Summary
Chlamydia trachomatis is an obligate intracellular pathogen responsible for ocular and genital infections of significant public health importance. C. trachomatis undergoes a biphasic developmental cycle alternating between two distinct forms: the infectious elementary body (EB), and the replicative but non-infectious reticulate body (RB). The molecular basis for these developmental transitions and the metabolic properties of the EB and RB forms are poorly understood as these bacteria have traditionally been difficult to manipulate through classical genetic approaches. Using two-dimensional liquid chromatography – tandem mass spectrometry (LC/LC-MS/MS) we performed a large-scale, label-free quantitative proteomic analysis of C. trachomatis LGV-L2 EB and RB forms. Additionally, we carried out LC-MS/MS to analyze the membranes of the pathogen-containing vacuole (“inclusion”). We developed a label-free quantification approaches to measure protein abundance in a mixed-proteome background which we applied for EB and RB quantitative analysis. In this manner, we catalogued the relative distribution of >54% of the predicted proteins in the C. trachomatis LGV-L2 proteome. Proteins required for central metabolism and glucose catabolism were predominant in the EB, whereas proteins associated with protein synthesis, ATP generation and nutrient transport were more abundant in the RB. These findings suggest that the EB is primed for a burst in metabolic activity upon entry, whereas the RB form is geared towards nutrient utilization, a rapid increase in cellular mass, and securing the resources for an impending transition back to the EB form. The most revealing difference between the two forms was the relative deficiency of cytoplasmic factors required for efficient Type III secretion (T3S) in the RB stage at 18 hpi, suggesting a reduced T3S capacity or a low frequency of active T3S apparatus assembled on a “per organism” basis. Our results show that EB and RB proteomes are streamlined to fulfill their predicted biological functions: maximum infectivity for EBs and replicative capacity for RBs.
Keywords: Chlamydia, Pathogenesis, Elementary body, Reticulate body, quantitative proteomics
Introduction
The obligate intracellular bacterium Chlamydia trachomatis is an important human pathogen that causes a variety of ocular and genital infections of significant clinical and public health importance. C. trachomatis is the most common bacterial cause of sexually transmitted diseases (WHO, 2001, Miller et al., 2004) and trachoma, the leading cause of infectious blindness worldwide (WHO, 2003, Burton & Mabey, 2009). A significant proportion of genital infections caused by C. trachomatis are left untreated due to their asymptomatic nature (Stamm, 1999), leading to serious complications including pelvic inflammatory disease, ectopic pregnancies and infertility, primarily in young women (Bebear & de Barbeyrac, 2009, Haggerty et al., 2010). Additionally, neonates exposed to this bacterium in the birth process can develop complications in the first months of life such as conjunctivitis and/or pneumonia (Darville, 2005).
As all bacteria within the family Chlamydiaceae, C. trachomatis undergoes a unique, biphasic developmental cycle alternating between two forms with clear functional differences. The elementary body (EB) is environmentally stable, metabolically dormant, infectious, and smaller in size compared to the reticulate body (RB), which is environmentally labile, highly replicative and non-infectious. Shortly after attachment, the EB secretes proteins into epithelial cells to promote their uptake (Clifton et al., 2004, Dautry-Varsat et al., 2005). Once inside the cell, the EB transitions to the RB form which replicates within a membrane-enclosed parasitopherous vacuole termed an “inclusion”. As the inclusion expands, replication becomes asynchronous and RBs begin to differentiate back into EBs. This differentiation involves a transition form, the intermediate body (IB), which can be observed under the electron microscope as an RB with a condensed nucleoid (Phillips et al., 1984). Throughout the infectious cycle, the bacterium exports an arsenal of effector proteins both to the inclusion membrane and to the host cytosol in order to manipulate host processes [(Betts et al., 2009) and references therein]. Notably, Chlamydia successfully avoids lysosomal fusion with the inclusion while selective interactions with other cellular compartments are maintained to grant the bacteria access to essential nutrients and allow intracellular survival [(Valdivia, 2008, Saka & Valdivia, 2010) and references therein]. At the end of the cycle, the inclusion occupies most of the host cell cytoplasm and EBs are released to the extracellular environment where other cells can be targeted for infection (Hybiske & Stephens, 2007). The transition from EB to RB, and back, all constitute obligatory steps in replication and dissemination of Chlamydiae and are likely achieved by functional partitioning of distinct sets of bacterial proteins into these developmental forms.
In the past decade, the genomes of numerous Chlamydia species and strains have been sequenced, providing a wealth of information as to the basic biology of these organisms (Stephens et al., 1998, Read et al., 2000, Read et al., 2003, Thomson et al., 2008). Also, transcriptional profiling of Chlamydia at various stages during the infectious cycle or during stress-induced persistence has provided a basic blueprint as to when each gene is activated (Albrecht et al., 2010, Belland et al., 2003). However, lack of information on when proteins are expressed and how their abundance changes during developmental transitions has limited our understanding of the physiology and pathogenesis of these experimentally fastidious organisms. Mass spectrometry (MS)-based methods for peptide identification and sequencing techniques allow the assessment of large-scale surveys of the repertoire of proteins expressed by an organism (“proteomics”) and have been applied for the study of microbial pathogenesis (Bhavsar et al., 2010). Indeed, several proteomics studies for Chlamydia have been published (Shaw et al., 2002, Mukhopadhyay et al., 2004, Wehrl et al., 2004, Skipp et al., 2005, Liu et al., 2010, Vandahl et al., 2002a, Vandahl et al., 2001), although they primarily relied on protein identification. Recent developments in MS coupled with powerful database search algorithms enable reliable and highly reproducible identification and quantification of up to thousands of proteins in highly complex mixtures (Nilsson et al., 2010, Walther & Mann, 2010). Therefore, quantitative proteomics offers a unique opportunity to address the biology of genetically intractable pathogens like C. trachomatis.
However, quantitative proteomic analyses on obligate intracellular pathogens like Chlamydia faces the unique challenge that isolation of pure bacterial protein extracts without host protein contaminants is nearly impossible, which can introduce a significant quantitative bias especially when the contribution of host proteins to the total sample vary among developmental forms and experimental samples. To overcome this, we developed an extension of the “Absolute Quantification” approach of Silva and Geromanos (Silva et al., 2006b, Silva et al., 2006a, Geromanos et al., 2009). We applied this experimental approach to perform a detailed proteomic comparison of EB and RB forms by LC/LC-MS/MS (with both, data-dependent and data-independent acquisition modes) (Delahunty & Yates, 2005, Silva et al., 2006b). Additionally, we determined the compendium of chlamydial proteins associated to inclusion membranes by LC-MS/MS. Results from this study underscore how the differential composition of EB and RB proteomes reflects the adaptation of these two developmental forms to fulfill their main biological functions, which are to achieve maximum infectivity for EBs and replicative capacity for RBs.
Results and Discussion
Proteomic analysis of C. trachomatis developmental forms by LC/LC-MS/MS
We chose a plaque purified C. trachomatis, biovar LGV, serovar L2, 434/Bu isolate for large-scale preparations and proteomic analysis. The developmental cycle of L2 434/Bu has been extensively characterized (Figure S1A) (Nicholson et al., 2003). Within 2 to 6 h post internalization, the EB differentiates into the RB form within nascent inclusions. Over the following hours, the RB increases in size and divides by binary fission, reaching maximum RB to EB ratio about 18-24 hours post-infection. At this stage, chlamydial replication becomes asynchronous and RBs initiate their differentiation back to the EB form with maximal production of infectious progeny and cell lysis occurring about 44-48 hpi (Nicholson et al., 2003). For C. trachomatis L2, 18 hpi is the point of the cycle when the inclusion consists mostly of RBs with very few EBs, and some RB forms beginning their developmental transition back to the EB (Nicholson et al., 2003). Earlier time-points (i.e. 6 or 12 hpi) did not yield enough material for LC/LC-MS/MS and later time-points (i.e. 30 or 36 hpi) would require the efficient separation of EBs and RBs at different stages of differentiation, which is currently not technically feasible. Therefore, to achieve maximal yields of each developmental form with minimal cross-contamination we purified RBs at 18 hpi and EBs at 44 hpi by sequential density gradients (Figure S1B). A potential caveat to this approach is that the RB fraction we analyzed represents a single time point during infection and its protein composition may not necessarily reflect that of RBs existing at earlier or later stages in the infectious cycle.
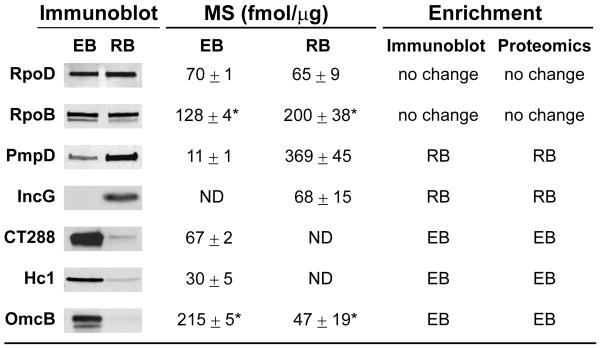
Gradient purified bacteria were analyzed by fluorescence microscopy and the EB-fraction primarily showed small particles less than 0.5 μm in size, compared to the RB-rich fraction, which showed larger (∼1 μm) cocci-shaped bacteria (Figure S1C). The purified fractions were used to prepare total EB and RB protein lysates (Figure S1D) and subjected to immunoblot analysis. Consistent with previous reports, the histone-like protein Hc1 which binds to the EB nucleoid (Wagar & Stephens, 1988) and the large cysteine-rich periplasmic protein OmcB (Watson et al., 1994), were highly enriched in EB fractions whereas the early inclusion membrane protein IncG (Scidmore-Carlson et al., 1999) was only detected in the RB form (Figure 1). EB and RB total protein lysates were then digested with trypsin and the resulting peptides subjected to reverse phase LC/LC-MS/MS, allowing for the implementation of label-free, MS-based determination of protein abundance as previously described (Delahunty & Yates, 2005, Silva et al., 2006b).
Figure 1. Immunoblot analysis of selected developmental stage-specific C. trachomatis proteins.
The relative abundance in EBs and RBs for a subset of chlamydial proteins was determined by immunoblot analysis and compared to mass spectrometry (MS)-based quantification. Quantitative trends were defined as “enrichment in EB” or “enrichment in RB” when EB to RB fold change was ≥ 2 and ≤ -2; otherwise they were considered as “no change”. Proteins that were below MS detection limits are indicated with “ND” (not detected). Asterisks added to RpoB and OmcB quantitative values represent a “quantitative flag”, indicating that mass spectrometry-based quantification may be less accurate due to differences in the tryptic peptide profiles observed for those proteins.
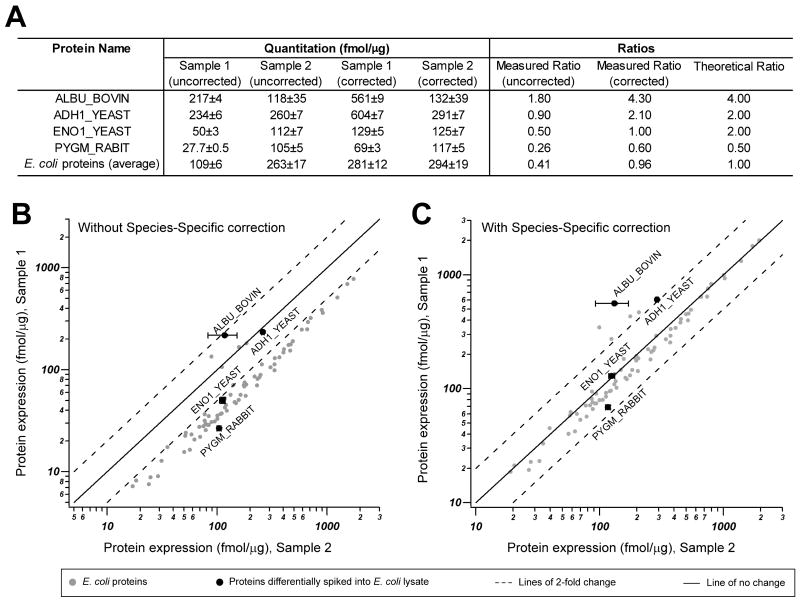
Despite extensive density gradient-based purification of bacteria, a total of 4360 unique peptides of human origin were detected, representing HeLa proteins that co-purified in our gradients. These “mixed-species” proteomes challenge label-free quantification methods, especially when the relative contribution of each species to the resulting proteome varies significantly among samples that are to be compared directly (e.g. RB vs. EB). To measure protein abundance in complex peptide mixtures containing a single target species and one or more “contaminating” species, we modified a previously-described “absolute quantification” approach (Silva et al., 2006b), so that even high levels of interference from other proteomes can be normalized out of the final quantitative measurement. Briefly, the methodology consisted of first calculating the fmol/μg of each protein followed by normalization to the proteome of interest (“species-specific” correction). We validated this approach by analyzing one sample representing a “mixed proteome” (Sample 1, Escherichia coli and mouse brain lysates, 50% each) and another representing a “pure proteome” (Sample 2, E. coli lysate). These samples were spiked with four selected proteins at pre-defined amounts and ratios prior to performing LC-MS/MS analysis and label-free quantification (Figure 2A). There was a systematic bias in the E. coli and spiked-in proteins, which was corrected upon applying the experimentally-derived “species-specific” correction factor (Figure 2B and C). Having accounted for these biases on quantification protocols, we proceeded to carry out the MS analysis of C. trachomatis EB and RB samples with differing degrees of human protein contamination. In addition, to directly compare EB and RB, samples were normalized on a “per organism” basis, as detailed in the Experimental procedures section.
Figure 2. Validation of label-free protein quantification of a complex mixture of proteins from two different species (“mixed proteomes”).
A. We generated an artificial E. coli – mouse brain model system to assess the accuracy of MS-based label free quantification of proteins in a multi-species protein lysate. In addition, four exogenous proteins were spiked at pre-defined ratios as indicated (theoretical ratio) into 1:1 E. coli:mouse brain lysate (Sample 1, representing a “mixed proteome”) or E. coli lysates (Sample 2, representing a single proteome). After trypsin digestion and LC-MS/MS analysis, the quantitative measurements (fmol/μg) of proteins were assessed with and without species-specific correction. When species-specific correction was applied, the corrected measured ratio was almost identical to the theoretical ratio. B-C. Lower panels show a graphic representation of the direct quantitative comparison of all proteins from Sample 1 and Sample 2, without and with species-specific correction, as indicated. Note that when all proteins in the sample are considered and no correction is applied (B), an obvious quantitative bias towards lower quantities in Sample 1 by approximately 2× is observed, as expected. When using the total micrograms of only the identified E. coli proteins to normalize protein concentration as means of “species-specific” correction (C), the result is that the correct ratio of E. coli and spiked-in proteins are reproduced. ADH1_YEAST, yeast alcohol dehydrogenase; ENO1_YEAST, yeast enolas; ALBU_BOVIN, bovine albumin; PYGM_RABBIT, rabbit glycogen phosphorylase. A logarithmic scale was used for x and y axis.
The C. trachomatis proteome
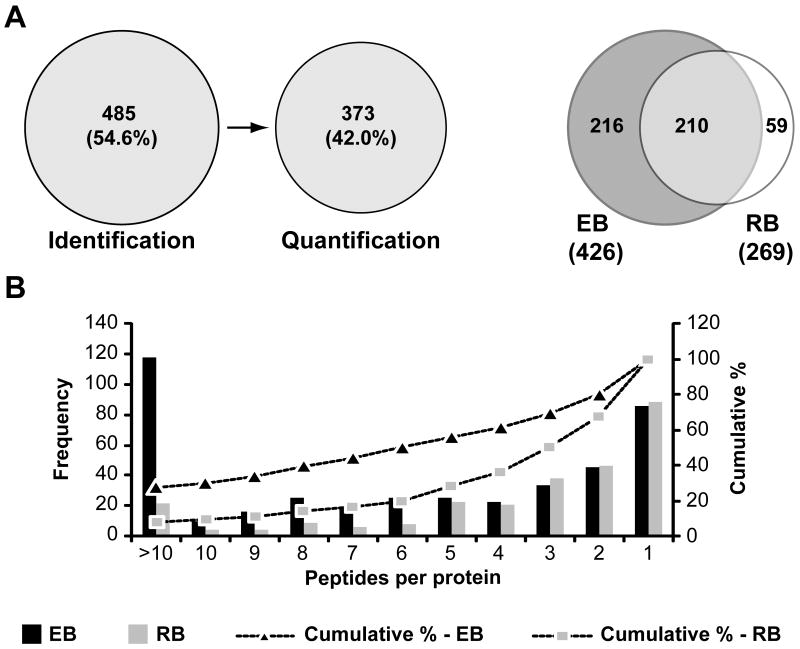
The MS analysis of C. trachomatis resulted in the identification of 4516 unique chlamydial peptides mapping to 485 non-redundant chlamydial proteins distributed between the two developmental forms and representing about 54% of the predicted proteome (Figure 3A). The majority of identifications relied on multiple peptides per protein with 67.3% and 80.6% of proteins having 2 or more peptides detected in RB and EB forms, respectively (Figure 3B). The maximum number of unique peptides for the most abundant protein was >100 (Table S1) and the average number of peptides per protein was ∼9 for EBs and ∼5 for RBs (data not shown). This is in contrast with a previous report where ∼320 chlamydial proteins were identified in EBs and RBs combined but ∼52% of them with only one peptide to match (Skipp et al., 2005). The identification of multiple peptides per protein allowed the quantification of 373 non-redundant chlamydial proteins (Figure 3A) with 97.6% of the proteins quantified with a relative standard deviation of less than 50% (Table S1 and data not shown).
Figure 3. Identification and quantification of the C. trachomatis L2 proteome.
(A) Venn diagrams indicating the number of proteins identified and quantified by LC/LC-MS/MS (left panel) and the overlap in protein identification among the two C. trachomatis developmental forms (right panel). (B) Frequency histogram displays the distribution of the number of unique peptides per protein. The primary y axis (bars) shows the number of proteins detected for each particular number of peptides per protein in EB and RB samples. The secondary y axis (dotted lines) is a representation of the cumulative percentage of proteins detected along decreasing number of unique peptides per protein; 67.3% and 80.6% of proteins in EBs and RBs, respectively, were identified with 2 or more peptides to match.
We independently verified the MS-based quantification performed by immunoblot analysis of selected proteins. For example, CT288 a putative inclusion membrane protein (Inc) of unknown function, was determined by MS to be exclusively in EBs (Figure 1). We generated antibodies against CT288 and confirmed that, like Hc1, this protein was mainly present in the EB, suggesting that this putative Inc protein, unlike other Inc proteins that are expressed during the RB-stage (Giles et al., 2006), performs a function early upon EB invasion. Similarly, quantitative MS determined a higher abundance of PmpD in the RB form, which was also verified by immunoblots (Figure 1). Other “house-keeping” proteins, such as the core components of RNA polymerase, RpoD and RpoB, were present in equal amounts in the two developmental forms. Overall, we found a very good agreement between MS-based quantification and immunoblot analysis. However, a small subset (4%) of proteins identified displayed significant differences in the relative abundance of individual tryptic peptides generated between the EB and RB forms. Consequently, for this subset of proteins the top 3 most abundant peptides were not entirely overlapped between EB and RB. Because the protein quantification algorithms used rely on a similar tryptic peptide profile being generated under the two experimental conditions, the comparative MS-based analysis for this protein subset may be less accurate. These proteins have been “flagged” in our datasets (Table S1).
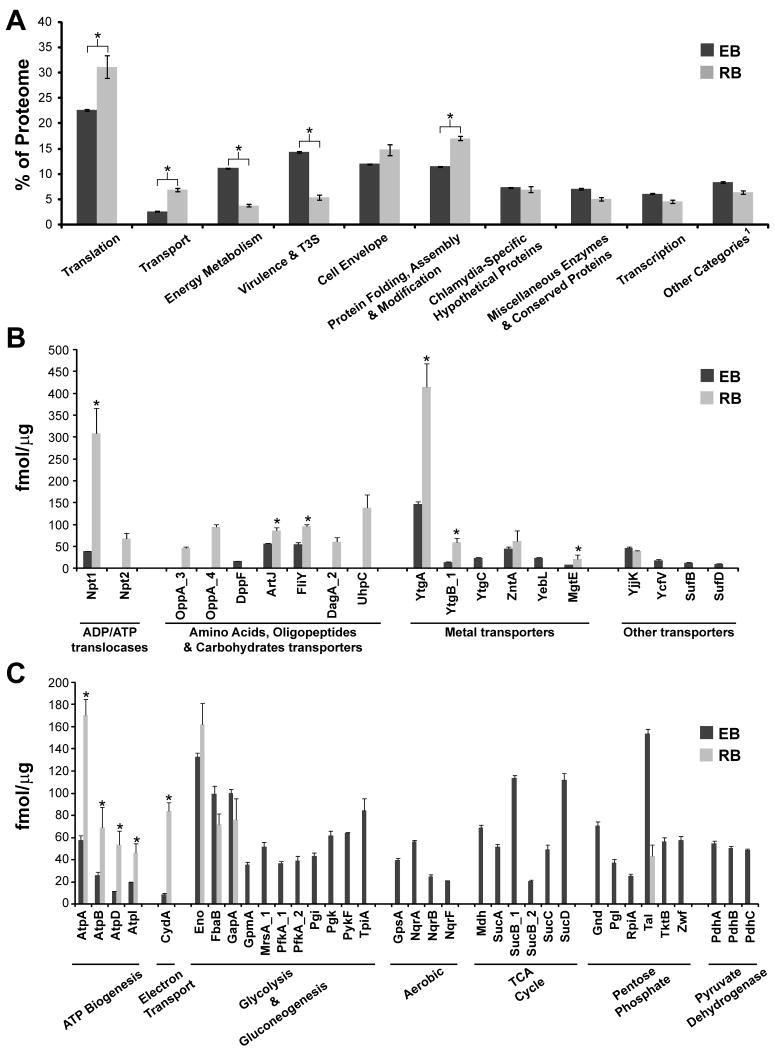
We annotated all proteins identified by MS, placed them in functional categories (Table S1), and determined the quantitative contribution of each category to the total EB and RB proteomes (Table 1). Notably, the six more prominent categories accounted for 76.4% and 80.5% of the total of EB and RB quantitated proteins, respectively (data not shown) and five out of these six top categories were shared between the two developmental forms (Table 1). However, their relative contribution to the total mass of EBs and RBs differed, particularly in the “Translation”, “Energy Metabolism”, “Transport”, “Protein Folding” and “Virulence & T3S” categories. Below we discuss how this detailed MS-based analysis of the chlamydial proteome revealed new insights into Chlamydia biology.
Table 1. Quantitative distribution of functional groups in EB and RB proteomes.
| Functional groups | EB - fmol/μg, (Rank) | RB - fmol/μg, (Rank) |
|---|---|---|
| Translation | 4661 ± 45, (1) | 6440 ± 462, (1) |
| Virulence & T3S-related | 2903 ± 37, (2) | 1310 ± 115, (6) |
| Cell Envelope | 2463 ± 31, (3) | 3063 ± 225, (3) |
| Protein Folding, Assembly & Modification | 2362 ± 72, (4) | 3513 ± 85, (2) |
| Energy Metabolism | 2300 ± 21, (5) | 777 ± 42, (9) |
| Chlamydia-Specific Hypothetical Proteins | 1503 ± 28, (6) | 1425 ± 115, (5) |
| Miscellaneous Enzymes&Conserved Proteins | 1449 ± 37, (7) | 1042 ± 58, (7) |
| Transcription | 1256 ± 12, (8) | 929 ± 65, (8) |
| Base & Nucleotide Metabolism | 530 ± 10, (9) | 208 ± 42, (12) |
| Transport | 516 ± 11, (10) | 1494 ± 70, (4) |
| DNA Replication, Modification, Repair & Recombination | 478 ± 27, (11) | 436 ± 28, (10) |
| Central Intermediary Metabolism | 182 ± 7, (12) | 58 ± 2, (15) |
| Standard Protein Secretion | 181 ± 10, (13) | 428 ± 83, (11) |
| Amino Acid Biosynthesis | 152 ± 7, (14) | 120 ± 16, (13) |
| Signal Transduction | 112 ± 5, (15) | 32 ± 6, (16) |
| Biosynthesis of Cofactors | 83 ± 4, (16) | 118 ± 9, (14) |
| Cell Division | 54 ± 4, (17) | 18 ± 3, (17) |
All C. trachomatis L2 proteins quantified were classified into functional categories as described in Experimental procedures. The total mass for each category in EB and RB forms was calculated (fmol/μg) and expressed as the mean ± standard deviation resulting from 4 determinations (2 biological replicates processed in duplicate). Functional groups were then ranked based on abundance. Protein groups ranked within the top 6, representing 76.4% and 80.5% of the total mass in EB and RB, respectively, are highlighted in bold.
The C. trachomatis cell envelope
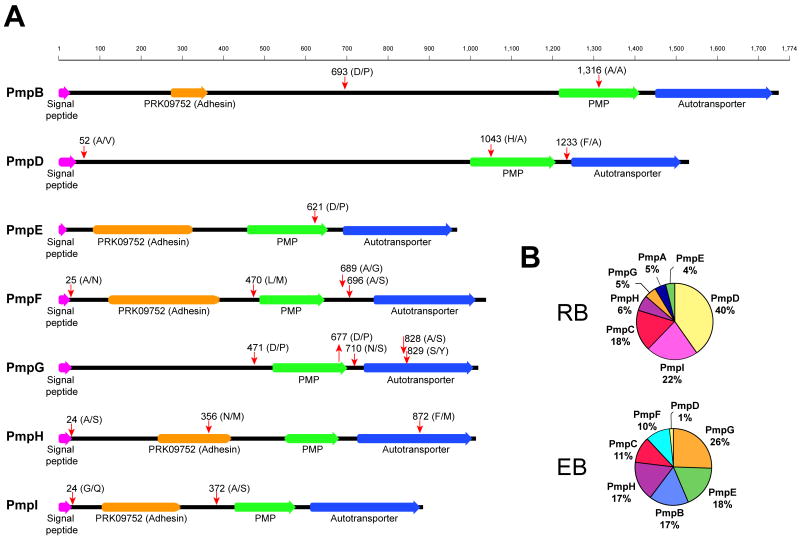
Proteins in the C. trachomatis envelope are attractive targets for the development of neutralizing vaccines (Howie et al., 2010, Peterson et al., 1991, Koehler et al., 1992, Zhang et al., 1987). Polymorphic membrane proteins (Pmps) are a diverse group of cell envelope proteins that are postulated to play a role in virulence, immunity and are leading candidates as targets for vaccine development (Gomes et al., 2006, Tan et al., 2010, Longbottom et al., 1998, Grimwood et al., 2001, Crane et al., 2006). All nine Pmps (PmpA to PmpI) encoded in the C. trachomatis genome (Stephens et al., 1998) were identified in our study with their relative abundance varying between developmental forms (Table S1 and Figure 4B), in agreement with previous studies (Skipp et al., 2005). PmpA and PmpI were only detected in the RB form whereas PmpF was only detected in the EB form; PmpB, E and G were more abundant in EBs as opposed to PmpC and D, which were more abundant in the RB form, and PmpH was expressed at similar levels in both developmental forms (Table S1). Pmps are classified as autotransporters with an N-terminal passenger domain and a C-terminal β-barrel autotransporter domain that allows translocation of proteins across the bacterial outer membrane (Henderson et al., 2004). The passenger domain can remain tethered to the outer membrane or be cleaved from the autotransporter domain and secreted (Henderson et al., 2004). We looked for evidence of Pmp processing by searching for semitryptic peptides (peptides not predicted to be generated by trypsin cleavage alone) in our MS/MS analysis, an approach that has been used to delineate proteolytically processed proteins (Thomas et al., 2010). We detected semitryptic peptides that mapped to all Pmps analyzed except for PmpA and PmpC. Several of these semitryptic peptides mapped to the predicted signal peptidase cleavage sites - to remove leader sequences of secreted proteins- and to sites adjacent to the autotransporter domain, especially for PmpD-G (Figure 4A and Table S2). In agreement with these findings, other investigators already suggested that posttranslational processing is an important feature of some Pmps. For instance, PmpB and D have been previously shown to be cleaved in C. trachomatis (Shaw et al., 2002, Kiselev et al., 2007, Kiselev et al., 2009) and evidence of Pmps processing has also been found in C. pneumoniae (Vandahl et al., 2002b, Grimwood et al., 2001). For PmpC we found no semitryptic peptides even though multiple peptides matching this protein were detected (16 in EB and 23 in RB, Table S1). Overall, these findings indicate that all Pmps are expressed, are targets of Sec-dependent transport, and undergo extensive proteolytic processing consistent with their function as autotransporters (Tan et al., 2010). Pmps are major components of the chlamydial outer membrane complex (COMC) and a recent proteomic study has focused on the COMC proteome to reveal new antigens for vaccination (Liu et al., 2010). We found that all proteins classified as being part of the COMC (Liu et al., 2010) were highly expressed in EBs, with MOMP, OmcB, CT623 and PmpG being the most abundant ones (Table S3). This is in agreement with previous reports documenting that these proteins are very abundant in the EB (Caldwell et al., 1981, Hatch et al., 1984, Tanzer & Hatch, 2001, Birkelund et al., 2009, Liu et al., 2010)
Figure 4. Semitryptic peptide analysis indicates that C. trachomatis polymorphic membrane proteins (PMPs) are extensively processed.
(A) Semitryptic peptides corresponding to PMPs were analyzed and mapped to the corresponding protein sequence. Potential cleavage sites identified are shown (red arrows). Cleavage motif, amino acid position, domains and signal peptides are shown. Signal peptides are indicated based on SignalP 3.0 prediction (http://www.cbs.dtu.dk/services/SignalP/) trained for Gram negative bacterial species. (B) Pie chart representing the relative abundance of different PMPs in the EB and RB forms, expressed as percentage of total mass corresponding to PMPs.
Proteins required for mRNA translation, protein transport and protein folding are abundant in the RB form
Factors required for mRNA translation constituted ∼22% of the mass of all proteins quantified in the EB and ∼31% in the RB form (Figure 5A), with the bulk of proteins corresponding to ribosomal proteins (Figure S2A). Out of 54 ribosomal proteins encoded in the C. trachomatis L2 434/Bu genome, 50 were detected in this study (Table S1), with the majority of them being expressed at higher levels in the RB form (Figure S2B), indicating that the RB devotes a significant proportion of its proteome to actively synthesizing proteins. Factors required for protein folding, assembly and post-translational modifications (chaperones, proteases, peptidases and isomerases -Table S1) were also prominent in the RB form, contributing to ∼ 17% of the proteome, compared to ∼ 11% in the EB (Figure 5A and Table 1).
Figure 5. Quantitative comparison of the EB and RB proteomes.
(A) Proteins were grouped into functional categories (as detailed in Table S1) and the total mass for each category was calculated. Values represent the mean contribution of each category expressed as percentage of the total proteome, resulting from four independent MS-based determinations. Error bars represent the standard deviation. Asterisks indicate statistically significant differences (p<0.01, Alternate Welch T-test not assuming equal standard deviation). 1. All protein categories that individually represented less than 3% of all quantified proteins both in EB and RB were grouped together as “Other”; these are “Base & Nucleotide Metabolism”, “DNA Replication, Modification, Repair & Recombination”, “Central Intermediary Metabolism”, “Standard Protein Secretion”, “Amino Acid Biosynthesis”, “Signal Transduction”, “Biosynthesis of Cofactors” and “Cell Division”. Expression levels for individual proteins for which quantitative values could be calculated within the category “Transport” (B) and “Energy Metabolism” (C) are represented and expressed as the mean (fmol/μg) resulting from four independent mass spectrometry-based determinations. Sub-categories are indicated. Error bars indicate the standard deviation. Asterisks indicate statistically significant differences (p<0.05, Alternate Welch T-test not assuming equal standard deviation).
Transporters, permeases and translocators grouped in the “Transport” category accounted for ∼ 7% of the total mass in the RB form, as compared to ∼ 2.5% in the EB (Figure 5A). Prominent among this category were nutrient transporters. C. trachomatis encodes two integral membrane proteins with ATP/ADP antiporter activity, Npt1 and Npt2 (Stephens et al., 1998, Tjaden et al., 1999). Npt1 mediates the import of host cell ATP coupled with chlamydial ADP export whereas Npt2 has a broader nucleotide transport capacity as it can transport ATP, CTP, GTP and UTP in a proton-dependent manner (Tjaden et al., 1999). Npt1 was ∼ 8 times more abundant in the RB than in the EB form, and Npt2, while abundant in the RB, was below the detection levels in EBs (Figure 5B and Table S1).
Amino acid, oligopeptide and sugar transporters were also preferentially found in the RB form. For instance, OppA_3 and OppA_4, OppB_2 and OppC_2, components of ABC-type oligopeptides transport systems and YccA (a predicted transporter of unknown substrate), were only detected in RBs (Table S1 and Figure 5B). A previous proteomics study comparing EB and RB, also detected OppA_4 only in the RB form (Skipp et al., 2005). Likewise, the Na+-linked D-alanine glycine permease DagA_2 and the hexose phosphate transport protein UhpC, were enriched in the RB form (Figure 5B). UhpC was also predicted to be enriched in the RB form in a previous transcriptome analysis of C. trachomatis (Albrecht et al., 2010). A number of metals transporters including YtgA, a strongly immunogenic periplasmic chlamydial protein (Raulston et al., 2007) possibly involved in iron transport (Miller et al., 2009), were also detected. YtgA and other components of the ytgAC operon were considerably more abundant in the RB than in the EB form (Figure 5B). The overrepresentation of proteins required for ATP, amino acids, oligopeptides, carbohydrate and metal transport is consistent with a high demand for nutrients and co-factors by actively replicating RBs. Overall, the prevalence of ribosomal proteins, nutrient transporters and posttranslational processing factors in the RB at 18 hpi is consistent with high metabolic activity in bacteria that are undergoing robust protein synthesis.
The RB accumulates proteins involved in ATP generation
C. trachomatis encodes a nearly complete tricarboxylic acid (TCA), glycolysis and pentose phosphate pathways for glucose catabolism (Stephens et al., 1998), and possesses the ability to synthesize ATP and generate an ion gradient across the cytoplasmic membrane (Stephens et al., 1998). Several proteins in these pathways, grouped in the “Energy metabolism” category, were present in EB and RB forms (Table S1). Paradoxically, this cluster of proteins accounted for 3.8% of all proteins in the metabolically active RB as compared to 11.0% in the metabolically “inert” EB (Figure 5A), primarily due to higher amounts of proteins required for glucose catabolism in the EB form (Figure 5C). This ability to use glucose as fuel suggests that the EB is equipped to cope with a high demand for energy at the very early stages post invasion, which has been suggested before by other investigators (Vandahl et al., 2001, Skipp et al., 2005, Sixt et al., 2011). At the RB stage, C. trachomatis switches to ATP synthesis by generating an ion gradient. This is likely achieved by using a eukaryotic-like vacuolar-(V-type) ATPase, as opposed to the flagellar (F)-type ATPases found in most eubacteria, mitochondria and chloroplasts (Stephens et al., 1998, Dean, 2006). V-type ATPases couple the transfer of protons or sodium ions across cytoplasmic membranes with ATP hydrolysis or synthesis (Forgac, 2007). Whereas in eukaryotic cells these enzymes are primarily involved in generating proton gradients across membranes, prokaryotic V-type ATPases are responsible for ATP synthesis (McClarty, 1999, Numoto et al., 2009). All six predicted V-type ATPase subunits (AtpA, AtpB, AtpD, AtpE, AtpI and AtpK) were expressed both in the EB and RB forms (Table S1) but at higher levels in latter (Figure 5C). Consistent with this, components of the cytochrome bd respiratory oxidase (e.g.CydA) (McClarty, 1999), which presumably helps transfer protons to reduce oxygen, were more abundant in the RB (Figure 5C). Overall, these findings indicate that replicating RBs generate ample pools of ATP both through transport from the host and through synthesis. This excess of ATP may fuel the replication of bacteria but also may account for the observation that EBs possess a large depot of stored ATP (Tipples & McClarty, 1993). Presumably ATP synthesis ramps up as the RB begins its transition to the EB form, and provides the invasive form of the bacterium with the fuel required to drive protein secretion during invasion, nascent inclusion biogenesis and to prime the developmental transition to the RB form.
The EB is pre-loaded with an abundant arsenal of type III secretion effectors and chaperones
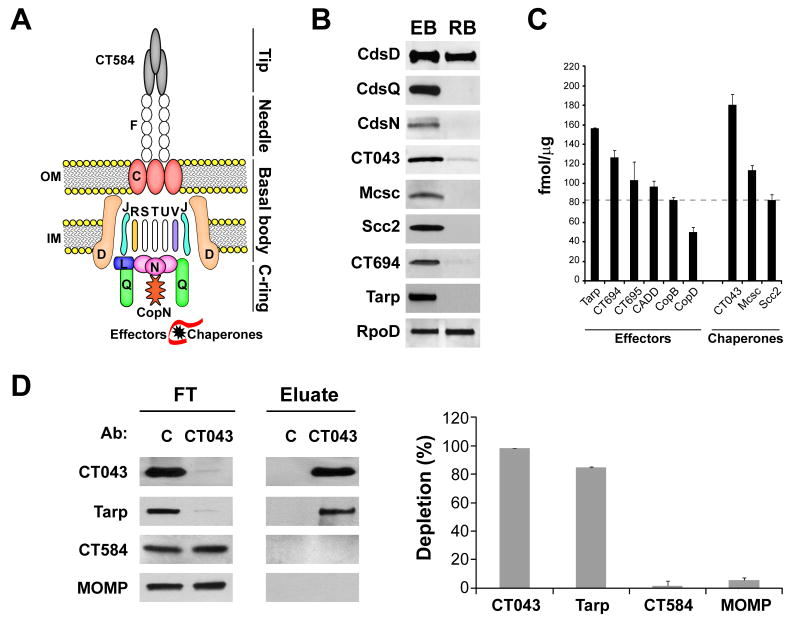
We observed significant differences in “Virulence & T3S” proteins between EB and RB forms. This protein group represented ∼ 14% of the EB proteome as opposed to ∼ 5% in the RB (Figure 5A). Most proteins within this group were related to Type III Secretion (T3S) system both in EB and RB (Table S1). The T3S apparatus is a conserved syringe-like multi-protein structure used by Gram-negative bacteria to inject protein effectors into eukaryotic host cells and as such, a functional T3S system is essential for virulence (Galan & Wolf-Watz, 2006, Beeckman & Vanrompay, 2010). The Chlamydiaceae encode a complete set of T3S genes (Hefty & Stephens, 2007, Peters et al., 2007), and assemble a T3S apparatus with a proposed architecture similar to that of other bacterial pathogens (Betts-Hampikian & Fields, 2010, Beeckman & Vanrompay, 2010). We identified most structural T3S-components (Figure 6A), 25 predicted T3S-effectors and 7 T3S-chaperones (Table S1). As shown in Figure 6B, we verified our proteomics results by immunoblot analysis of a subset of T3S-effectors (Tarp, CT694), T3S-components (basal body component CdsD and cytoplasmic accessory factors CdsQ and CdsN) and -chaperones (Mcsc, CT043, Scc2). T3S-chaperones and empirically verified T3S-effectors in EBs were among the top 25% most abundantly expressed proteins (Figure 6C). T3S chaperones stabilize secreted effectors, provide signals for efficient secretion and partially unfold effectors for translocation through the T3S needle (Yip et al., 2005). High expression of T3S-chaperones is consistent with proposed models where EBs are preloaded with abundant T3S-effectors (Peters et al., 2007). We determined that Tarp, an effector essential for invasion (Jewett et al., 2010), was the most abundant T3S-effector and CT043 (Spaeth et al., 2009) was the most abundant putative T3S-chaperone (Figure 6C) in EBs. We tested the hypothesis that the most abundant T3S-chaperone would bind to the most abundant effector by immunoprecipitating CT043 from EB total lysates and assessing the degree of Tarp co-immunoprecipitation. Anti-CT043 antisera depleted both CT043 and Tarp from EB lysates, but not CT584, an abundant predicted T3S needle-tip protein (Markham et al., 2009), or the major outer membrane protein MOMP (Figure 6D), underscoring that most Tarp in the EB is in complex with CT043. Similar results have been obtained for other abundant T3S effectors (Y. Chen and RH. Valdivia, unpublished).
Figure 6. T3S chaperones, cytoplasmic accessory factors and effectors are abundant in the EB form.
(A) Cartoon displays a schematic representation of the chlamydial T3S-system (Betts-Hampikian & Fields, 2010). Cds proteins are shown with letter designation only. Proteins for which expression was detected are shown in color. OM: outer membrane. IM: inner membrane. (B) Comparison of expression levels for selected T3S-components (basal body component CdsD and cytoplasmic accessory factors CdsQ and CdsN), -chaperones (Mcsc, CT043, Scc2), and effectors (Tarp, CT694) between EB and RB forms, as determined by immunoblots. (C) Expression levels for recognized T3S-effectors and –chaperones in EBs are represented as the mean (fmol/μg) resulting from four independent mass spectrometry-based determinations. Dotted line indicates the 75th percentile for the EB proteome (82.5 fmol/μg), highlighting that the shown effectors and chaperones rank in the top 25% of proteins by abundance. Error bars indicate the standard deviation. (D) The most abundant chaperone, CT043, is associated with the most abundant T3S effector, Tarp. Immunoblots correspond to immunoprecipitation experiments using rabbit-raised anti-CT043 antibody. The flow through (FT) and the bound material (Eluate) obtained after incubating EB lysates with beads crosslinked with either pre-immune serum (used as a control, C) or anti-CT043 sera, were blotted for CT043, Tarp, CT584 and MOMP (left panel). Quantitative immunoblots show the % depletion of Tarp, CT043, CT584 and MOMP compared to controls, after incubation with anti-CT043 crosslinked beads (right panel). Note co-depletion of Tarp by anti-CT043 antibodies. Result is representative of at least three independent experiments.
Interestingly, when we analyzed the expression levels of T3S-components, we found that whereas CdsD, a component of the basal body (Johnson et al., 2008), was equally abundant in the RB and EB forms, two components that are peripherally associated with the T3S basal apparatus, the C-ring component CdsQ (Johnson et al., 2008) and the ATPase CdsN (Stone et al., 2008), were largely absent in the RB (Table S1 and Figure 6B). In Shigella the C-ring component Spa33/SpaO and the ATPase Spa47 are required for the assembling of the needle and for the export of T3S-effectors (Jouihri et al., 2003, Morita-Ishihara et al., 2006). Both the C-ring component and the ATPase have been proposed to act as recognition platforms for T3S substrates (Beeckman & Vanrompay, 2010). Similarly, in the flagellar T3S system, the C-ring is required for proper assembly of the flagellum (Gonzalez-Pedrajo et al., 2006) and both the ATPase and the C-ring have been proposed to increase the efficiency of secretion of flagellar substrates (Erhardt & Hughes, 2010, Konishi et al., 2009). The relative absence of the cytoplasmic accessory factors CdsQ and CdsN in the RB forms suggest that additional factors may substitute for these components, as has been suggested for flagellar T3S system (Erhardt & Hughes, 2010, Konishi et al., 2009). The CdsN homologue CT717 could potentially substitute for CdsN at this stage. However, we were unable to detect CT717 by MS in either developmental form. Given that T3S chaperones are also markedly absent in RBs, an alternative interpretation for the absence of CdsQ and CdsN is that the RB stage has a reduced T3S capacity or that the number of active T3S apparatus is limited. Yet, we should highlight that this is unlikely to have an impact on secretion of non-T3S factors by RBs. Our results suggest that the RB can devote more resources to increase bacterial numbers and cellular mass by sharing the task of synthesizing and secreting virulence factors. As each RB begins its transition to the IB (∼20 hpi for C. trachomatis L2), we postulate that the synthesis of CdsQ, CdsN, T3S chaperones and EB-specific effectors ramps up to pre-pack the future EBs with these secretion enhancers.
Inclusion membrane proteomics
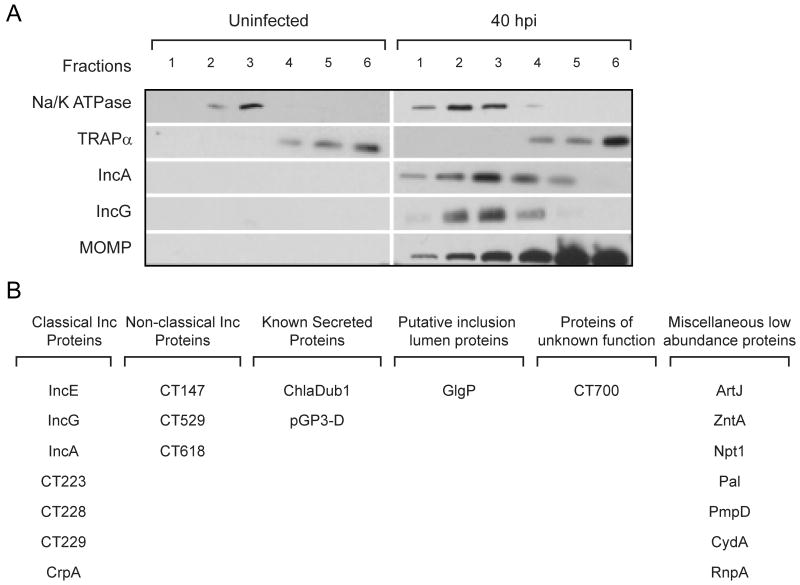
In addition to the EB and RB forms, the inclusion membrane itself is heavily modified by Chlamydia-derived proteins (Moorhead et al., 2010), representing a poorly explored subset of the chlamydial proteome in infected cells. Prominent among these are Inc proteins, putative T3S substrates that share a common bi-lobed hydrophobic motif predicted to direct their insertion into the inclusion membrane (Bannantine et al., 2000). To identify chlamydial proteins at the inclusion membrane, we fractionated membranes from infected cells on isopycnic density gradients and monitored fractions enriched for the well-characterized inclusion membrane proteins IncA and IncG (Li et al., 2008a) (Figure 7A). Total proteins were extracted from these fractions and subjected to LC-MS/MS analysis. Although the bulk of proteins (>99%) were of human origin, we were able to identify a number of chlamydial proteins which likely represent the top abundant proteins associated with the inclusion membrane. Within the chlamydial proteins identified, a small group of proteins expressed at very high levels in the EB form (mainly the components of the chlamydial outer membrane complex MOMP, PmpG and CT623, the T3S-component CdsD, the hypothetical protein CT875, the ABC transporter ytgA/CT067, few translation and initiation factors and one ribosomal protein– data not shown) were considered likely contaminants and removed from the analysis. This led to a final list of 21 chlamydial proteins over-represented in IncA/IncG enriched membrane fractions (Figure 7B). As expected, most proteins in this group consisted of predicted inclusion membrane proteins. In addition, we detected known secreted proteins like the virulence plasmid protein pGP3-D (Li et al., 2008b) and the predicted T3S-effector ChlaDub1/CT868 (Jehl et al., 2011), along with GlgP/CT248 involved in glycogen metabolism. Also, a number of miscellaneous proteins of low abundance in the EB but enriched in inclusion membrane fractions were detected, as detailed in Figure 7B. These included PmpD, which is was not identified as being enriched in the Chlamydia outer membrane complex in a recent publication (Liu et al., 2010), thus unlikely a contaminant in our inclusion membrane preparations. A surprising finding from this analysis is the presence of periplasmic and cytosolic membrane proteins in association with inclusion membrane fractions. These include an arginine transporter (ArtJ/CT381), a metal transporter (zntA/CT727), the ATP/ADP translocase (Npt1/CT065) and the cytochrome oxidase subunit I (CydA/CT013). Whether the enrichment of these proteins in the inclusion membrane fraction is nonspecific or they perform scavenging functions in the inclusion lumen or in association with inclusion membranes, remains to be determined.
Figure 7. Identification of C. trachomatis proteins associated with inclusion membranes.
(A) Immunoblots showing fractionation of total membranes extracted from uninfected cells and from cells infected with C. trachomatis L2 (40 hours post-infection). Na/K ATPase and TRAPα were used as markers for plasma and endoplasmic reticulum membranes, respectively. IncA and IncG were used as markers for inclusion membranes and MOMP indicates the fractions enriched in intact bacteria. Proteins from fractions 2 and 3, which were enriched for IncA and IncG, were extracted and analyzed by LC-MS/MS. (B) Compendium of C. trachomatis L2 proteins associated with inclusion membranes. We highlight previously identified inclusion membrane proteins (Li et al., 2008a) and proteins not abundant in EBs.
Conclusions
C. trachomatis, a pathogen with high impact on human health, has been experimentally difficult to study as traditional genetic and physiological approaches have been of limited use. With a quantitative proteomic approach, we provided molecular evidence of the different metabolic and virulence properties of the two developmental stages of C. trachomatis. We developed an approach to overcome the quantitative biases that arise from different levels of contamination of EB and RB extracts with host cell proteins. This approach can be applied to other studies where quantitative proteomics is carried out in the context of “mixed-proteomes”, such as host-pathogen and host-symbiont interactions.
It is important to note that for our proteomic study, the RB form was purified at 18 hpi, a stage at which RB replication is logarithmic and contamination with EB form (due to the asynchronous nature of EB to RB transition and eventually to a low number of dead EBs) is minimal (Nicholson et al., 2003). Our findings on RBs at this stage may not apply to RB forms at other stages of infection. The isolation and characterization of RB forms at different stages of infection for longitudinal studies is limited by the ability to isolate a pure population of these fragile Chlamydia forms and by the detection limits of current mass spectrometers. However, at the current rate of advances in MS-based detection and quantification methods such analysis may soon be possible.
Our quantitative proteomics study provided experimental confirmation for what had been suggested in transcriptional studies (Nicholson et al., 2003). The RB form at 18hpi was found to be primarily primed for robust protein synthesis, nutrient transport and the accumulation of ATP. Thus, the proteome of the RB at this stage is specialized for efficient replication and for the imminent transition to the EB form. In contrast, the EB is primed for high T3S capacity and for generating a burst of energy via glucose catabolism to fuel the EB to RB developmental transition. A novel observation from these studies is the relative lack in the RB form of T3S chaperones and cytoplasmic accessory factors required for efficient recognition of T3S substrates (Jouihri et al., 2003, Beeckman & Vanrompay, 2010, Morita-Ishihara et al., 2006). One possible interpretation of our findings is that as the EB transitions to the RB form there is a progressive reduced capacity in chaperone-assisted T3S by each individual RB. As infection progresses the need for T3S effector translocation could be incrementally compensated by the growing numbers of RBs within one inclusion. This would allow individual RBs to save resources for replication and EB generation and prevent a potential excess of T3S-effectors from being secreted by exponentially increasing numbers of RBs. Such excess of effectors could harm the host cell, disrupt the development of the inclusion or provide substrates for antigen presentation by the infected cell.
Experimental procedures
Culture conditions and purification of C. trachomatis EB and RB forms
C. trachomatis biovar LGV, serotype L2, strain 434/Bu was propagated in HeLa CCL2 monolayers (ATCC, Rockville, Maryland, USA) grown in Dulbecco's minimal essential medium (DMEM high glucose 1×) (Gibco/Invitrogen Life Technologies, Carlsbad, California, USA) supplemented with 10% fetal bovine serum (Mediatech, Inc., Manassas, Virginia, USA) at 37°C, 5% CO2 in a humidified atmosphere. Infections were carried out in T175 flasks (Sarstedt, Nümbrecht, Germany) by adding a suspension of EBs at a multiplicity of infection (MOI) of 10. C. trachomatis RB and EB forms were collected at 18 and 44 hpi, respectively. Purification of C. trachomatis EBs and RBs was performed by density gradient centrifugation essentially as previously described (Caldwell et al., 1981) with minor modifications. Briefly, Renografin was replaced by Omnipaque 350 (GE Healthcare, Princeton, New Jersey, USA) supplemented with NaCl 160 mM. The gradients were prepared by diluting Omnipaque 350-160 mM NaCl in SPG buffer (3.8 mM KH2PO4, 7.2 mM K2HPO4, 4.9 mM L-glutamic acid, 218 mM sucrose, pH=7.4) such that final concentrations of Omnipaque 350 were 28.5%, 38.0%, 41.8% and 51.3% in successive layers. To assure purity of LGV-L2, the strain was plaque purified twice (O'Connell & Nicks, 2006) before generating EB and RB forms used for proteomic analysis. All strains and cell lines were Mycoplasma -free as determined by a previously described PCR method (van Kuppeveld et al., 1992).
Preparation of C. trachomatis total protein extracts and normalization strategy
To prepare total protein extracts for LC/LC-MS/MS, EBs or RBs were suspended in lysis buffer (20 mM Tris.HCl pH 8.0, 5 mM EDTA, 50 mM NaCl, 15 mM DTT, 0.06 mM 2-Mercaptoethanol, 1 mM Na3VO4, 50 mM NaF, 100 mM NH4HCO3, 0.5% RapiGest SF Surfactant [Waters Corp., Milford, Maryland, USA] in 100 mM NH4HCO3), briefly sonicated on ice, heated at 65°C for 10 min and then boiled for 5 min in water. Lysates were centrifuged at 25,000 × g (10 min., 4°C) to remove insoluble debris and supernatants were stored at -80°C. Total protein content in the extracts was estimated by Bradford assay (Bradford, 1976). To visualize the band pattern (Figure S1D), 20 μg of EB and RB protein extracts were loaded onto SDS-PAGE 4-15% gradient gels (Bio-Rad, Hercules, California, USA) and stained with SYPRO Orange (Invitrogen Life Technologies, Carlsbad, California, USA), scanned in a Typhoon 9410 Variable Image Phosphor Imager and processed with ImageQuant 5.2 (GE Healthcare, Princeton, New Jersey, USA). For two preparations each of EB and RB samples, a total of 53 μg and 16.5 μg of protein respectively was digested with trypsin: briefly, samples concentrations were normalized to approximately 1 μg/μl (using BSA-calibrated Bradford assay) in 50 mM ammonium bicarbonate supplemented with 0.25% v/v RapiGest SF Surfactant (Waters Corp., Milford, Maryland, USA). Protein was reduced with 10 mM dithiothreitol at 80°C for 15 minutes, alkylated with 20 mM iodoacetamide at room temperature in the dark for 1 hr, and digested with trypsin (20 ng/μg, Promega Gold) overnight at 37°C. RapiGest SF Surfactant (Waters Corp., Milford, Maryland, USA) was hydrolyzed with the addition of trifluoroacetic acid to 0.5% v/v final and heating at 60°C for 2 hrs. The samples were then diluted 1:1 with 200 mM ammonium formate (pH 10.0) to raise the pH to 10. Six μl injections (approximately 3 μg protein) were utilized for LC/LC-MS/MS analysis. We took into account the different proportion of human content in both samples (as detailed in “Protein Quantification by Mass Spectrometry”) and calculated the yield of bacterial proteins in EB and RB extracts. To normalize these samples on a “per organism” basis, serial dilutions of gradient-purified EBs and RBs were suspended in 1xPBS, stained with Bodipy TR C5 Ceramide (BODIPY TR) following manufacturer's instructions (Invitrogen Life Technologies, Carlsbad, California, USA), and the numbers of bacteria per μl in EB and RB suspensions used to prepare protein lysates were determined by fluorescence microscopy and DIC microscopy and referred to protein concentration obtained in EB and RB extracts. The “mass per particle” values obtained for EB and RB forms were very similar (34.3 and 37.9 fg/particle, respectively), allowing for a direct comparison of protein quantitative values (expressed as fmol/μg of chlamydial protein) between EB and RB samples.
Antibodies, western blot and immunoprecipitations
Anti-CT288, -CT694, -CT584, -CdsQ and -Tarp antibodies were generated in our laboratory by immunization of female White New Zealand rabbits with GST-purified fusions expressed in Escherichia coli as previously described (Spaeth et al., 2009). Rabbit polyclonal anti-CT043, -CdsN, -Scc2 and -Mcsc antibodies were also generated in our laboratory using recombinant His-tagged proteins. Additional antisera include: rabbit plyclonal anti-IncA (Cocchiaro et al., 2008), anti-MOMP and anti-CdsD (K. Fields, U. of Miami, USA); anti-RpoB and anti-RpoD (M. Tan, UC Irvine, USA); anti-Hc1 and anti-IncG (T. Hackstadt, Rocky Mountains Laboratories, USA); anti-PmpD (fraction 2) (M. Lampe, U. of Washington, USA), anti-OmcB (T. Hatch, University of Tennessee Health Science Center, USA) anti TRAPα (C. Nicchitta, Duke University) and mouse monoclonal anti Na-K ATPase (hybridoma bank, University of Iowa, USA). For western blots, protein extracts were loaded onto SDS-PAGE 4-15% gradient gels, transferred to 0.45 μM nitrocellulose membranes using a Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad, Hercules, California, USA), blocked in 5% non-fat powder milk in TBST (50 mM Tris-Base, 150 mM NaCl, 0.1% Tween 20 pH 7.4) and incubated with primary antibodies diluted in 5% non-fat powder milk in TBST, followed by incubation with goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Sigma-Aldrich, St. Louis, Missouri, USA). A chemoluminescence reaction (Pierce, Rockford, Illinois, USA) and ECL-hyperfilm (GE Healthcare, Princeton, New Jersey, USA) were used to detect immunoreactive material. Immunoprecipitation assays were performed using Pierce Crosslink Immunoprecipitation Kit (Pierce, Rockford, Illinois, USA). IgGs against CT043 or pre-immune sera were cross-linked to protein A/G resins according to manufacturer's instructions. Approximately 5 × 109 inclusion forming units of EBs were lysed in Pierce IP lysis buffer (1% Nonidet P40, 5% glycerol in TNE pH 7.4) supplemented with EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland), 1 mM DTT and 1 mM PMSF. After sonication, lysates were cleared by centrifuging at 25,000 × g (10 min., 4°C) and the supernatants were then incubated with IgG- resins for 2 h at 4°C. Bound proteins were eluted with Pierce elution buffer (pH 2.8). Lysates after binding were also collected (flow through, FT). Both samples were subjected to SDS-PAGE electrophoresis and analyzed by western blot. The percentage of depletion in the FT vs. control was calculated in quantitative western blots carried out essentially as described above but using Alexa Fluor 680-labelled goat anti-Rabbit (Invitrogen Life Technologies, Carlsbad, California, USA) as secondary antibody and developing the membranes in a LI-COR Odyssey instrument (Lincoln, New England, USA).
Microscopy
Gradient-purified EBs and RBs were fixed onto glass coverslips with 2% formaldehyde – 0.025% glutaraldehyde. For indirect immunofluorescence, fixed EBs and RBs were blocked with 5% bovine serum albumin (BSA) in phosphate buffer saline solution (PBS) and incubated with polyclonal anti-MOMP antibody on ice for 1 h. Immunoreactive material was detected with Alexafluor conjugated secondary antibodies (Invitrogen Life Technologies, Carlsbad, California, USA). Coverslips were mounted in Mowiol mounting medium and images were acquired with a Leica TCS SL confocal microscope and processed with Leica imaging software. Gradient-purified EBs and RBs were also suspended in 1xPBS and stained with Bodipy TR C5 Ceramide (BODIPY TR) following manufacturer's instructions (Invitrogen Life Technologies, Carlsbad, California, USA). Fluorescent and differential interference contrast microscopy images of BODIPY TR-stained EBs and RBs were acquired with a Zeiss Axioscope epifluorescence microscope equipped with a Hamimatsu CCD camera and processed with Axiovision v3.0 imaging software.
LC/MS Data collection
Peptide digests from each of the samples (EB and RB) were analyzed using a nanoAcquity UPLC system with 2D Technology coupled to a Synapt HDMS mass spectrometer (Waters Corp, Milford, MA), using a method which generated five fractions at pH 10 with approximately equal loading in each fraction, similar to previously described (Gilar et al., 2005a, Gilar et al., 2005b, Dowell et al., 2008). Approximately 3 μg of digested EB or RB sample was first trapped at 2 μl/min at 97/3/0.1 v/v/v water/acetonitrile (MeCN)/formic acid on a 5 μm XBridge BEH130 C18 300 μm × 50 mm column. Four-minute steps at 2 μl/min to the following % MeCN were used to generate the 5 fractions at pH 10, with the composition returning to 97/3/0.1 composition during the analytical second dimension: 10.8%, 14.0%, 16.7%, 20.4%, and 65.0%. At each condition, the flowthrough from the first dimension was diluted online 10-fold with 99.8/0.1/0.1 v/v/v water/MeCN/formic acid and trapped on a 5 μm Symmetry C18 300 μm × 180 mm trapping column. Separations for each fraction were then performed on a 1.7 μm Acquity BEH130 C18 75 μm × 250 mm column (Waters) using a 90-min gradient of 5 to 40% MeCN with 0.1% formic acid at a flow rate of 0.3 μl/min and 45°C column temperature. We conducted a one five-fraction data-independent analysis (MSE) analysis and one five-fraction data-dependent analysis (DDA) of each sample (two EB and two RB), for a total of 8 sample injections and forty 90-minute LC-MS acquisitions. MSE runs of samples obtained from different sections were performed in random order, and used 0.9 sec cycle time alternating between low collision energy (6 V) and high collision energy ramp (15 to 40 V). The data-dependent analysis (DDA) mode utilized a 0.9 sec MS scan followed by MS/MS acquisition on the top 3 ions with charge greater than 1. MS/MS scans for each ion used an isolation window of approximately 3 Da, a maximum of 4 seconds per precursor, and dynamic exclusion for 120 seconds within 1.2 Da of the selected precursor.
LC-MS Data Processing
For the purposes of label-free peptide quantitation, EB and RB samples were processed independently because of the large difference in sample composition. For robust peak detection and label-free alignment across sample injections (20 for EB and 20 for RB), the commercial package Rosetta Elucidator v3.3 (Rosetta Biosoftware, Inc., Seattle, WA) with multidimensional PeakTeller algorithm was utilized, in a similar manner to a number of recent publications (Paweletz et al., 2010, Meng et al., 2007). Alignment between samples across each fraction is accomplished first, followed by summation of the five fractions to obtain aggregate data for each sample. Feature intensities for each injection were subjected to robust median scaling (top and bottom 10% excluded) to generate a single intensity measurement for each feature (accurate mass and retention time pair) in each sample. As previously reported (Dowell et al., 2008), the High/Low pH RPLC configuration generated highly unique fractions, with 89% and 87% of peptides eluting in only a single fraction for EB and RB samples, respectively.
We utilized both DDA and MSE to generate peptide identifications. For DDA acquisition files, .mgf searchable files were produced in Rosetta Elucidator and searches were then submitted to and retrieved from the Mascot v2.2 (Matrix Sciences, Inc) search engine in an automated fashion. For MSE data, ProteinLynx Global Server 2.4 (Waters Corporation) was used to generate searchable files which were then submitted to the IdentityE search engine (Waters Corporation, Milford, MA) (Geromanos et al., 2009, Li et al., 2009). Results files were then imported back into Elucidator. A total of 96,306 and 73,222 rank 1 search results were obtained for the EB and RB samples, respectively. To enable global spectra scoring across results from both search engines, all search results were concurrently validated using the PeptideProphet and ProteinProphet algorithms in Elucidator using independent reverse decoy database validation (Keller et al., 2002, Nesvizhskii et al., 2003). The PeptideProphet score was set such that the aggregate data set has a 1% peptide FDR, which corresponded to a score of 0.77 and 0.82 for EB and RB datasets, respectively. Each peptide identified was allowed to be assigned to a single protein entry, and these assignments were made by ProteinProphet according to the rules of parsimony. For the EB samples there were 754 human peptides and 3916 chlamydial peptides identified, and 14 peptides identified which were in both human and chlamydia databases. For the RB samples there were 4025 human peptides and 1274 chlamydial peptides identified, and 8 peptides shared between human and chlamydia databases.
Both DDA and MSE data were searched against an aggregate database of Swissprot human (http://www.uniprot.org, 20328 unique entries) and NCBI Chlamydia (http://www.ncbi.nlm.nih.gov/pubmed/, 5063 unique entries), both downloaded in Sept 24, 2009. Full Chlamydia database was utilized as opposed to only C. trachomatis L2 434/Bu to enable the possibility of detecting proteins not yet placed in the L2 434/Bu-specific database, or detecting additional amino acid substitutions not yet sequenced in the strain. The database was appended with full 1× reverse database for peptide false discovery rate determination, and duplicates were removed using Protein Digest Simulator Basic (http://omics.pnl.gov/software). Precursor ion mass tolerance was 20 ppm for both PLGS and Mascot searches, and product ion tolerance was 0.04 Da for Mascot and 40 ppm for PLGS. Enzyme specificity was set to tryptic for Mascot and PLGS 2.4 searches, with the exception of the specific semitryptic data analysis, which utilized only the DDA data files for Mascot semitryptic enzyme searches. A maximum of 2 missed cleavages were allowed. Carbamidomethyl cysteine was included as a fixed modification, and variable modifications included oxidized methionine and deamidated asparagine and glutamine.
LC-MS Data Quality Control
Data quality control for the 20 analyses of each sample type was performed within the Rossetta Elucidator software package using principal components analysis (PCA). Three dimensional PCA indicated excellent reproducibility and tight grouping of technical replicates both for EB and RB samples (Figure S3).
Protein Quantification by Mass Spectrometry
We employed a novel adaptation of the previously described “Absolute Quantitation” approach (Delahunty & Yates, 2005, Silva et al., 2006b), with several modifications to address mixed-species proteomic samples. First, peptides identified that are homologous between Chlamydia and human are removed, as the species origin of these peptides cannot be defined (n=14 peptides in EBs, n=8 peptides in RBs). Next, the peptides were ranked according to their average intensity across the study and the average intensity of the top 3 peptides for each protein was calculated as a measurement of each protein's relative abundance in the sample, as previously described (Silva et al., 2006b). We also calculate this average for proteins with two peptides to match, which also gives a reasonable estimation of protein abundance (Reidel et al., 2010), and is necessary because of the low molecular weight of many Chlamydia proteins. Proteins with only one peptide to match were eliminated from the quantitative analysis. Next, based on the historic response factor of the instrument utilized (2050 ± 400 counts/fmol, n=117) we calculated the fmol of each protein in the sample. Using each protein's molecular weight from the database, this fmol quantity was converted to nanograms, and by summing these values for each analysis we calculate the total ng in each sample. The total protein loading calculated in this manner is highly comparable to the 3 μg measured by BSA-calibrated Bradford assay for each sample; the EB value was 2760±160 ng for 4 analyses and RB value was 2570±170 ng for 4 analyses. The average Chlamydia protein content in EB was 2450±290 ng (89%) and in RB was 578±94 ng (22%). Finally, to calculate the species-specific protein expression for each sample, the fmol value for each protein was divided by the ng sum for only the species of interest, and scaled by 1000 to yield fmol/μg. The fmol/μg and standard deviation across four measurements for all proteins in EB and RB are reported in Table S1. This quantitative procedure was vetted prior to deployment on EB/RB samples using a model system where an E. coli lysate (Waters MassPrep) was differentially spiked with exogenous proteins (MassPrep Standard yeast enolase, bovine albumin, yeast alcohol dehydrogenase, and rabbit glycogen phosphorylase) in predefined ratios (Figure 2). One of the samples was then mixed 50/50 with mouse brain lysate digest, and 1 μg total of each sample was analyzed in triplicate by single dimension LC-MS/MS. Instrumentation and quantitative methodology was identical to described above, except only a single dimension of separation was used. Figure 2 shows that without species-specific correction (i.e. using sum of all protein ng as denominator to express fmol/μg quantity), there is a systematic bias against the E. coli and spiked-in proteins. Nevertheless, this bias was efficiently removed when the “species-specific” correction was applied.
Purification and proteomics analysis of inclusion membrane proteins
HeLa cells were infected with L2 434/Bu EBs (MOI∼10) for 40 h and harvested in Hanks balanced salt solution (HBSS) containing complete EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland) and 2 mM PMSF. The cell suspension was sonicated at 50 watts (3 times, 30 sec) and the cell lysate centrifuged at 500 × g for 10 min at 4 °C. The post nuclear supernatant was then transferred to ultra-clear ultracentrifuge tubes (Beckman Coulter Inc., Brea, California, USA) and centrifuged at 30,000 × g for 30 min at 4 °C (swinging bucket SW 41 Ti Rotor, Beckman L8-70M ultracentrifuge). The pellet was collected in ice-cold HBSS, briefly sonicated and the homogenous suspension was overlaid on 8 ml of 30% Omnipaque 350 (GE Healthcare, Princeton, New Jersey, USA). After centrifugation (30,000 × g, 30 min, 4 °C) the floating membrane fraction (seen as a band on top of the 30% Omnipaque 350) was collected carefully, diluted 8-10× with HBSS and sedimented (100,000 × g, 1 h, 4 °C). This membrane pellet was resuspended and homogenized in 2ml of 25% OptiPrep (Sigma-Aldrich, St. Louis, Missouri, USA) in TNE buffer (20 mM Tris·HCl pH=7.5, 150 mM NaCl, 1 mM EDTA) and placed at bottom of ultra-clear ultracentrifuge tubes to layer successively 20.0%, 17.5%, 15.0%, 12.5%, 10.0% and 0% OptiPrep in TNE. Following ultracentrifugation (150,000 × g, 16 h, 4 °C), membrane fractions at the 17.5%, 15.0%, 12.5% and 10.0% OptiPrep interfaces were collected, washed with 100 mM Na2CO3 (pH=11.5) and dissolved in 0.5% Triton ×100/TNE. Proteins in these samples were acetone precipitated and dissolved in 0.5% RapiGest SF Surfactant (Waters Corp., Milford, Maryland, USA) in 100 mM NH4HCO3. The pellets that were enriched in inclusion membranes (fractions 2 and 3) were analyzed by by the same method as above except with a single dimension of LC-MS/MS and utilizing 1 ug extracts.
Protein category analysis
For comparisons between EB and RB proteomes, all proteins detected were annotated (Table S1) and grouped into functional groups and sub-groups. To define the functional groups, we essentially used the classification described in the first C. trachomatis genome (Stephens et al., 1998) and updated it based on information available in the public databases NCBI (http://www.ncbi.nlm.nih.gov/sites/entrez), Uniprot (http://www.uniprot.org), CMR (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi) and ChlamydiaeDB (http://www.chlamydiaedb.org/portal/web/chlamydiaedb), as well as in specific publications when available (see Table S1). For the “Virulence & T3S” category, we included T3S-effectors (Jehl et al., 2011), -components and -chaperones (Hefty & Stephens, 2007, Betts-Hampikian & Fields, 2010), inclusion membrane proteins (Li et al., 2008a) and few other proteins originally considered as “Pathogenesis-Associated” in previous publications (CT153 (Taylor et al., 2010), SodA/CT294, PapQ/CT601, AhpC/CT603 (Stephens et al., 1998)), as shown in Table S1. Most of the proteins were classified in only one functional group and a few in two or more, as indicated in Table S1. For quantitative analysis of functional categories comparing EBs and RBs, the mass corresponding to proteins within each group was summed to obtain the total mass for that group. Only proteins identified with 2 or more peptides were quantified and included in the category analysis. If a given protein belonged to two or more functional groups, it was counted two or more times, accordingly.
Protein identification numbers and proteomic data repository
Identification numbers for all 485 proteins analyzed in this study as well as their corresponding primary locus in reference strain used in our experiments (C. trachomatis serovar L2 434/Bu) are provided in Table S1 and were retrieved from NCBI public database (http://www.ncbi.nlm.nih.gov/protein/). For convenience, equivalent primary loci corresponding to the widely used reference strain C. trachomatis serovar D UW-3/CX were also provided (Table S1) and eventually used as the primary name of the protein when no other specific name was available. The unfiltered MS datasets can be downloaded from: https://discovery.genome.duke.edu/express/resources/1635/1635_RB_exportwithMaxScores_042511.xlsx (for RB proteome) and https://discovery.genome.duke.edu/express/resources/1635/1635_EB_exportwithMaxScores_042511.xlsx (for EB proteome).
Supplementary Material
Acknowledgments
We thank K. Fields and M. Horn for comments and feedback on this manuscript, A. Xavier, M. Tan, K. Fields, H. Caldwell and T. Hackstadt for reagents. Special thanks to Dr. Keith Fadgen and Dr. Martha Stapels of Waters Corporation for technical assistance and method development with the 2DLC, and to Waters Corporation for providing the 2D nanoAcquity. This work was supported by grants from the NIH (AI096320 and 1AI068032-04S1) and the Burroughs Wellcome Trust Program in Infectious Diseases. The Duke Proteomics Core Facility is supported in part by Duke University's CTSA grant 10L1 RR024128-01 from NCRR/NIH. H.A. Saka was supported by a fellowship from the Pew Latin American Fellows Program in the Biomedical Sciences.
References
- Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 2010;38:868–877. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Griffiths RS, Viratyosin W, Brown WJ, Rockey DD. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2000;2:35–47. doi: 10.1046/j.1462-5822.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- Bebear C, de Barbeyrac B. Genital Chlamydia trachomatis infections. Clin Microbiol Infect. 2009;15:4–10. doi: 10.1111/j.1469-0691.2008.02647.x. [DOI] [PubMed] [Google Scholar]
- Beeckman DS, Vanrompay DC. Bacterial secretion systems with an emphasis on the chlamydial Type III secretion system. Curr Issues Mol Biol. 2010;12:17–41. [PubMed] [Google Scholar]
- Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci U S A. 2003;100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts-Hampikian HJ, Fields KA. The chlamydial type III secretion mechanism: revealing cracks in a tough nut. Frontiers in Microbiology. 2010;1:1–13. doi: 10.3389/fmicb.2010.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts HJ, Wolf K, Fields KA. Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal. Current Opinion in Microbiology. 2009;12:81–87. doi: 10.1016/j.mib.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Bhavsar AP, Auweter SD, Finlay BB. Proteomics as a probe of microbial pathogenesis and its molecular boundaries. Future Microbiol. 2010;5:253–265. doi: 10.2217/fmb.09.114. [DOI] [PubMed] [Google Scholar]
- Birkelund S, Morgan-Fisher M, Timmerman E, Gevaert K, Shaw AC, Christiansen G. Analysis of proteins in Chlamydia trachomatis L2 outer membrane complex, COMC. FEMS Immunol Med Microbiol. 2009;55:187–195. doi: 10.1111/j.1574-695X.2009.00522.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burton MJ, Mabey DC. The global burden of trachoma: a review. PLoS Negl Trop Dis. 2009;3:e460. doi: 10.1371/journal.pntd.0000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc Natl Acad Sci U S A. 2008;105:9379–9384. doi: 10.1073/pnas.0712241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane DD, Carlson JH, Fischer ER, Bavoil P, Hsia RC, Tan C, Kuo CC, Caldwell HD. Chlamydia trachomatis polymorphic membrane protein D is a species-common pan-neutralizing antigen. Proc Natl Acad Sci U S A. 2006;103:1894–1899. doi: 10.1073/pnas.0508983103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darville T. Chlamydia trachomatis infections in neonates and young children. Semin Pediatr Infect Dis. 2005;16:235–244. doi: 10.1053/j.spid.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Dautry-Varsat A, Subtil A, Hackstadt T. Recent insights into the mechanisms of Chlamydia entry. Cell Microbiol. 2005;7:1714–1722. doi: 10.1111/j.1462-5822.2005.00627.x. [DOI] [PubMed] [Google Scholar]
- Dean D. Lessons and Challenges Arising from the “First Wave” of Chlamydia Genome Sequencing. In: Bavoil Patrik M, Wyrick Priscilla B., editors. Chlamydia Genomics and Pathogenesis. Wymondham, Norfolk, U.K.: Horizon Bioscience; 2006. pp. 1–25. [Google Scholar]
- Delahunty C, Yates JR., 3rd Protein identification using 2D-LC-MS/MS. Methods. 2005;35:248–255. doi: 10.1016/j.ymeth.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Dowell JA, Frost DC, Zhang J, Li L. Comparison of two-dimensional fractionation techniques for shotgun proteomics. Anal Chem. 2008;80:6715–6723. doi: 10.1021/ac8007994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Hughes KT. C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Mol Microbiol. 2010;75:376–393. doi: 10.1111/j.1365-2958.2009.06973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- Geromanos SJ, Vissers JP, Silva JC, Dorschel CA, Li GZ, Gorenstein MV, Bateman RH, Langridge JI. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009;9:1683–1695. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- Gilar M, Olivova P, Daly AE, Gebler JC. Orthogonality of separation in two-dimensional liquid chromatography. Anal Chem. 2005a;77:6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- Gilar M, Olivova P, Daly AE, Gebler JC. Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J Sep Sci. 2005b;28:1694–1703. doi: 10.1002/jssc.200500116. [DOI] [PubMed] [Google Scholar]
- Giles DK, Whittimore JD, LaRue RW, Raulston JE, Wyrick PB. Ultrastructural analysis of chlamydial antigen-containing vesicles everting from the Chlamydia trachomatis inclusion. Microbes Infect. 2006;8:1579–1591. doi: 10.1016/j.micinf.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Gomes JP, Nunes A, Bruno WJ, Borrego MJ, Florindo C, Dean D. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J Bacteriol. 2006;188:275–286. doi: 10.1128/JB.188.1.275-286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pedrajo B, Minamino T, Kihara M, Namba K. Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol Microbiol. 2006;60:984–998. doi: 10.1111/j.1365-2958.2006.05149.x. [DOI] [PubMed] [Google Scholar]
- Grimwood J, Olinger L, Stephens RS. Expression of Chlamydia pneumoniae polymorphic membrane protein family genes. Infect Immun. 2001;69:2383–2389. doi: 10.1128/IAI.69.4.2383-2389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty CL, Gottlieb SL, Taylor BD, Low N, Xu F, Ness RB. Risk of sequelae after Chlamydia trachomatis genital infection in women. J Infect Dis. 2010;201 2:S134–155. doi: 10.1086/652395. [DOI] [PubMed] [Google Scholar]
- Hatch TP, Allan I, Pearce JH. Structural and polypeptide differences between envelopes of infective and reproductive life cycle forms of Chlamydia spp. J Bacteriol. 1984;157:13–20. doi: 10.1128/jb.157.1.13-20.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefty PS, Stephens RS. Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J Bacteriol. 2007;189:198–206. doi: 10.1128/JB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie SE, Horner PJ, Horne AW, Entrican G. Immunity and vaccines against sexually transmitted Chlamydia trachomatis infection. Curr Opin Infect Dis. 2010 doi: 10.1097/QCO.0b013e3283421081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci U S A. 2007;104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehl MA, Arnold R, Rattei T. Effective--a database of predicted secreted bacterial proteins. Nucleic Acids Res. 2011;39:D591–595. doi: 10.1093/nar/gkq1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett TJ, Miller NJ, Dooley CA, Hackstadt T. The conserved Tarp actin binding domain is important for chlamydial invasion. PLoS Pathog. 2010;6:e1000997. doi: 10.1371/journal.ppat.1000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DL, Stone CB, Mahony JB. Interactions between CdsD, CdsQ, and CdsL, three putative Chlamydophila pneumoniae type III secretion proteins. J Bacteriol. 2008;190:2972–2980. doi: 10.1128/JB.01997-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouihri N, Sory MP, Page AL, Gounon P, Parsot C, Allaoui A. MxiK and MxiN interact with the Spa47 ATPase and are required for transit of the needle components MxiH and MxiI, but not of Ipa proteins, through the type III secretion apparatus of Shigella flexneri. Mol Microbiol. 2003;49:755–767. doi: 10.1046/j.1365-2958.2003.03590.x. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kiselev AO, Skinner MC, Lampe MF. Analysis of pmpD expression and PmpD post-translational processing during the life cycle of Chlamydia trachomatis serovars A, D, and L2. PLoS One. 2009;4:e5191. doi: 10.1371/journal.pone.0005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev AO, Stamm WE, Yates JR, Lampe MF. Expression, processing, and localization of PmpD of Chlamydia trachomatis Serovar L2 during the chlamydial developmental cycle. PLoS One. 2007;2:e568. doi: 10.1371/journal.pone.0000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler JE, Birkelund S, Stephens RS. Overexpression and surface localization of the Chlamydia trachomatis major outer membrane protein in Escherichia coli. Mol Microbiol. 1992;6:1087–1094. doi: 10.1111/j.1365-2958.1992.tb01545.x. [DOI] [PubMed] [Google Scholar]
- Konishi M, Kanbe M, McMurry JL, Aizawa S. Flagellar formation in C-ring-defective mutants by overproduction of FliI, the ATPase specific for flagellar type III secretion. J Bacteriol. 2009;191:6186–6191. doi: 10.1128/JB.00601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GZ, Vissers JP, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- Li Z, Chen C, Chen D, Wu Y, Zhong Y, Zhong G. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect Immun. 2008a;76:2746–2757. doi: 10.1128/IAI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen D, Zhong Y, Wang S, Zhong G. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect Immun. 2008b;76:3415–3428. doi: 10.1128/IAI.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Afrane M, Clemmer DE, Zhong G, Nelson DE. Identification of Chlamydia trachomatis outer membrane complex proteins by differential proteomics. J Bacteriol. 2010;192:2852–2860. doi: 10.1128/JB.01628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longbottom D, Russell M, Dunbar SM, Jones GE, Herring AJ. Molecular cloning and characterization of the genes coding for the highly immunogenic cluster of 90-kilodalton envelope proteins from the Chlamydia psittaci subtype that causes abortion in sheep. Infect Immun. 1998;66:1317–1324. doi: 10.1128/iai.66.4.1317-1324.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham AP, Jaafar ZA, Kemege KE, Middaugh CR, Hefty PS. Biophysical characterization of Chlamydia trachomatis CT584 supports its potential role as a type III secretion needle tip protein. Biochemistry. 2009;48:10353–10361. doi: 10.1021/bi901200y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarty G. Chlamydial Metabolism as Inferred from the Complete Genome Sequence. In: Stephens RS, editor. Chlamydia: Intracellular Biology, Pathogenesis and Immunity. Washington, D.C., U.S.A.: American Society for Microbiology; 1999. pp. 69–98. [Google Scholar]
- Meng F, Wiener MC, Sachs JR, Burns C, Verma P, Paweletz CP, Mazur MT, Deyanova EG, Yates NA, Hendrickson RC. Quantitative analysis of complex peptide mixtures using FTMS and differential mass spectrometry. J Am Soc Mass Spectrom. 2007;18:226–233. doi: 10.1016/j.jasms.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Miller JD, Sal MS, Schell M, Whittimore JD, Raulston JE. Chlamydia trachomatis YtgA is an iron-binding periplasmic protein induced by iron restriction. Microbiology. 2009;155:2884–2894. doi: 10.1099/mic.0.030247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WC, Ford CA, Morris M, Handcock MS, Schmitz JL, Hobbs MM, Cohen MS, Harris KM, Udry JR. Prevalence of chlamydial and gonococcal infections among young adults in the United States. JAMA. 2004;291:2229–2236. doi: 10.1001/jama.291.18.2229. [DOI] [PubMed] [Google Scholar]
- Moorhead AM, Jung JY, Smirnov A, Kaufer S, Scidmore MA. Multiple host proteins that function in phosphatidylinositol-4-phosphate metabolism are recruited to the chlamydial inclusion. Infect Immun. 2010;78:1990–2007. doi: 10.1128/IAI.01340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita-Ishihara T, Ogawa M, Sagara H, Yoshida M, Katayama E, Sasakawa C. Shigella Spa33 is an essential C-ring component of type III secretion machinery. J Biol Chem. 2006;281:599–607. doi: 10.1074/jbc.M509644200. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Miller RD, Summersgill JT. Analysis of altered protein expression patterns of Chlamydia pneumoniae by an integrated proteome-works system. J Proteome Res. 2004;3:878–883. doi: 10.1021/pr0400031. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Nicholson TL, Olinger L, Chong K, Schoolnik G, Stephens RS. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J Bacteriol. 2003;185:3179–3189. doi: 10.1128/JB.185.10.3179-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Mann M, Aebersold R, Yates JR, 3rd, Bairoch A, Bergeron JJ. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- Numoto N, Hasegawa Y, Takeda K, Miki K. Inter-subunit interaction and quaternary rearrangement defined by the central stalk of prokaryotic V1-ATPase. EMBO Rep. 2009;10:1228–1234. doi: 10.1038/embor.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell CM, Nicks KM. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology. 2006;152:1601–1607. doi: 10.1099/mic.0.28658-0. [DOI] [PubMed] [Google Scholar]
- Paweletz CP, Wiener MC, Bondarenko AY, Yates NA, Song Q, Liaw A, Lee AY, Hunt BT, Henle ES, Meng F, Sleph HF, Holahan M, Sankaranarayanan S, Simon AJ, Settlage RE, Sachs JR, Shearman M, Sachs AB, Cook JJ, Hendrickson RC. Application of an end-to-end biomarker discovery platform to identify target engagement markers in cerebrospinal fluid by high resolution differential mass spectrometry. J Proteome Res. 2010;9:1392–1401. doi: 10.1021/pr900925d. [DOI] [PubMed] [Google Scholar]
- Peters J, Wilson DP, Myers G, Timms P, Bavoil PM. Type III secretion a la Chlamydia. Trends Microbiol. 2007;15:241–251. doi: 10.1016/j.tim.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Peterson EM, Cheng X, Markoff BA, Fielder TJ, de la Maza LM. Functional and structural mapping of Chlamydia trachomatis species-specific major outer membrane protein epitopes by use of neutralizing monoclonal antibodies. Infect Immun. 1991;59:4147–4153. doi: 10.1128/iai.59.11.4147-4153.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DM, Swenson CE, Schachter J. Ultrastructure of Chlamydia trachomatis infection of the mouse oviduct. J Ultrastruct Res. 1984;88:244–256. doi: 10.1016/s0022-5320(84)90122-9. [DOI] [PubMed] [Google Scholar]
- Raulston JE, Miller JD, Davis CH, Schell M, Baldwin A, Ferguson K, Lane H. Identification of an iron-responsive protein that is antigenic in patients with Chlamydia trachomatis genital infections. FEMS Immunol Med Microbiol. 2007;51:569–576. doi: 10.1111/j.1574-695X.2007.00336.x. [DOI] [PubMed] [Google Scholar]
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Myers GS, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, Holtzapple E, Khouri H, Federova NB, Carty HA, Umayam LA, Haft DH, Peterson J, Beanan MJ, White O, Salzberg SL, Hsia RC, McClarty G, Rank RG, Bavoil PM, Fraser CM. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 2003;31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidel B, Thompson JW, Farsiu S, Moseley MA, Skiba NP, Arshavsky VY. Proteomic Profiling of a Layered Tissue Reveals Unique Glycolytic Specializations of Photoreceptor Cells. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M110.002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka HA, Valdivia RH. Acquisition of nutrients by Chlamydiae: unique challenges of living in an intracellular compartment. Curr Opin Microbiol. 2010;13:4–10. doi: 10.1016/j.mib.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scidmore-Carlson MA, Shaw EI, Dooley CA, Fischer ER, Hackstadt T. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol Microbiol. 1999;33:753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- Shaw AC, Gevaert K, Demol H, Hoorelbeke B, Vandekerckhove J, Larsen MR, Roepstorff P, Holm A, Christiansen G, Birkelund S. Comparative proteome analysis of Chlamydia trachomatis serovar A, D and L2. Proteomics. 2002;2:164–186. doi: 10.1002/1615-9861(200202)2:2<164::aid-prot164>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Silva JC, Denny R, Dorschel C, Gorenstein MV, Li GZ, Richardson K, Wall D, Geromanos SJ. Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol Cell Proteomics. 2006a;5:589–607. doi: 10.1074/mcp.M500321-MCP200. [DOI] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006b;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- Sixt BS, Heinz C, Pichler P, Heinz E, Montanaro J, Op den Camp HJ, Ammerer G, Mechtler K, Wagner M, Horn M. Proteomic analysis reveals a virtually complete set of proteins for translation and energy generation in elementary bodies of the amoeba symbiont Protochlamydia amoebophila. Proteomics. 2011;11:1868–1892. doi: 10.1002/pmic.201000510. [DOI] [PubMed] [Google Scholar]
- Skipp P, Robinson J, O'Connor CD, Clarke IN. Shotgun proteomic analysis of Chlamydia trachomatis. Proteomics. 2005;5:1558–1573. doi: 10.1002/pmic.200401044. [DOI] [PubMed] [Google Scholar]
- Spaeth KE, Chen YS, Valdivia RH. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 2009;5:e1000579. doi: 10.1371/journal.ppat.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm WE. Chlamydia trachomatis infections: progress and problems. J Infect Dis. 1999;179 2:S380–383. doi: 10.1086/513844. [DOI] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Stone CB, Johnson DL, Bulir DC, Gilchrist JD, Mahony JB. Characterization of the putative type III secretion ATPase CdsN (Cpn0707) of Chlamydophila pneumoniae. J Bacteriol. 2008;190:6580–6588. doi: 10.1128/JB.00761-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C, Hsia RC, Shou H, Carrasco JA, Rank RG, Bavoil PM. Variable expression of surface-exposed polymorphic membrane proteins in in vitro-grown Chlamydia trachomatis. Cell Microbiol. 2010;12:174–187. doi: 10.1111/j.1462-5822.2009.01389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer RJ, Hatch TP. Characterization of outer membrane proteins in Chlamydia trachomatis LGV serovar L2. J Bacteriol. 2001;183:2686–2690. doi: 10.1128/JB.183.8.2686-2690.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LD, Nelson DE, Dorward DW, Whitmire WM, Caldwell HD. Biological characterization of Chlamydia trachomatis plasticity zone MACPF domain family protein CT153. Infect Immun. 2010;78:2691–2699. doi: 10.1128/IAI.01455-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Weintraub ST, Hakala K, Serwer P, Hardies SC. Proteome of the large Pseudomonas myovirus 201 phi 2-1: delineation of proteolytically processed virion proteins. Mol Cell Proteomics. 2010;9:940–951. doi: 10.1074/mcp.M900488-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson NR, Holden MT, Carder C, Lennard N, Lockey SJ, Marsh P, Skipp P, O'Connor CD, Goodhead I, Norbertzcak H, Harris B, Ormond D, Rance R, Quail MA, Parkhill J, Stephens RS, Clarke IN. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 2008;18:161–171. doi: 10.1101/gr.7020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipples G, McClarty G. The obligate intracellular bacterium Chlamydia trachomatis is auxotrophic for three of the four ribonucleoside triphosphates. Mol Microbiol. 1993;8:1105–1114. doi: 10.1111/j.1365-2958.1993.tb01655.x. [DOI] [PubMed] [Google Scholar]
- Tjaden J, Winkler HH, Schwoppe C, Van Der Laan M, Mohlmann T, Neuhaus HE. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J Bacteriol. 1999;181:1196–1202. doi: 10.1128/jb.181.4.1196-1202.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr Opin Microbiol. 2008;11:53–59. doi: 10.1016/j.mib.2008.01.003. [DOI] [PubMed] [Google Scholar]
- van Kuppeveld FJ, van der Logt JT, Angulo AF, van Zoest MJ, Quint WG, Niesters HG, Galama JM, Melchers WJ. Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl Environ Microbiol. 1992;58:2606–2615. doi: 10.1128/aem.58.8.2606-2615.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandahl BB, Birkelund S, Christiansen G. Proteome analysis of Chlamydia pneumoniae. Methods Enzymol. 2002a;358:277–288. doi: 10.1016/s0076-6879(02)58095-2. [DOI] [PubMed] [Google Scholar]
- Vandahl BB, Birkelund S, Demol H, Hoorelbeke B, Christiansen G, Vandekerckhove J, Gevaert K. Proteome analysis of the Chlamydia pneumoniae elementary body. Electrophoresis. 2001;22:1204–1223. doi: 10.1002/1522-2683()22:6<1204::AID-ELPS1204>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Vandahl BB, Pedersen AS, Gevaert K, Holm A, Vandekerckhove J, Christiansen G, Birkelund S. The expression, processing and localization of polymorphic membrane proteins in Chlamydia pneumoniae strain CWL029. BMC Microbiol. 2002b;2:36. doi: 10.1186/1471-2180-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagar EA, Stephens RS. Developmental-form-specific DNA-binding proteins in Chlamydia spp. Infect Immun. 1988;56:1678–1684. doi: 10.1128/iai.56.7.1678-1684.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Mann M. Mass spectrometry-based proteomics in cell biology. J Cell Biol. 2010;190:491–500. doi: 10.1083/jcb.201004052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson MW, Lambden PR, Everson JS, Clarke IN. Immunoreactivity of the 60 kDa cysteine-rich proteins of Chlamydia trachomatis, Chlamydia psittaci and Chlamydia pneumoniae expressed in Escherichia coli. Microbiology. 1994;140(Pt 8):2003–2011. doi: 10.1099/13500872-140-8-2003. [DOI] [PubMed] [Google Scholar]
- Wehrl W, Meyer TF, Jungblut PR, Muller EC, Szczepek AJ. Action and reaction: Chlamydophila pneumoniae proteome alteration in a persistent infection induced by iron deficiency. Proteomics. 2004;4:2969–2981. doi: 10.1002/pmic.200400917. [DOI] [PubMed] [Google Scholar]
- WHO. Global prevalence and incidence of selected curable sexually transmitted infections: overview and estimates. [Accessed 6 August 2010];2001 Available: http://www.who.int/hiv/pub/sti/who_hiv_aids_2001.02.pdf.
- WHO. Report of the 2nd global scientific meeting on trachoma WHO/PBD/GET 03.1. [Accessed August 6 2010];2003 Available: http://www.who.int/blindness/2nd%20GLOBAL%20SCIENTIFIC%20MEETING.pdf.
- Yip CK, Finlay BB, Strynadka NC. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat Struct Mol Biol. 2005;12:75–81. doi: 10.1038/nsmb879. [DOI] [PubMed] [Google Scholar]
- Zhang YX, Stewart S, Joseph T, Taylor HR, Caldwell HD. Protective monoclonal antibodies recognize epitopes located on the major outer membrane protein of Chlamydia trachomatis. J Immunol. 1987;138:575–581. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.