Abstract
Huntington’s disease (HD) is a neurodegenerative genetic disorder that affects the brain. Atrophy of deep grey matter structures has been reported and it is likely that underlying pathologic processes occur before, or in concurrence with, volumetric changes. Measurement of metabolite concentrations in these brain structures has the potential to provide insight into pathological processes. We aim to gain understanding of metabolite changes with respect to the disease stage and pathophysiological changes. We studied five brain regions using magnetic resonance spectroscopy (MRS) using a 7-Tesla MRI scanner. Localized proton spectra were acquired to obtain six metabolite concentrations. MRS was performed in the caudate nucleus, putamen, thalamus, hypothalamus, and frontal lobe in 44 control subjects, premanifest gene carriers and manifest HD. In the caudate nucleus, HD patients display lower NAA (p = 0.009) and lower creatine concentration (p = 0.001) as compared to controls. In the putamen, manifest HD patients show lower NAA (p = 0.024), lower creatine concentration (p = 0.027), and lower glutamate (p = 0.013). Although absolute values of NAA, creatine, and glutamate were lower, no significant differences to controls were found in the premanifest gene carriers. The lower concentrations of NAA and creatine in the caudate nucleus and putamen of early manifest HD suggest deficits in neuronal integrity and energy metabolism. The changes in glutamate could support the excitotoxicity theory. These findings not only give insight into neuropathological changes in HD but also indicate that MRS can possibly be applied in future clinical trails to evaluate medication targeted at specific metabolic processes.
Keywords: Huntington’s disease, MRI, MRS, Energy metabolism
Introduction
Huntington’s disease (HD) is a neurodegenerative autosomal dominant disorder. The causative gene mutation is located on the short arm of chromosome 4 and consists of an expanded cytosine–adenine–guanine (CAG)-repeat within the Htt-gene. This expansion results in the synthesis of an abnormal huntingtin protein that causes neuronal damage, brain atrophy, and ultimately leading to functional disturbances of motor, cognition, and behavior.
HD research has revealed widespread changes throughout the brain [5]. Controversy remains as to which structures are affected at different disease stages. Atrophy of the striatum is regarded as the hallmark of the pathologic findings in HD [22]. MRI studies demonstrate that the caudate nucleus and putamen begin to show atrophy up to a decade before clinical manifestations occur [2, 29, 34]. Structures such as the thalamus, hypothalamus, the frontal lobe, white matter, and cortical grey matter have all been implicated to some degree at this pre-manifest stage, although findings differ [1, 3, 5, 18].
The pathophysiological mechanism leading to neuronal damage remains unclear. Currently, the two most accepted hypotheses describe impaired energy metabolism and the excitotoxicity of neurons [8, 23]. Both hypotheses can potentially be explored by means of non-invasive in vivo measurements of metabolites such as creatine and glutamate, using localized magnetic resonance spectroscopy (MRS). However, previous studies measuring the changes in the metabolite levels related to the damaging processes in HD have reported conflicting results. In one study, a lower level of N-acetylaspartate (NAA) was confirmed post-mortem in the putamen and cortex in manifest HD [7]. In vivo altered levels of NAA, creatine, choline, and glutamate have been reported in both premanifest and manifest HD in several brain structures, such as the striatum and thalamus using MRS [9, 11, 12, 24, 25, 31, 32]. However, in contrast, other studies using localized MRS did not detect changes in these metabolite levels in either manifest or premanifest HD [6, 21, 35].
One of the factors that might explain discrepancies between previous studies is the relatively poor spatial and/or spectral resolution of localized spectra. Improvement in MRS methodology in terms of spectral resolution in combination with small voxel size and total scanning time can be achieved using a high-field MRI scanner [14, 33]. This allows metabolite quantification in small, well-defined anatomical structures, such as the caudate nucleus and putamen.
The major aims of the present study were to assess metabolite differences between manifest HD patients or premanifest HD gene carriers and controls and to assess the association between these metabolite differences and clinical measures of disease severity in order to obtain a greater understanding of the pathophysiological changes in HD with respect to the disease stage. The hypotheses were as follows; as the neuronal integrity and the energy metabolism would be compromised, we expected the NAA and creatine to be lower in premanifest and manifest HD, especially in the striatum. Possible changes in glutamate could be expected based on the excitotoxicity theory, with higher levels in both premanifest and manifest HD.
Materials and methods
Subjects
Participants were recruited from the outpatient neurology clinic of the Leiden University Medical Center. Inclusion criteria for the early manifest HD consisted of genetic confirmation with a CAG repeat ≥39, and the presence of motor disturbances as measured by the Unified Huntington’s Disease Rating Scale ’99 (UHDRS) defined as a total motor score (TMS) >5 and a total functional capacity (TFC) ≥7. All participants received a diagnostic confidence rating of 4, representing "motor abnormalities that are unequivocal signs of HD (≥99% confidence)". Inclusion criteria for the premanifest HD participants consisted of genetic confirmation with a CAG repeat ≥39, and the absence of motor disturbances on the UHDRS defined as a TMS ≤5. Healthy gene-negative family members or partner/spouses were recruited as control subjects in an effort to reduce influences of environmental factors. Exclusion criteria for all participants consisted of significant (neurological) comorbidity, a major psychiatric diagnosis, history of severe head injury, or incompatibility for MRI. In total, 44 subjects were recruited to undergo MRS in one or more regions of interest (ROI). Time constraints and HD-related issues (such as patient motion) accounted for not all participants completing the MRS protocol in all five of the ROIs. The study was approved by the local Medical Ethical Committee of the Leiden University Medical Center. All participants provided written informed consent to participate in the study.
Clinical measures
Clinical evaluation for all subjects consisted of the UHDRS motor scale (score 0–124) and TFC, a global scale of impairment in daily life activities, score 0–13. Furthermore, a short cognitive battery was administered consisting of the Mini Mental State Exam (MMSE), Stroop test word reading card (Stoop-II), the Symbol Digit Modality Test (SDMT), Trail making Test part B (TMT-B), the Wechsler Memory Scale (MQ) and premorbid IQ estimation using the Dutch Adult Reading Test (DART) [26]. Behavioral disturbances were evaluated with the Beck Depression Inventory 2nd version (BDI-II) [4] and the Problem Behaviour Assessment short version (PBA-s). Predicted years to onset were calculated from current age and CAG repeat length using the formula by Langbehn et al. [13].
MRI/MRS acquisition
MRI and MRS were performed on a Philips 7-Tesla Achieva whole-body scanner (Philips Healthcare, Best, The Netherlands) with a NOVA Medical quadrature transmit coil and 16-channel receive coil array. For accurate planning, a high-resolution, three-dimensional T1-weighted GRE scan was acquired (TR/TE = 11/5.4 ms, voxel size 0.44 × 0.44 × 0.84 mm, total scan time 1:46). Localized proton spectra were acquired using a stimulated echo acquisition mode (STEAM) sequence. The voxel was placed within the region of interest with the maximum volume containing only tissue from the intended structure, minimizing the contribution from surrounding tissue and also partial volume effects. Regions of interest consisted of the caudate nucleus, putamen, thalamus, hypothalamus, and the prefrontal white and cortical grey matter. For the prefrontal voxel, positioning included approximately 50% white and 50% cortical grey matter. Only for the hypothalamus voxel was some cerebral spinal fluid (CSF) necessarily present since the voxel was placed bilaterally. For each voxel in each ROI, a water reference signal was acquired for adequate quantification. The following scan parameters were used for the STEAM spectra: TR/TE/TM = 2,000/19/25 ms, BW 4 kHz, 2,048 complex data points and 128 signal averages, giving a data acquisition time of approximately 5 min per spectrum. The chemical shift artifact was minimal (<1 mm), which is shown in Fig. 1. Water suppression was performed using a frequency-selective RF pulse and gradient spoiling, and six saturation bands were additionally applied to suppress the signal from the surrounding tissue. All scans included a reference scan without water suppression for quantification. When an MRS scan failed (for example due to motion), it was standard protocol to try the same ROI again: however, in some cases this resulted in insufficient time for the full protocol to be completed, since the time for each patient in the magnet was strictly limited to 1 h.
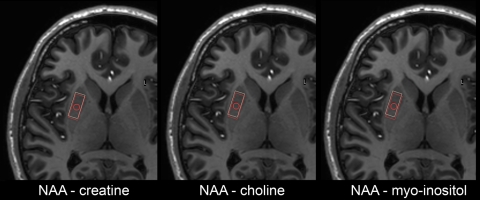
Fig. 1.
Chemical shift at 7-Tesla using STEAM. Chemical shifts provided for an actual planning. Red box NAA, white box metabolite as stated below each figure. Only a minimal chemical shift exists. The chemical shift of water is almost zero, as the separate water file is planned according to the NAA-voxel. NAA N-acetylaspartate
MRS post-processing
MRS data were analyzed with LCModel [19, 20] using the unsuppressed water as an internal reference to calculate the concentrations of metabolites. Concentrations of choline, creatine, glutamine + glutamate (Glx), total NAA (N-acetylaspartylglutamate + NAA), myo-inositol and lactate were calculated for analysis.
Stringent quality control was enforced to exclude data with high estimated errors. A signal-to-noise ratio of ≤3 and very high values of residual errors in the LCModel fitting were applied as initial spectral exclusion criteria. Furthermore, the LCModel fit for metabolites was required to have a Cramér–Rao lower bound (CRLB) of 20% or less. This was required for all metabolites within a single spectrum, with the exception of lactate. For lactate, CRLB above 100% were excluded from further analysis. If lactate was not quantified with CRLB <100% in at least 50% of analyzed spectra, the analysis of lactate was deemed not detectable; this method was first presented by Tkac et al. [33].
In total, 30 spectra were included in the analysis for the hypothalamus, 36 of the thalamus, 28 of the caudate nucleus, 27 of the putamen, and 32 subjects of the prefrontal region.
Statistics
Statistical analyses were performed using SPSS for Windows (version 17.0, SPSS Inc, Chicago, IL, USA). All clinical variables were assessed between groups with a one-way analysis of variance (ANOVA), with post hoc testing. To compare all metabolites in the five brain regions between the manifest and premanifest HD group versus the control group, an ANOVA with planned comparisons was performed. The nature of this study is exploratory, and therefore in order to prevent inflation of type II errors and avoid important results going unreported, no correction for multiple testing was performed, as is accepted practice in such types of study [17]. To examine the relationship between metabolic concentrations and clinical measures in gene carriers (manifest and premanifest HD), partial Pearson correlation analysis corrected for age was performed.
Results
Demographics
No differences in age, education level, or IQ were present between the manifest HD and control group, or between the premanifest HD and control group. No significant difference exists in CAG repeat length between the premanifest and the manifest HD group. For all other clinical assessments variables, we refer to the results in Table 1.
Table 1.
Demographic variables and scores on clinical assessments for the three groups
| Control n: 18 (♂: 9 ♀: 9) | Premanifest n: 14 (♂: 6 ♀: 8) | Manifest n:12 (♂: 5 ♀: 7) | ||||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
| Age | 47.7 | 7.4 | 42.9 | 11.0 | 48.6 | 7.0 |
| Years of education | 15.9 | 3.2 | 14.3 | 3.3 | 15.3 | 2.7 |
| IQ | 106.7 | 7.8 | 102.2 | 12.5 | 102.3 | 9.5 |
| CAG | 20.3c | 2.9 | 43.1 | 3.4 | 43.7 | 2.3 |
| Disease burden | na | na | 306.4 | 87.0 | 385.4e | 72.5 |
| Predicted YTO | na | na | 10.4 | 8.2 | na | na |
| UHDRS TMS | 2.0 | 2.4 | 2.6 | 1.3 | 20.9a | 14.1 |
| UHDRS TFC | 12.9 | 0.2 | 12.6 | 0.6 | 11.3a | 1.9 |
| PBA-s | 5.9 | 1.4 | 5.0 | 1.3 | 13.2a | 3.8 |
| BDI-II | 3.9 | 4.6 | 5.2 | 5.5 | 6.4 | 6.1 |
| MMSE | 29.5 | 0.7 | 28.5 | 1.7 | 28.3b | 1.6 |
| SDMT | 59.2 | 8.9 | 48.5 | 10.8 | 37.3d | 6.7 |
| TMT-B | 46.7 | 15.1 | 67.8 | 27.8 | 101.4a | 51.2 |
| Stroop II | 104.0 | 15.4 | 95.1 | 19.4 | 79.3b | 19.9 |
| MQ | 127.3 | 11.5 | 115.5 | 23.2 | 99.7b | 13.2 |
SD Standard deviation, CAG CAG repeat length of larger allele, predicted YTO predicted years to disease onset, UHDRS TMS Unified Huntington’s Disease Rating Scale Total Motor Score, PBA-s Problem Behaviour Assessment short version, TFC Total Functional Capacity, BDI-II Beck Depression Inventory 2nd version, MMSE Mini Mental State Exam, TMT Trail Making Test part B, SDMT Symbol Digit Modality Test, MQ Memory Quotient, Stroop II Stroop word reading card, na not applicable
aSignificant differences between manifest HD compared to both premanifest and controls
bSignificant differences between manifest HD and controls
cSignificant differences between control and both premanifest and manifest HD
dSignificant differences between all three groups
eSignificant difference between premanifest and manifest HD
Voxel planning
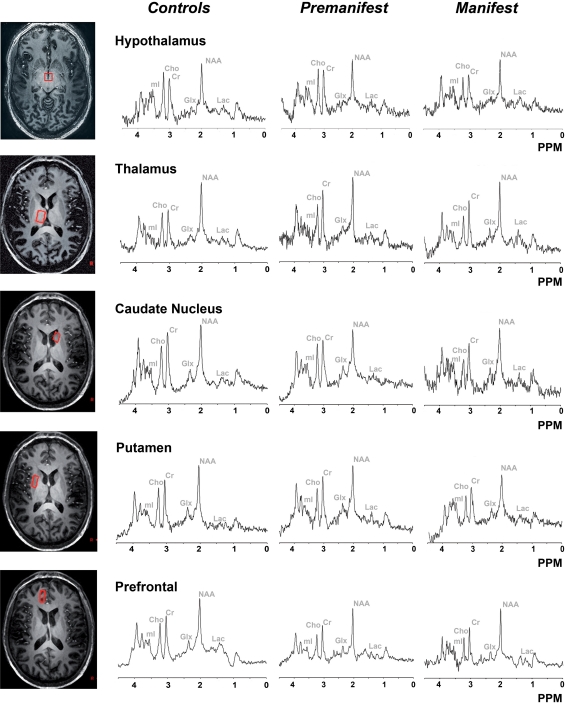
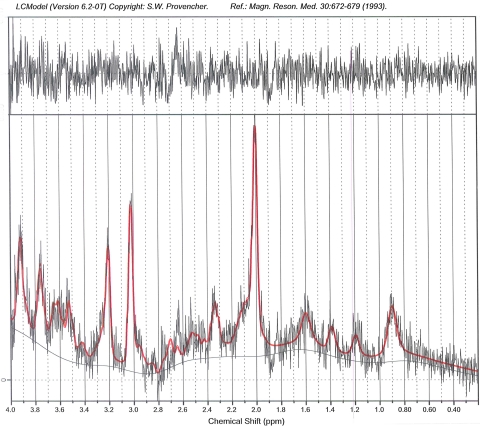
Typical examples of localized proton spectra for the five examined regions, along with metabolite identification, are shown in Fig. 2. The voxel location is shown on the left side. For clarity, the voxel is only shown in one orientation. Actual planning of the voxel required oblique planning in all three dimensions. Figure 3 provides a typical example spectrum and fit from the LCmodel.
Fig. 2.
Localized proton MR spectra from different regions of the brain. On the left side the voxel is displayed in the transverse direction, a typical spectrum of that structure is shown on the right side for the three groups. Five different regions are displayed: hypothalamus, thalamus, caudate nucleus, putamen, prefrontal region. Cho choline, Cr creatine, NAA N-acetylaspartate, mI myo-inositol, Glx glutamate + glutamine, Lac lactate, PPM parts per million. A Gaussian filter of 4 Hz was applied
Fig. 3.
LCModel output example. This figure depicts the output provided by the LCModel analysis. The black line is the raw spectrum and the red line is the fit by LCmodel. PPM parts per million
Voxel sizes were individually set based upon the different sizes of structures in different individuals, although the voxel size was never below 1.0 ml. With these sizes, it was possible in all cases to include only the intended (grey matter) structure, without contamination of other (white matter) structures, the exception being the prefrontal region. The voxel size ranged from 1.0 ml in the caudate nucleus for a manifest HD participant to a maximum of 3.4 ml for the putamen of a participant in the control group.
Metabolite analysis
The concentrations of the six metabolites (NAA, creatine, choline, glutamate, lactate, and myo-inositol) in the five brain regions are shown in Table 2 for the three groups. In the caudate nucleus, the manifest HD group demonstrated significantly lower NAA (F = 8.12, p = 0.009) and lower creatine concentration (F = 13.60, p = 0.001) compared to the control subjects. In the putamen, manifest HD showed significantly lower NAA (F = 5.81, p = 0.024), lower creatine concentration (F = 5.58, p = 0.027), and glutamate/glutamine (F = 7.21, p = 0.013) compared to controls. No significant differences were found in the hypothalamus, thalamus, or prefrontal region. Comparison of the premanifest gene carrier group and the control group did not show any significant differences in any of the regions. However, the absolute values of NAA, creatine, and glutamate were lower in the caudate nucleus and putamen in the premanifest gene carrier group, but did not reach significance.
Table 2.
Metabolite concentrations from five brain regions in HD
| Control | Premanifest | Manifest | |||||||
|---|---|---|---|---|---|---|---|---|---|
| n | Mean | SD | n | Mean | SD | n | Mean | SD | |
| Hypothalamus | 11 | 10 | 9 | ||||||
| Creatine | 10.7 | 2.1 | 9.4 | 2.0 | 9.3 | 1.7 | |||
| Choline | 3.7 | 0.7 | 3.4 | 0.6 | 3.3 | 0.8 | |||
| NAA | 12.1 | 1.9 | 11.9 | 1.5 | 11.3 | 2.1 | |||
| Glx | 12.5 | 3.0 | 12.0 | 4.0 | 11.0 | 4.8 | |||
| Myo-inositol | 13.4 | 4.8 | 12.7 | 3.1 | 13.3 | 2.8 | |||
| Lactate | 1.5 | 1.0 | 1.1 | 1.1 | 1.0 | 1.4 | |||
| Thalamus | 14 | 13 | 9 | ||||||
| Creatine | 12.7 | 3.5 | 12.8 | 3.6 | 12.1 | 1.6 | |||
| Choline | 3.1 | 0.8 | 3.0 | 0.7 | 3.1 | 0.4 | |||
| NAA | 17.8 | 6.0 | 15.8 | 2.6 | 15.9 | 1.6 | |||
| Glu/Gln | 14.8 | 5.1 | 15.1 | 3.4 | 12.8 | 3.4 | |||
| Myo-inositol | 8.5 | 4.1 | 7.3 | 2.6 | 9.5 | 4.4 | |||
| Lactate | 1.1 | 1.0 | 0.5 | 0.7 | 1.8 | 0.9 | |||
| Caudate Nucleus | 12 | 11 | 5 | ||||||
| Creatine | 12.5 | 1.7 | 11.7 | 2.2 | 8.9* | 1.2 | |||
| Choline | 3.1 | 0.7 | 2.8 | 0.5 | 2.6 | 0.6 | |||
| NAA | 12.1 | 2.3 | 10.8 | 2.0 | 9.0** | 1.1 | |||
| Glx | 15.4 | 5.2 | 13.3 | 5.7 | 10.8 | 1.6 | |||
| Myo-inositol | 6.2 | 1.4 | 7.1 | 2.0 | 7.9 | 2.0 | |||
| Lactate | 1.2 | 0.9 | 1.5 | 0.9 | 0.5 | 0.8 | |||
| Putamen | 13 | 9 | 5 | ||||||
| Creatine | 13.6 | 2.3 | 12.1 | 2.2 | 10.9* | 0.9 | |||
| Choline | 3.4 | 0.8 | 3.1 | 0.5 | 3.0 | 0.4 | |||
| NAA | 14.8 | 1.6 | 14.4 | 1.7 | 12.8* | 0.9 | |||
| Glx | 16.3 | 3.4 | 14.2 | 3.4 | 11.4* | 3.8 | |||
| Myo-inositol | 7.3 | 2.1 | 6.5 | 3.1 | 10.5 | 4.0 | |||
| Lactate | 0.8 | 0.6 | 1.0 | 1.3 | 0.3 | 0.4 | |||
| Prefrontal | 13 | 9 | 10 | ||||||
| Creatine | 9.1 | 2.4 | 9.2 | 1.6 | 9.9 | 2.1 | |||
| Choline | 2.3 | 0.5 | 2.4 | 0.4 | 2.3 | 0.5 | |||
| NAA | 11.7 | 2.4 | 11.6 | 1.5 | 11.9 | 3.0 | |||
| Glx | 11.3 | 2.8 | 9.8 | 2.2 | 13.3 | 4.0 | |||
| Myo-inositol | 9.5 | 2.4 | 8.8 | 1.9 | 11.9 | 4.9 | |||
| Lactate | 1.2 | 1.1 | 1.5 | 1.5 | 0.8 | 0.9 | |||
Concentrations of metabolites in the three groups. Since the water concentration and the T 1 and T 2 relaxation times of the individual metabolites in the specific voxels of interest are unknown at the field strength of 7 Tesla, data are expressed as relative values to the water peak in arbitrary units (AU)
NAA N-acetylaspartate, Glx glutamate + glutamine
* p < 0.05
** p < 0.005
Relationship to clinical measures
The association of metabolite levels with clinical measures is shown in Table 3. TFC was associated with creatine levels in caudate nucleus and putamen and with NAA in the putamen, showing decreased metabolite levels corresponding to poorer clinical scores. The NAA concentration in the putamen was associated with higher scores on the UHDRS TMS, indicating lower NAA to correspond to more motor disturbances. Lower glutamate/glutamine related to poorer scores on the cognitive tests SDMT and TMT. Finally, a higher PBA score, indicating more behavioral disturbances, corresponded to lower creatine in the caudate nucleus.
Table 3.
Relationship of the clinical measures with metabolite concentration in the caudate nucleus and putamen
| Caudate nucleus | Putamen | ||||
|---|---|---|---|---|---|
| Creatine | NAA | Creatine | NAA | Glx | |
| Disease burden | |||||
| R | −0.266 | −0.473 | 0.168 | 0.284 | −0.400 |
| p | 0.339 | 0.075 | 0.584 | 0.347 | 0.157 |
| UHDRS TMS | |||||
| R | −0.280 | −0.228 | −0.324 | −0.415 | −0.255 |
| p | 0.157 | 0.253 | 0.106 | 0.035* | 0.200 |
| PBA-s | |||||
| R | −0.545 | −0.207 | −0.063 | 0.064 | 0.052 |
| p | 0.003* | 0.300 | 0.758 | 0.754 | 0.797 |
| TFC | |||||
| R | 0.409 | 0.106 | 0.440 | 0.556 | 0.095 |
| p | 0.034* | 0.598 | 0.025* | 0.003* | 0.636 |
| BDI-II | |||||
| R | −0.256 | −0.022 | 0.005 | 0.206 | 0.152 |
| p | 0.197 | 0.914 | 0.983 | 0.323 | 0.457 |
| MMSE | |||||
| R | −0.196 | −0.106 | −0.070 | −0.108 | −0.223 |
| p | 0.328 | 0.598 | 0.735 | 0.601 | 0.265 |
| TMT-B | |||||
| R | 0.310 | 0.270 | 0.185 | 0.049 | 0.438 |
| p | 0.115 | 0.173 | 0.366 | 0.813 | 0.022* |
| SDMT | |||||
| R | 0.171 | 0.133 | 0.239 | 0.125 | 0.555 |
| p | 0.404 | 0.518 | 0.250 | 0.552 | 0.003* |
| MQ | |||||
| R | 0.151 | 0.078 | −0.049 | −0.013 | 0.326 |
| p | 0.451 | 0.700 | 0.811 | 0.951 | 0.097 |
| Stroop II | |||||
| R | −0.196 | −0.106 | −0.070 | −0.108 | −0.223 |
| p | 0.328 | 0.598 | 0.735 | 0.601 | 0.265 |
Correlation analysis of metabolite concentration with clinical measures
UHDRS TMS Unified Huntington’s Disease Rating Scale Total Motor Score, PBA-s Problem Behaviour Assessment short version, TFC Total Functional Capacity, BDI-II Beck Depression Inventory 2nd version, MMSE Mini Mental State Exam, TMT Trail Making Test part B, SDMT Symbol Digit Modality Test, MQ Memory Quotient, Stroop II Stroop word reading card, R partial r correlation coefficient, p p value
* p < 0.05
Discussion
The major findings of this study are lower concentrations of creatine and NAA in the caudate nucleus and the putamen, and a reduction of glutamate in the putamen in manifest HD. A relationship between the differences in these metabolic levels and clinical measures of disease severity, especially global functioning, was demonstrated. No statistically significant differences in any metabolite concentration were observed when the premanifest group was compared to controls, although the absolute lower values could indicate a subtle decline.
As creatine is considered an important marker for brain energy metabolism [10], the finding of lower creatine levels suggests impaired energy metabolism in manifest HD. In the healthy population, the concentration of creatine in the brain is considered to be fairly stable, however, specific pathology has been shown to influence creatine concentrations [10]. The finding of lower creatine in putamen and caudate nucleus is supported by findings of Sanchez et al. [25], who observed a reduction of both creatine and NAA in the striatum. Reynolds et al. [21] went on to propose creatine as a possible biomarker, when they demonstrated lower levels in the caudate nucleus in premanifest HD. Contrary to our findings, Reynolds et al. [21] did not report altered creatine levels in the putamen. Results from the TRACK-HD study reported by Sturrock et al. [27] show similar lower values of creatine in the putamen as our study. Jenkins et al. [11, 12] found additional evidence for impaired energy metabolism as displayed by elevated lactate levels in HD within the occipital cortex. However, our study did not show elevated lactate in any of the examined structures. This finding can possibly be explained by the fact that Jenkins et al. [11] used a more severely affected HD population with a lower mean TFC score of 7.3. Moreover, the region analyzed by Jenkins et al. [11, 12] did not consist of the basal ganglia, but of the occipital cortex, which is a very metabolically active area of the cortex and is also severely atrophied in HD [29]. Although, these studies are not completely comparable on the basis of methodology and results, they do all point towards the importance of assessment of metabolic changes and specifically the energy metabolism markers such as creatine.
NAA is a marker for the integrity of neurons and axons [10] and lower NAA levels suggest a decrease in neuronal integrity. As atrophy of numerous brain structures is apparent in manifest HD, it is likely that neurons are affected and a decrease in NAA may occur. This hypothesis was previously confirmed within the striatum [6, 11, 25]. Again, the recent report from Sturrock et al. [27] is similar to our findings of lower NAA in the putamen. Our study confirms the finding of decreased NAA in the striatum (caudate nucleus plus putamen), and more importantly, our data show that neuronal damage may be present in both the caudate nucleus and putamen separately. The ability to acquire spectra from relatively small voxels is an important advantage of higher magnetic fields in localized MR spectroscopy.
Glutamate is of interest in light of the excitotoxicity theory, which states that an overstimulation of neurons causes cell damage and eventually cell death. In neurons, this can occur either due to increased levels of glutamate (and/or its precursor glutamine) or due to an increase in sensitivity of the glutamate receptors, both resulting in the same effect [8, 23]. Taylor et al. described increased levels of glutamate, supporting this theory [31, 32]. However, the glutamate levels in these studies were expressed as a ratio to creatine, and could also be influenced by changes of the creatine level. Our study demonstrates reduced glutamate levels in the putamen. An explanation may be that the number of viable neurons is decreased to an extent where glutamate is lowered along with the neuron count. Also, the sensitivity of glutamate receptors could be altered, resulting in altered levels of glutamate. However, without further investigation, these propositions are highly speculative.
In the premanifest HD group, no changes in any of the examined metabolites levels could be demonstrated. This was an unexpected finding, as previous studies have reported structural abnormalities, e.g., atrophy, at this stage [5, 30, 34] and some reports exist of lowered levels of metabolites in premanifest HD gene carriers [21, 25]. Sanchez-Pernaute et al. [25] did report lower NAA and lower creatine in the striatum in four premanifest gene carriers; however, the article already stated that there were soft motor signs present in two out of four in this group. Reynolds et al. [21] reported on lower values of creatine, NAA, yet concluded that there was no pathognomonic profile in metabolite changes, but stated that there was great heterogeneity in this respect. Nonetheless, we did hypothesize that premanifest HD would show altered levels of metabolites as atrophy (which is already present more than a decade before disease onset [2, 15, 30, 34]) is logically the result of underlying processes. We must therefore conclude that either we cannot (yet) measure these changes or the processes involved are more complex than simple linear correlation between metabolite levels and disease severity. For instance, the excitotoxicity theory can possibly be a process measured by increased (damaging effects) or decreased (loss of healthy neurons) levels of glutamate depending on the individual disease stage. When performing group analysis, these measurements can become diluted. Individual assessment of longitudinal changes could shed more light on these changes. An explanation as to why no significant differences in our premanifest population were found, may be that our premanifest group was too heterogeneous in terms of proximity to disease onset. When taking into consideration the "disease burden score", a proven correlate of striatal damage and thereby proximity to onset [16], a range of 121.5–450.5 exists within our premanifest HD sample. This large range suggests a great deal of variability in striatal damage and therefore of possible metabolite differences. Even so, despite this large range in disease burden score, a decline in absolute values of NAA and creatine was observed in the premanifest group, although this did not reach statistical significance. A second explanation could be our very stringent inclusion criteria. Only premanifest participants were included when there was no evidence of motor symptoms, as quantified by an UHDRS motor score of 5 or less, whereas other studies may have allowed participation of premanifest gene carriers with a higher degree of clinical abnormalities.
Our data show that metabolite levels are associated with clinical measures of disease severity. This finding highlights the feasibility of MRS in clinical trails, whereby metabolite levels could be target outcome measures. Endeavors using creatine as a treatment for HD with MRS as a monitoring tool have demonstrated the feasibility of this method [28].
This study shows the clinical application of high-field 7-Tesla MRI. The spectral resolution in combination with small voxel size and total scanning time is a clear improvement in MRS methodology [14, 33]. This allows the examination of metabolites in small, well-defined anatomical structures, such as the caudate nucleus and putamen for HD, and can be used in the examination of many other disorders where spectral and spatial resolution is of great importance.
A limitation of our study was the fact that the premanifest group was not uniformly distributed according to expected disease onset, which may have lead to a relatively large spread of the metabolite concentrations. A more homogeneous group close to onset could possibly reveal more significant results already in the premanifest stage of the disease. Also, the small number of participants included in the manifest groups could be seen as a limitation. However, despite the small amount of participants, the results were still significant. Furthermore, the hypothalamus region (bilaterally) included some CSF in the voxel, especially in the manifest HD group, as a result of atrophy, which could account for the less reliable measurements. This was not a problem for any of the other regions. Finally, the frontal region consisted of approximately 50% of both grey and white matter, which could lead to false-negative findings if grey or white matter would be unequally affected.
In conclusion, in manifest HD, lower NAA was found in the caudate nucleus and putamen, supporting the theory of impaired energy metabolism as part of the pathophysiology of Huntington’s disease. Glutamate levels were lowered in the putamen, however, to which extent this finding supports the excitotoxicity theory remains unclear. The relationship with clinical measures of function makes MRS a potential disease monitor and could also possibly be used to evaluate therapeutics targeted at metabolic processes in HD.
Conflict of interest
None.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- AU
Arbitrary units
- BDI-II
Beck Depression Inventory 2nd version
- CAG
Cytosine–adenine–guanine
- DART
Dutch Adult Reading Test
- FWHM
Full width half maximum
- Glx
Glutamate + glutamine
- HD
Huntington’s disease
- MMSE
Mini Mental State Exam
- NAA
N-acetylaspartate
- PMGC
Premanifest gene carrier
- STEAM
Stimulated echo acquisition mode
- TFC
Total functional capacity
- TMS
Total motor score
- UHDRS
Unified Huntington’s Disease Rating Scale
Footnotes
S. J. A. van den Bogaard and E. M. Dumas contributed equally to this work.
References
- 1.Aylward EH, Anderson NB, Bylsma FW, Wagster MV, Barta PE, Sherr M, Feeney J, Davis A, Rosenblatt A, Pearlson GD, Ross CA. Frontal lobe volume in patients with Huntington’s disease. Neurology. 1998;50:252–258. doi: 10.1212/wnl.50.1.252. [DOI] [PubMed] [Google Scholar]
- 2.Aylward EH, Sparks BF, Field KM, Yallapragada V, Shpritz BD, Rosenblatt A, Brandt J, Gourley LM, Liang K, Zhou H, Margolis RL, Ross CA. Onset and rate of striatal atrophy in preclinical Huntington disease. Neurology. 2004;63:66–72. doi: 10.1212/01.wnl.0000132965.14653.d1. [DOI] [PubMed] [Google Scholar]
- 3.Aziz NA, Pijl H, Frolich M, van der Graaf AW, Roelfsema F, Roos RA. Increased hypothalamic-pituitary-adrenal axis activity in Huntington’s disease. J Clin Endocrinol Metab. 2009;94:1223–1228. doi: 10.1210/jc.2008-2543. [DOI] [PubMed] [Google Scholar]
- 4.Beck AT, Steer RA, Brown GK. BDI-II, Beck Depression Inventory: Manual. 2. San Antonio: Harcourt Brace; 1996. [Google Scholar]
- 5.Bohanna I, Georgiou-Karistianis N, Hannan AJ, Egan GF. Magnetic resonance imaging as an approach towards identifying neuropathological biomarkers for Huntington’s disease. Brain Res Rev. 2008;58(1):209–225. doi: 10.1016/j.brainresrev.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Clarke CE, Lowry M, Quarrell OWJ (1998) No change in striatal glutamate in Huntington’s disease measured by proton magnetic resonance spectroscopy. Parkinsonism Relat Disord 4(3):123–127 [DOI] [PubMed]
- 7.Dunlop DS, Mc Hale DM, Lajtha A. Decreased brain N-acetylaspartate in Huntington’s disease. Brain Res. 1992;580:44–48. doi: 10.1016/0006-8993(92)90925-Y. [DOI] [PubMed] [Google Scholar]
- 8.Estrada Sanchez AM, Mejia-Toiber J, Massieu L. Excitotoxic neuronal death and the pathogenesis of Huntington’s disease. Arch Med Res. 2008;39:265–276. doi: 10.1016/j.arcmed.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Gomez-Anson B, Alegret M, Munoz E, Sainz A, Monte GC, Tolosa E. Decreased frontal choline and neuropsychological performance in preclinical Huntington disease. Neurology. 2007;68:906–910. doi: 10.1212/01.wnl.0000257090.01107.2f. [DOI] [PubMed] [Google Scholar]
- 10.Gujar SK, Maheshwari S, Bjorkman-Burtscher I, Sundgren PC. Magnetic resonance spectroscopy. J Neuroophthalmol. 2005;25:217–226. doi: 10.1097/01.wno.0000177307.21081.81. [DOI] [PubMed] [Google Scholar]
- 11.Jenkins BG, Koroshetz WJ, Beal MF, Rosen BR. Evidence for impairment of energy metabolism in vivo in Huntington’s disease using localized 1H NMR spectroscopy. Neurology. 1993;43:2689–2695. doi: 10.1212/wnl.43.12.2689. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins BG, Rosas HD, Chen YC, Makabe T, Myers R, MacDonald M, Rosen BR, Beal MF, Koroshetz WJ. 1H NMR spectroscopy studies of Huntington’s disease: correlations with CAG repeat numbers. Neurology. 1998;50:1357–1365. doi: 10.1212/wnl.50.5.1357. [DOI] [PubMed] [Google Scholar]
- 13.Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR. A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin Genet. 2004;65:267–277. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- 14.Mekle R, Mlynarik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med. 2009;61:1279–1285. doi: 10.1002/mrm.21961. [DOI] [PubMed] [Google Scholar]
- 15.Paulsen JS, Langbehn DR, Stout JC, Aylward E, Ross CA, Nance M, Guttman M, Johnson S, McDonald M, Beglinger LJ, Duff K, Kayson E, Biglan K, Shoulson I, Oakes D, Hayden M. Detection of Huntington’s disease decades before diagnosis: the predict HD study. J Neurol Neurosurg Psychiatry. 2007;79(8):874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penney JB, Jr, Vonsattel JP, MacDonald ME, Gusella JF, Myers RH. CAG repeat number governs the development rate of pathology in Huntington’s disease. Ann Neurol. 1997;41:689–692. doi: 10.1002/ana.410410521. [DOI] [PubMed] [Google Scholar]
- 17.Perneger TV. What’s wrong with Bonferroni adjustments. BMJ. 1998;316:1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Politis M, Pavese N, Tai YF, Tabrizi SJ, Barker RA, Piccini P. Hypothalamic involvement in Huntington’s disease: an in vivo PET study. Brain. 2008;131:2860–2869. doi: 10.1093/brain/awn244. [DOI] [PubMed] [Google Scholar]
- 19.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 20.Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14:260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds NC, Jr, Prost RW, Mark LP. Heterogeneity in 1H-MRS profiles of presymptomatic and early manifest Huntington’s disease. Brain Res. 2005;1031:82–89. doi: 10.1016/j.brainres.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 22.Roos RAC. Neuropathology of Huntington’s chorea. In: Vinken PJ, Bruyn GW, Klawans HL, editors. Handbook of clinical neurology, extrapyramidal disorders. Amsterdam: Elsevier Science Publishers; 1986. [Google Scholar]
- 23.Roze E, Saudou F, Caboche J. Pathophysiology of Huntington’s disease: from huntingtin functions to potential treatments. Curr Opin Neurol. 2008;21:497–503. doi: 10.1097/WCO.0b013e328304b692. [DOI] [PubMed] [Google Scholar]
- 24.Ruocco HH, Lopes-Cendes I, Li LM, Cendes F. Evidence of thalamic dysfunction in Huntington disease by proton magnetic resonance spectroscopy. Mov Disord. 2007;22:2052–2056. doi: 10.1002/mds.21601. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Pernaute R, Garcia-Segura JM, del Barrio AA, Viano J, de Yebenes JG. Clinical correlation of striatal 1H MRS changes in Huntington’s disease. Neurology. 1999;53:806–812. doi: 10.1212/wnl.53.4.806. [DOI] [PubMed] [Google Scholar]
- 26.Schmand B, Bakker D, Saan R, Louman J. The Dutch reading test for adults: a measure of premorbid intelligence level. Tijdschr Gerontol Geriatr. 1991;22:15–19. [PubMed] [Google Scholar]
- 27.Sturrock A, Laule C, Decolongon J, Dar SR, Coleman AJ, Creighton S, Bechtel N, Reilmann R, Hayden MR, Tabrizi SJ, MacKay AL, Leavitt BR. Magnetic resonance spectroscopy biomarkers in premanifest and early Huntington disease. Neurology. 2010;75(19):1702–1710. doi: 10.1212/WNL.0b013e3181fc27e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabrizi SJ, Blamire AM, Manners DN, Rajagopalan B, Styles P, Schapira AH, Warner TT. Creatine therapy for Huntington’s disease: clinical and MRS findings in a 1-year pilot study. Neurology. 2003;61:141–142. doi: 10.1212/01.wnl.0000070186.97463.a7. [DOI] [PubMed] [Google Scholar]
- 29.Tabrizi SJ, Langbehn DR, Leavitt BR, Roos RA, Durr A, Craufurd D, Kennard C, Hicks SL, Fox NC, Scahill RI, Borowsky B, Tobin AJ, Rosas HD, Johnson H, Reilmann R, Landwehrmeyer B, Stout JC. Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8(9):791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabrizi SJ, Langbehn DR, Leavitt BR, Roos RA, Durr A, Craufurd D, Kennard C, Hicks SL, Fox NC, Scahill RI, Borowsky B, Tobin AJ, Rosas HD, Johnson H, Reilmann R, Landwehrmeyer B, Stout JC. Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8(9):791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor-Robinson SD, Weeks RA, Bryant DJ, Sargentoni J, Marcus CD, Harding AE, Brooks DJ. Proton magnetic resonance spectroscopy in Huntington’s disease: evidence in favour of the glutamate excitotoxic theory. Mov Disord. 1996;11:167–173. doi: 10.1002/mds.870110209. [DOI] [PubMed] [Google Scholar]
- 32.Taylor-Robinson SD, Weeks RA, Sargentoni J, Marcus CD, Bryant DJ, Harding AE, Brooks DJ. Evidence for glutamate excitotoxicity in Huntington’s disease with proton magnetic resonance spectroscopy. Lancet. 1994;343:1170. doi: 10.1016/S0140-6736(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 33.Tkac I, Oz G, Adriany G, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at high magnetic fields: metabolite quantification at 4T vs. 7T. Magn Reson Med. 2009;62:868–879. doi: 10.1002/mrm.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Bogaard SJ, Dumas EM, Acharya TP, Johnson H, Langbehn DR, Scahill RI, Tabrizi SJ, van Buchem MA, van der Grond J, Roos RA. Early atrophy of pallidum and accumbens nucleus in Huntington’s disease. J Neurol. 2010;258(3):412–420. doi: 10.1007/s00415-010-5768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Oostrom JC, Sijens PE, Roos RA, Leenders KL. 1H magnetic resonance spectroscopy in preclinical Huntington disease. Brain Res. 2007;1168:67–71. doi: 10.1016/j.brainres.2007.05.082. [DOI] [PubMed] [Google Scholar]