). EMSAs with anti-Arnt 2 antibody and normal rabbit IgG were included as isotype controls (A). Nuclear extracts from t-BHQ treated (50 μM) HepG2 cells were incubated with the labeled −2698ARE-CBR3 oligonucleotide and antibodies as described in the text. Comparative densitometric analysis for specific complexes A, B and C (B. See text for details). EMSA experiments were repeated 3 times to evaluate reproducibility. In all cases, band intensity values were compared against the band intensity of the “no antibody” reaction, which was set arbitrarily at 1. Data represent the mean ±SD. Asterisks indicate significant differences between band intensity values (*, p<0.05; **, p<0.01; ***, p<0.001). The plus symbol (+) indicates no significant differences (p>0.05).
4. DISCUSSION
A recent study by Ebert et al. on cancer cell lines elegantly demonstrated that Nrf2 is a key mediator for the induction of CBR3 expression in response to prototypical activators of the Nrf2/ARE pathway [14]. Here, we documented significant induction of CBR3 mRNA and CBR3 protein in cells treated with the Nrf2 activator t-BHQ (Fig. 2). Thus, our main goal was to identify functional AREs in the promoter of human CBR3. The results of this study suggest that the distal −2698ARE in the CBR3 promoter acts as a bona fide ARE by mediating the transcriptional regulation of CBR3 through the Nrf2/ARE pathway. Two main lines of evidence lend support to the potential functional role of −2698ARE. First, the gene reporter experiments with engineered CBR3 promoter constructs suggest that −2698ARE may contribute to: 1) sustain basal CBR3 promoter activity, and 2) mediate the transcriptional induction of CBR3 by t-BHQ via Nrf2 activation in two cellular contexts (HepG2 and MCF-7 cells). Second, the binding of specific nuclear protein complexes to −2698ARE-CBR3 oligonucleotides was increased when performing EMSA with nuclear extracts from t-BHQ treated cells. Competitive EMSA experiments with a modified oligonuleotide lacking −2698ARE further highlighted the importance of the core motif (5′-TGACnnnGC-3′) for mediating the binding of Nrf2-containing nuclear protein complexes to DNA [19, 20]. EMSA experiments with anti-Nrf2 antibodies further suggested that Nrf2 may be present in specific complexes B and C. Nuclear factors other than Nrf2 (e.g., Nrf1 and Nrf3) can bind to conserved AREs, and additional studies are needed to determine whether these factors also mediate induction of human CBR3 in response to prototypical activators [26, 27].
Variable basal CBR3 mRNA expression as well as induction by Nrf2 activators has been observed in a panel of cell lines that included HepG2 cells [14]. Based on these observations, Ebert et al. speculated that constitutive differences in the Nrf2/ARE pathway (e.g., mutations in the Nrf2 partner Keap 1) and/or gene silencing mechanisms (e.g., CBR3 promoter hypermethylation) may explain differences in CBR3 expression among model cell lines. Our data from gene reporter experiments showing marked differences in basal and inducible CBR3 promoter activity in HepG2 and MCF7 cells lend further support to these notions. For example, deletion of −2698ARE abrogated the induction of promoter activity by t-BHQ in HepG2 cells (Figs. 3D and E). This result suggests that −2698ARE is a predominant cis-regulatory element for the induction of CBR3 in HepG2 cells. In MCF-7 cells, the lack of −2698ARE did not completely abrogate the induction of gene promoter activity in response to t-BHQ. Thus, additional cis- and/or trans- regulatory elements may mediate induction of CBR3 expression in MCF-7 cells (Figs. 3F, G and H). In fact, we have identified another conserved ARE (5′-GTGACAGAGCA-3′) located at 1607 bp upstream of the translation initiation codon (A+1TG) of CBR3. It is possible to hypothesize that both AREs (i.e., distal and proximal) may operate in concert to modulate CBR3 expression in specific cellular contexts.
The CBR3 promoter region contains additional DNA motifs with potential regulatory roles [10]. For example, there are 2 conserved xenobiotic response elements (XREs) located at 1851 and 5484 bp upstream of the translation initiation codon of CBR3 (5′-CACGCnA/T-3′; Cheng and Blanco, unpublished observation). In line, incubations with the bi-functional Nrf2/AhR ligand β-naphthoflavone induced CBR3 mRNA levels in HepG2 by 8- (50 μM, 18 h) and 14-fold (100 μM, 18 h), respectively (p<0.05. Supplemental Fig. S1). Ebert et al. also observed induction of CBR3 mRNA expression in HepG2 cells after treatment with the bi-functional Nrf2/AhR inducer benzo[k]fluoranthene [14]. Therefore, studies are warranted to determine whether CBR3 expression is co-regulated by Nrf2 and AhR in a manner similar to the NQO1 gene [20].
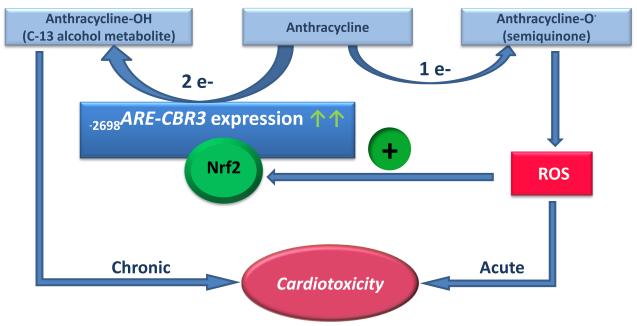
Recent findings suggest that the expression of polymorphic CBR3 impacts the variable pharmacodynamics of the anticancer anthracyclines doxorubicin and daunorubicin [6-9]. The use of anthracyclines for cancer chemotherapy is hampered by the development of acute and/or chronic dose-limiting cardiotoxicity in some patients [28]. Biochemical and genetic evidence suggest that the anthracycline alcohol metabolites synthesized by CBR activity (e.g., doxorubicinol and daunorubicinol) mediate the chronic type of cardiotoxicity [29-31]. On the other hand, studies have shown that the parent anthracyclines induce acute cardiotoxicity through mechanisms that invoke oxidative stress and the production of radical oxidative species (ROS) [32, 33]. It is known that ROS induce the translocation of Nrf2 into the cell nucleus and the concomitant expression of a battery of ARE containing target genes [19]. Thus, we propose that the Nrf2/ARE pathway may provide a potential mechanistic connection between the acute cardiotoxicity mediated mostly by ROS and the chronic type of cardiotoxicity induced by anthracycline C-13 alcohol metabolites. That is, parent anthracyclines induce the production of ROS and the translocation of Nrf2 into the nucleus, which in turn up-regulates CBR3 expression and increases the synthesis of cardiotoxic anthracycline alcohol metabolites (Fig. 7). Furthermore, it is known that common dietary phytochemicals (e.g., sulforane, lycopene, resveratrol, and capsaicin) induce the nuclear translocation of Nrf2 and the expression of its target genes [34]. Recent epidemiological studies suggest that dietary antioxidants (e.g., sulforane and lycopene) cause unwanted phytochemical–anticancer-drug interactions that compromise therapeutic efficacy and enhance toxicity [35, 36]. Therefore, our proposed model provides the rationale to examine whether induction of polymorphic CBR3 expression by the Nrf2/ARE pathway impacts the pharmacodynamics of anthracyclines in cancer patients consuming dietary phytochemicals (Figure 7).
Figure 7.
The potential role of human CBR3 during the pathogenesis of anthracycline-related cardiotoxicity. ROS induce Nrf2 nuclear translocation and binding to the functional −2698ARE in the CBR3 gene promoter, which in turn increases CBR3 activity and the synthesis of cardiotoxic anthracycline alcohol metabolites
This study identifies a conserved ARE in the promoter region of human CBR3 that mediates gene induction through the Nrf2/ARE pathway in 2 model cell lines: HepG2 and MCF-7. Studies to elucidate the impact of variable CBR3 expression via the Nrf2/ARE pathway during the pathogenesis of anthracycline-related cardiotoxicity are warranted.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institute of General Medical Sciences [GM073646]. James L. Kalabus is a Daiichi Sankyo fellow.
Abbreviations
- CBR3
carbonyl reductase 3
- ARE
antioxidant response element
- Nrf2
nuclear factor [erythroid-derived 2]-like 2 (official symbol: NFE2L2)
- RT-PCR
reverse transcription-polymerase chain reaction
- DMSO
dimethyl sulfoxide
- bp
base pairs
- EMSA
electrophoretic mobility shift assay
- ROS
reactive oxygen species
- GPXs
glutathione peroxidases
- NQO1
NAD(P)H:quinone oxidoreductase-1
- NQO2
NRH:quinone oxidoreductase 2
- SOD
superoxide dismutase
- GSTs
glutathione S-transferases
- GPXs
glutathione peroxidases
- GCLC
glutamate-cysteine ligase catalytic subunit
- GCLM
glutamate cysteine ligase modifier subunit.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. REFERENCES
- [1].Oppermann U. Carbonyl reductases: the complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology. Annu Rev Pharmacol Toxicol. 2007;47:293–322. doi: 10.1146/annurev.pharmtox.47.120505.105316. [DOI] [PubMed] [Google Scholar]
- [2].Hoffmann F, Maser E. Carbonyl reductases and pluripotent hydroxysteroid dehydrogenases of the short-chain dehydrogenase/reductase superfamily. Drug Metab Rev. 2007;39:87–144. doi: 10.1080/03602530600969440. [DOI] [PubMed] [Google Scholar]
- [3].Chen Z, Kastaniotis AJ, Miinalainen IJ, Rajaram V, Wierenga RK, Hiltunen JK. 17beta-hydroxysteroid dehydrogenase type 8 and carbonyl reductase type 4 assemble as a ketoacyl reductase of human mitochondrial FAS. Faseb J. 2009;23:3682–91. doi: 10.1096/fj.09-133587. [DOI] [PubMed] [Google Scholar]
- [4].Pilka ES, Niesen FH, Lee WH, El-Hawari Y, Dunford JE, Kochan G, et al. Structural basis for substrate specificity in human monomeric carbonyl reductases. PLoS One. 2009;4:e7113. doi: 10.1371/journal.pone.0007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Miura T, Nishinaka T, Terada T. Different functions between human monomeric carbonyl reductase 3 and carbonyl reductase 1. Mol Cell Biochem. 2008;315:113–21. doi: 10.1007/s11010-008-9794-5. [DOI] [PubMed] [Google Scholar]
- [6].Blanco JG, Leisenring WM, Gonzalez-Covarrubias VM, Kawashima TI, Davies SM, Relling MV, et al. Genetic polymorphisms in the carbonyl reductase 3 gene CBR3 and the NAD(P)H:quinone oxidoreductase 1 gene NQO1 in patients who developed anthracycline-related congestive heart failure after childhood cancer. Cancer. 2008;112:2789–95. doi: 10.1002/cncr.23534. [DOI] [PubMed] [Google Scholar]
- [7].Bains OS, Karkling MJ, Lubieniecka JM, Grigliatti TA, Reid RE, Riggs KW. Naturally occurring variants of human CBR3 alter anthracycline in vitro metabolism. J Pharmacol Exp Ther. 2010;332:755–63. doi: 10.1124/jpet.109.160614. [DOI] [PubMed] [Google Scholar]
- [8].Fan L, Goh BC, Wong CI, Sukri N, Lim SE, Tan SH, et al. Genotype of human carbonyl reductase CBR3 correlates with doxorubicin disposition and toxicity. Pharmacogenet Genomics. 2008;18:623–31. doi: 10.1097/FPC.0b013e328301a869. [DOI] [PubMed] [Google Scholar]
- [9].Blanco JG, Sun C-L, Landier W, Chen L, Esparza-Duran D, Leisenring W, et al. Anthracycline-related Cardiomyopathy after Childhood Cancer: Role of Polymorphisms in Carbonyl-Reductase Genes - A Report from the Children’s Oncology Group. J Clin Oncol. 2011 doi: 10.1200/JCO.2011.34.8987. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang J, Blanco JG. Identification of the Promoter of Human Carbonyl Reductase 3 (CBR3) and Impact of Common Promoter Polymorphisms on Hepatic CBR3 mRNA Expression. Pharm Res. 2009;26:2209–15. doi: 10.1007/s11095-009-9936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cao J, Gonzalez-Covarrubias V, Straubinger RM, Wang H, Duan X, Yu H, et al. A rapid, reproducible, on-the-fly orthogonal array optimization method for targeted protein quantification by LC/MS and its application for accurate and sensitive quantification of carbonyl reductases in human liver. Anal Chem. 2010;82:2680–9. doi: 10.1021/ac902314m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, et al. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (-/-) mice. Cancer Lett. 2006;243:170–92. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- [13].Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- [14].Ebert B, Kisiela M, Malatkova P, El-Hawari Y, Maser E. Regulation of human carbonyl reductase 3 (CBR3; SDR21C2) expression by Nrf2 in cultured cancer cells. Biochemistry. 2010;49:8499–511. doi: 10.1021/bi100814d. [DOI] [PubMed] [Google Scholar]
- [15].Adair-Kirk TL, Atkinson JJ, Griffin GL, Watson MA, Kelley DG, DeMello D, et al. Distal airways in mice exposed to cigarette smoke: Nrf2-regulated genes are increased in Clara cells. Am J Respir Cell Mol Biol. 2008;39:400–11. doi: 10.1165/rcmb.2007-0295OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hubner RH, Schwartz JD, De Bishnu P, Ferris B, Omberg L, Mezey JG, et al. Coordinate control of expression of Nrf2-modulated genes in the human small airway epithelium is highly responsive to cigarette smoking. Mol Med. 2009;15:203–19. doi: 10.2119/molmed.2008.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shen G, Xu C, Hu R, Jain MR, Gopalkrishnan A, Nair S, et al. Modulation of nuclear factor E2-related factor 2-mediated gene expression in mice liver and small intestine by cancer chemopreventive agent curcumin. Mol Cancer Ther. 2006;5:39–51. doi: 10.1158/1535-7163.MCT-05-0293. [DOI] [PubMed] [Google Scholar]
- [18].Niture SK, Kaspar JW, Shen J, Jaiswal AK. Nrf2 signaling and cell survival. Toxicol Appl Pharmacol. 2010;244:37–42. doi: 10.1016/j.taap.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dhakshinamoorthy S, Long DJ, 2nd, Jaiswal AK. Antioxidant regulation of genes encoding enzymes that detoxify xenobiotics and carcinogens. Curr Top Cell Regul. 2000;36:201–16. doi: 10.1016/s0070-2137(01)80009-1. [DOI] [PubMed] [Google Scholar]
- [20].Nioi P, Hayes JD. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2004;555:149–71. doi: 10.1016/j.mrfmmm.2004.05.023. [DOI] [PubMed] [Google Scholar]
- [21].Ellis EM. Reactive carbonyls and oxidative stress: Potential for therapeutic intervention. Pharmacology & Therapeutics. 2007;115:13–24. doi: 10.1016/j.pharmthera.2007.03.015. [DOI] [PubMed] [Google Scholar]
- [22].Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- [23].Macari ER, Lowrey CH. Induction of human fetal hemoglobin via the NRF2 antioxidant response signaling pathway. Blood. 2011;117:5987–97. doi: 10.1182/blood-2010-10-314096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang W, Jaiswal AK. Nuclear factor Nrf2 and antioxidant response element regulate NRH:quinone oxidoreductase 2 (NQO2) gene expression and antioxidant induction. Free Radic Biol Med. 2006;40:1119–30. doi: 10.1016/j.freeradbiomed.2005.10.063. [DOI] [PubMed] [Google Scholar]
- [25].Yokota S-i, Higashi E, Fukami T, Yokoi T, Nakajima M. Human CYP2A6 is regulated by nuclear factor-erythroid 2 related factor 2. Biochemical Pharmacology. 2011;81:289–94. doi: 10.1016/j.bcp.2010.09.020. [DOI] [PubMed] [Google Scholar]
- [26].Sankaranarayanan K, Jaiswal AK. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. J Biol Chem. 2004;279:50810–7. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- [27].Yang H, Magilnick N, Lee C, Kalmaz D, Ou X, Chan JY, et al. Nrf1 and Nrf2 regulate rat glutamate-cysteine ligase catalytic subunit transcription indirectly via NF-kappaB and AP-1. Mol Cell Biol. 2005;25:5933–46. doi: 10.1128/MCB.25.14.5933-5946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wouters KA, Kremer LCM, Miller TL, Herman EH, Lipshultz SE. Protecting against anthracycline-induced myocardial damage: a review of the most promising strategies. British Journal of Haematology. 2005;131:561–78. doi: 10.1111/j.1365-2141.2005.05759.x. [DOI] [PubMed] [Google Scholar]
- [29].Olson RD, Mushlin PS, Brenner DE, Fleischer S, Cusack BJ, Chang BK, et al. Doxorubicin cardiotoxicity may be caused by its metabolite, doxorubicinol. Proc Natl Acad Sci U S A. 1988;85:3585–9. doi: 10.1073/pnas.85.10.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Olson LE, Bedja D, Alvey SJ, Cardounel AJ, Gabrielson KL, Reeves RH. Protection from Doxorubicin-Induced Cardiac Toxicity in Mice with a Null Allele of Carbonyl Reductase 1. Cancer Res. 2003;63:6602–6. [PubMed] [Google Scholar]
- [31].Forrest GL, Gonzalez B, Tseng W, Li X, Mann J. Human carbonyl reductase overexpression in the heart advances the development of doxorubicin-induced cardiotoxicity in transgenic mice. Cancer Res. 2000;60:5158–64. [PubMed] [Google Scholar]
- [32].Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: Molecular Advances and Pharmacologic Developments in Antitumor Activity and Cardiotoxicity. Pharmacol Rev. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- [33].Menna P, Salvatorelli E, Minotti G. Cardiotoxicity of Antitumor Drugs. Chem Res Toxicol. 2008;21:978–89. doi: 10.1021/tx800002r. [DOI] [PubMed] [Google Scholar]
- [34].Surh YJ, Kundu JK, Na HK. Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. Planta Med. 2008;74:1526–39. doi: 10.1055/s-0028-1088302. [DOI] [PubMed] [Google Scholar]
- [35].Hardy ML, Lawenda BD, Kelly KM, Ladas EJ, Sagar SM, Vickers A, et al. Dietary supplement use in cancer care: help or harm. Should supplemental antioxidant administration be avoided during chemotherapy and radiation therapy? Hematol Oncol Clin North Am. 2008;22:581–617. vii. [Google Scholar]
- [36].Lawenda BD, Kelly KM, Ladas EJ, Sagar SM, Vickers A, Blumberg JB. Should supplemental antioxidant administration be avoided during chemotherapy and radiation therapy? J Natl Cancer Inst. 2008;100:773–83. doi: 10.1093/jnci/djn148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.