Abstract
The pathogenic origin of autoimmune diseases can be traced to both genetic susceptibility and epigenetic modifications arising from exposure to the environment. Epigenetic modifications influence gene-expression and alter cellular functions without modifying the genomic sequence. CpG-DNA methylation, histone-tail modifications, and micro-RNAs (miRNAs) are the main epigenetic mechanisms of gene regulation. Understanding the molecular mechanisms that are involved in the pathophysiology of autoimmune diseases is essential for the introduction of effective, target-directed, and tolerated therapies. In this review, we summarize recent findings that signify the importance of epigenetic modifications in autoimmune disorders while focusing on systemic lupus erythematosus (SLE). We discuss future directions in basic research, autoimmune diagnostics, and applied therapy.
The concept of epigenetics and its involvement in autoimmune diseases
Despite the multitude of approaches utilized for determining the origin of autoimmune diseases, a definitive understanding of the pathogenic mechanisms involved remains poorly defined. Although Mendelian inheritance has been demonstrated for rare disorders, most autoimmune diseases are associated with polymorphisms in susceptibility genes and transmission rates are significantly lower than expected [1]. In these cases, environmental factors appear to influence disease onset, progression, and outcome [2].
Recently, the influence of epigenetic mechanisms in autoimmune diseases has been investigated in multiple studies. Epigenetic modifications can influence gene expression, thereby altering cellular function without modifying the genomic sequence [3]. They play a central role in controlling tissue and signal specific gene expression and are responsible for the determination of defined gene expression profiles of tissues and cellular subsets, such as tissue specific cytokine expression of the T helper subsets [3]. It is becoming clear that epigenetic mechanisms contribute to a variety of disorders, including autoimmune diseases. Environmental factors can modify epigenetic marks, influencing disease onset and progression. Three main epigenetic processes are in the center of epigenetic control: DNA methylation, nucleosome repositioning by histone modifications, and micro-RNAs (miRNAs) [4].
Systemic lupus erythematosus (SLE) is a multi-factorial autoimmune disease with a wide range of clinical manifestations and severity. SLE is characterized by multi-system involvement, phases of remission and relapse, and the presence of autoantibodies against nuclear, cytoplasmic, and cell surface antigens [7]. Despite years of study, the mechanisms responsible for loss of immune tolerance remain to be elucidated. Dysregulation of B- and T-lymphocyte function [6, 7], transcription factor and cytokine expression, and antigen presentation have been reported in a disease activity-dependent manner [5, 7]. In some patients, the pathogenic process has been attributed to single gene effects, such as complement factor deficiencies [8] or TREX1 mutations [9]. Still, the majority of SLE patients exhibit disease that is multigenic and/or multifactorial in origin. Predisposing genetic variations include well-established susceptibility alleles in the major histocompatibility complex region (HLA*DRB1503 [10]), interferon-regulatory-factor 5 (IRF5), signal transducer and activator of transcription 4 (STAT4), tyrosine protein kinase (BLK), B-cell scaffold protein with ankyrin-repeats (BANK1), programmed-cell-death 1 (PDCD1), methyl-CpG-Binding Protein 2 (MECP2), and tumor-necrosis-factor superfamily member 4 (TNFSF4) genes [11].
Regardless of a growing number of known predisposing alleles, not all carriers develop clinical disease. Patients with the same or similar alleles may present with different disease course, and the penetrance of SLE in monozygotic twins is 25–45% [10, 11]. This suggests that additional environmental factors, including diet, drugs, exposure to toxins, or a history of infections, play important roles in disease onset and progression.
Additional autoimmune disorders, including rheumatoid arthritis, scleroderma, type 1 diabetes, and multiple sclerosis have been associated with epigenetic alterations (Box 1). At this point, data elucidating the involvement and pathophysiologic relevance of epigenetic patterns in these autoimmune diseases is limited, as compared to SLE. Therefore, we discuss the involvement of the main epigenetic mechanisms in the pathogenesis of autoimmune diseases while focusing on SLE. Understanding epigenetic mechanisms in the pathophysiology of autoimmune diseases will help to establish more target-directed therapy with reduced systemic side effects. It will allow clinicians to better address inter-individual differences in the pathophysiology and clinical presentation of variable diseases, such as SLE.
Box 1. Further autoimmune diseases associated with epigenetic alterations.
In addition to SLE, other autoimmune diseases have been shown to be associated with epigenetic alterations and impaired gene expression. Several complex diseases are currently the focus of intensive research. Epigenetics will presumably gain even more importance in understanding multifactorial disorders and fill the gap between the genomic predisposition of an individual and the development of a specific disease phenotype.
Rheumatoid arthritis is a systemic autoimmune disease that results in the destruction of affected joints. Similar to SLE, the pathogenesis is complex and not completely understood. It has been suggested that genetic risk factors (HLA-DRB1*0401, *0404, PTPN22) in concert with environmentally induced (epigenetic) alterations result in immune-dysregulation [12–14].
Systemic scleroderma is a rare condition of unknown origin that shows characteristics of autoimmune diseases with over-expression of pro-inflammatory cytokines, progressive vasculopathy, and excessive collagen-deposition [13]. Because a high prevalence of systemic scleroderma in specific geographic locations has been reported, environmental factors, particularly inhaled chemicals, are suggested to play a role in disease pathogenesis[15].
Multiple sclerosis is a chronic inflammatory disease that results in myelin destruction and subsequent neurodegeneration. Etiology and pathophysiology are complex with genetic predisposition (MHC complex), polygenic inheritance with incomplete penetrance, environmental risk factors, and temporal and special dynamics[16–18, 28].
Type 1 diabetes is a T lymphocyte mediated autoimmune disease. While susceptibility genes have been reported (MHC class II, insulin, PTPN22, CTLA4, IL-rRA), increasing evidence support the idea that additional, namely, environmental factors are involved [13, 28].
Sjogren’s syndrome is an autoimmune disease that affects salivary and lacrimal glands, causing symptoms of xerophthalmia and/or xerostomia. A subset of patients with SLE, rheumatoid arthritis, or scleroderma exhibit clinical features that overlap with Sjogren’s syndrome (secondary Sjogren’s syndrome). Regardless of a large number of attempts to find genetic and environmental factors that cause the disease, the pathogenesis remains unknown [13].
DNA methylation
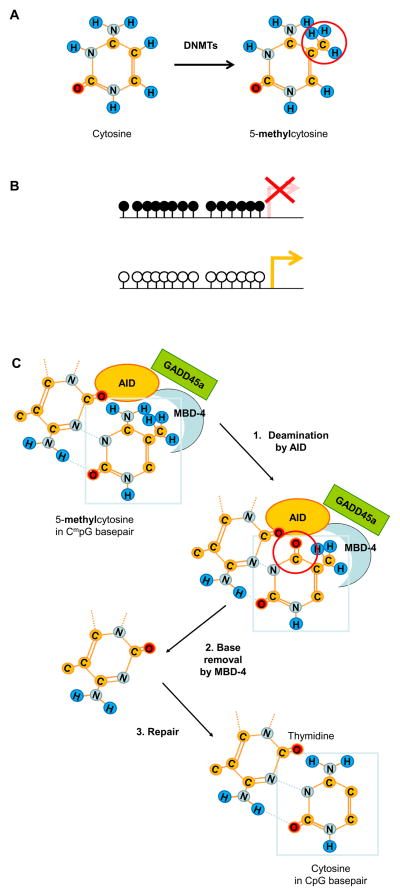
Transcription factors need to bind to corresponding cis-DNA sequences to regulate gene transcription. Thus, transcription factors require an accessible DNA structure, and the most efficient way of gene silencing is to prevent transcription factor binding to DNA. One way to prevent binding is the addition of methyl groups by DNA methyltransferases (DNMTs) to the 5′ carbon position of cytosine within cytosine-phosphate-guanosine (CpG)-dinucleotides (Figure 1A) [3, 4]. DNMT1 is responsible for re-methylation of hemi-methylated CpGs during cell-division and thereby maintains DNA methylation patterns; DNMT3a and b produce de-novo methylation. Dysregulation of DNA methylation has been linked to the expression of multiple diseases, depending on the genomic region and the genetic background of an individual [3].
Figure 1.
A) Structure of cytosine and 5-methyl-cytosine. DNA methyltransferases (DNMTs) methylate cytosine groups in CpG sequences.
B) Schematic representation of DNA methylation patterns in a silenced region (upper panel), compared to a transcriptionally active region (lower panel). Open circles represent un-methylated, filled circles methylated CpGs.
C) Methylated CpG (CmpG) demethylation by GADD45a. DNA demethylation may occur through a two-step enzymatic process, promoted by GADD45a. The first step involves deamination of CmpG nucleotides by activation-induced deaminase (AID), generating a thymine product and a G:T mismatch. The second step involves thymine base removal by methyl-CpG-binding domain 4 (MBD-4). In the following, the missing base gets replaced by a non-methylated cytosine group, resulting in demethylation of CmpG dinucleotides [36].
In SLE, a generally hypomethylated state of T and B lymphocyte genes has been reported [4, 19]. CD4+ but not CD8+ T lymphocytes from SLE patients have recently been reported to display gene hypomethylation when compared with normal controls [4, 19]. The degree of hypomethylation correlates with disease activity, and numerous methylation-sensitive genes are overexpressed in lupus CD4+ T cells: cytokine genes (IL4 [20], IL6 [21], IL10, and IL13 [22]) and co-stimulatory molecules (CD70/CD26L [23], CD6 [24], CD11A [25], and CD40L/CD154 [26]). SLE-associated DNA hypomethylation results in an overexpression of CD8+ T lymphocyte and NK cell-specific perforin (PRF1) [27, 28], stimulatory and inhibitory killer-cell-Ig-like-receptor (KIR) [29], and the serin/threonine protein-phosphatase gene PP2A [30, 31]. The expression of CD5 on the surface of B lymphocytes is reduced in SLE, secondary to hypomethylation of an intracellularly expressed truncated CD5 variant (CD5-E1B). Reduced CD5 expression on the cellular surface promotes autoreactivity [32]. The involvement of DNA methylation in X-chromosomal silencing and sex chromosomal balance provides a potential explanation for the female predominance in SLE [32]. Females are characterized by partial suppression of X-chromosomal activity by methylation of specific regions (Figure 1B) [33]. This mechanism contributes to the balancing of copy number differences of X-chromosomal genes between females and males. CD40 ligand is a B lymphocyte costimulatoy molecule, and is encoded on the X-chromosome. It is overexpressed in CD4+ T lymphocytes from female SLE patients, and contributes to the expression of pathogenic antibodies. Hypomethylation of the inactivated X-chromosome in CD4+ T cells of female SLE patients, and experimental demethylation with 5-azacytidine, results in over-expression of CD40 ligand. CD4+ T lymphocytes from male SLE patients and 5-acazytidine treated CD4+ T lymphocytes from men did not show increased CD40 ligand expression due to the fact that the male X-chromosome is demethylated under physiological conditions [26].
Further evidence for the involvement of reduced DNA methylation in SLE has been provided by a study that investigated disease discordant monocygotic twins [18]. Diseased twins presented with significantly reduced CpG-DNA methylation when compared to their healthy siblings. This was accompanied with changes in DNA methylation and the expression of ribosomal RNA genes that were independent of changes in repetitive sequences [18].
Sequences derived from human endogenous retrovial elements (HERV) account for about 8% of the human genome (while 3% of the human genome encodes for genes indispensable for life). Integration of HERVs in the human genome occurred millions of years ago through exogenous retroviral infections. Over multiple generations, transmission became vertical and Mendelian. Retro-elements are flanked by long terminal repeats and inactivated by DNA methylation, supercoiled conformation, or mutations [4, 28]. Recently, methylation-levels of the HERV long interspersed repetitive element-1 (LINE-1) in CD4+, and CD8+ T lymphocytes and B lymphocytes of SLE patients were reported to be decreased [34]. Activation of HERVs presumably results in a general disturbance of gene expression, in particular of genes that may contribute to the development of autoimmune diseases [4, 28, 34].
Clinical experience had shown that tuberculosis patients treated with hydralazine or procainamide develop SLE- and RA-like symptoms. The ability to reproduce similar symptoms in mice treated with hydralazine allowed researchers to investigate the role of DNA methylation in these autoimmune disorders. Hydralazine and procainamide de-methylate CpG-DNA sequences through direct impairment of DNMT1-activity (hydralazine) or the inhibition of the extracellular-signal regulated kinase (ERK) pathway (procainamide), which is central for the induction of DNMT1 and DNMT3 expression [28]. Various in vitro and in vivo studies using de-methylating agents support the hypothesis that DNA hypomethylation plays a pathophysiological role in SLE [4, 19]. Studies have shown that these agents induce lupus-like features in both animal models and human T lymphocytes. Adoptive transfer of demethylated T lymphocytes into young (< 12 weeks) DBA/2 mice induced a lupus-like disease with glomerulonephritis and autoantibody production [4, 19]. Demethylated CD4+ T lymphocytes from healthy individuals exhibit gene expression patterns that are similar to those of SLE T cells. T lymphocytes become auto-reactive, begin to spontaneously lyse syngeneic macrophages, and induce B lymphocyte activation and antibody-production [4, 19]. Further evidence for the role of DNA demethylation in SLE was provided by studies in lupus-prone MRL/lpr mice. DNA methylation levels in the thymus and axillary lymph nodes of 20-week-old diseased animals are significantly reduced, when compared to 4-week-old asymptomatic animals [4, 19].
However, the molecular mechanisms that underlie DNA hypomethylation in SLE remain unclear and represent the focus of current research. An interesting clue to help explain T lymphocyte hypomethylation in SLE patients is the finding that the mitogen-activated protein kinase (MAPK)/ERK pathway can regulate DNMT levels [4, 19]. The application of ERK pathway inhibitors, such as hydralazine, to B and T lymphocytes result in overexpression of methylation-sensitive genes. It has been further demonstrated that a defect in protein kinase C δ (PKCδ) induction in T lymphocytes results in downregulation of the ERK pathway and a subsequent lack of DNMT1-activation in SLE T lymphocytes [4, 19, 35]. This is supported by the finding that PKCδ-deficient mice develop SLE-like symptoms including B lymphocyte expansion, autoantibody production, IL-6 overexpression, and spontaneous germinal-center formation [28].
At least three additional molecules have been suggested to be involved in DNA demethylation in SLE: growth arrest and DNA damage-inducible protein alpha (GADD45a), activation-induced deaminase (AID), and methyl-CpG-binding domain 4 (MBD-4) related G:T glycosylase. GADD45a is suspected to promote demethylation by interacting with AID and MBD-4. Thus, GADD45a directly participates in DNA-demethylation through a mechanism which involves 5-methyl-cytosine-deaminases and a G:T mismatch-specific thymine glycosylase [19, 36] (Figure 1C). GADD45a is overexpressed in CD4+ T cell from SLE patients [37]. Interestingly, GADD45a mRNA has been reported to be upregulated in SLE CD4+ T cells in response to UV-irradiation and may explain the sun exposure-related autoimmune phenomena observed in SLE patients [19, 37]. Nevertheless, studies on DNMT-expression in SLE patients have produced conflicting results. DNMT1 and DNMT3a have been reported to be downregulated in SLE CD4+ T lymphocytes in some [38, 39], but not all, studies [40, 41]. Conflicting results may be due to variability in disease activity as well as discrepancies between DNMT-expression levels and protein activity. The fact that DNA demethylation can occur in a tissue-, cell-, or region-specific manner and that measurements of gross expression levels may be strongly influenced by the sample further complicates the resolution of this issue.
Histone modifications
A nucleosome is the basic subunit of chromatin and comprises 146 base pairs of DNA coiled around a histone octamer. The histone octamer consists of two copies each of the histone proteins H2A, H2B, H3 and H4. Histone complexes possess flexible N-terminal tails that are accessible to post-translational modifications and strongly impact the functional capacities of nucelosomes [4]. Modifications include histone acetylation, methylation, ubiquitination, phosphorylation, sumoylation, deimination (or citrullination), ADP-ribosylation and proline-isomerization [42]. Each histone-tail modification facilitates specific changes in nucleosome arrangement and chromatin-structure. Histone H3 lysine 9 (H3K9) acetylation, for example, is associated with transcriptional activation, and H3K9 methylation represses transcriptional activity [43]. DNA methylation and histone modifications are often coincident, and are interconnected by a multitude of mechanisms [44]. The aforementioned methyl-CpG-binding domain (MBD) proteins selectively bind methylated DNA and recruit histone deacetylase (HDAC) and/or histone methyltransferases (Figure 2) [45]. Tri-methylation of H3K4, a transcription-activating variant, blocks the binding of DNMT3a and suppresses DNA methylation in these regions [46].
Figure 2.
A) Nucleosome arrangement in transcriptionally inactive heterochromatin and active euchromatin. Compact heterochromatin is characterized by dense nucleosome packing, repressive histone modifications and high degrees of DNA methylation (purple circles). Euchromatin is characterized by de-compaction of nucleosome fibers, permissive histone modifications and a low degree of DNA methylation. In accessible regions, transcription regulatory factors can bind to DNA motifs and induce transcription. In these regions, DNA is accessible to DNases (DNase hypersensitivity site).
B) Schematic of the (hypothetical) model depicting the interplay between DNA methyltransferases (DNMTs) and histone methyltransferases (using the example of G9a). HP1 is an adapter complex that promotes the interplay between DNMTs and histone methyltransferases. Thereby, HP1 promotes DNA methylation by DNMTs. The association of DNMTs with G9a could, in turn, allow a direct impact of DNA methylation on H3K9 methylation states [44].
The histone modification patterns that have been reported in SLE are complex. There is evidence that tissue-specific acetylation in some regions is associated with disease activity, whereas histone acetylation in other regions seems to have protective effects [47, 48, 49]. Experimental data is available for the TNF-α promoter in SLE, and histone hyperacetylation has been shown to be associated with increased monocyte maturation and TNF-α expression in SLE patients [48]. However, it remains unclear if this represents a pathogenic step, or rather an effect of the pathophysiological changes in SLE. Treatment of T lymphocytes from healthy donors with HDAC inhibitors resulted in decreased CD3ζ chain expression, resembling signaling abnormalities observed in patients with SLE [48] which further supports the role of histone hyperacetylation in SLE. One example of a disease-related HDAC-regulated locus in SLE is IL2. The cAMP response element (CRE) modulator α (CREMα) is a ubiquitously expressed transcription factor belonging to the CREB family of transcription factors. CREM is overexpressed in SLE T cells [50] and is responsible for the termination of IL-2 expression in T lymphocytes. CREMα was shown to recruit HDAC to CRE sites in the IL2 promoter, causing histone de-acetylation and contributing to the silencing of IL2 in SLE T cells [51]. The involvement of histone modifications in the pathophysiology of SLE is further supported by studies in lupus-prone mice. The histone deacetylase Sirtuin-1 (Sirt1) is overexpressed in MRL/lpr CD4+ T lymphocytes, whereas E1A binding protein p300, P300/CBP-associated factor, and HDAC7 are expressed at lower levels when compared with MRL/MPJ mice [52]. Suppression of Sirt1 activity by anti-Sirt1 miRNA resulted in transient elevation of H3 and H4 acetylation, and reduction of anti-DNA titers, glomerular IgG deposition, and renal pathology scores [18]. In another study, the application of HDAC inhibitors resulted in downregulation of IL-6, IL-10, IL-12, and IFN-γ levels in MRL/lpr splenocytes in vitro and in improvement of proteinuria, glomerulonephritis, and spleen weight in vivo [47]. Considering that HDAC-treated cells exhibit autoantigens that strongly react to SLE-derived autoantibodies, the hypothesis has been put forward that hyperacetylation of genes related to apoptosis or the cell cycle play a role in SLE [52].
The involvement of histone modifications in the pathogenesis of autoimmune diseases is almost certain. Still, histone hyperacetylation and hypoacetylation seem to occur in both region- and tissue-specific fashions, and this may explain some of the antithetic in vivo and in vitro results discussed above. HDAC recruitment and hypoacetylation in some regions seem to coexist with hyperacetylation in other regions. The application of HDAC inhibitors, resulting in genome-wide acetylation of histones, seems to improve the symptoms in lupus-prone mice [47] and mice induced to develop rheumatoid arthritis [53], whereas hyperacetylation of various genetic loci in human SLE is associated with disease severity [48].
Epigenetic modifications enhance immunogenicity and auto-antibody production
The aforementioned components of the nucleosome are a major antigen source in SLE and other autoimmune diseases. Years prior to the development of clinical symptoms, SLE patients exhibit antinuclear antibodies and antibodies against extractable nuclear antigens [54]. Nucleosomes and histones as well as both single- and double-stranded DNA are detected in the serum of SLE patients and lupus-prone mice [55]. The presence of these intra-cellular components in the circulation has been explained by aberrant apoptosis and/or reduced clearance of apoptotic cells [19, 56]. SLE-specific auto-antigens were also detected in surface-blebs of late apoptotic cells [19]. Similarly, rheumatoid factor and anti-citrullinated antibodies precede the appearance of clinical rheumatoid arthritis by several years [12].
Thus, several authors have suggested that the hypomethylated state and de-methylated DNA fragments in the serum of SLE patients can mimic microbial DNA and induce the production of anti-DNA-antibodies that play a role in the pathophysiology of SLE [4, 19]. Furthermore, mice deficient in factors required for the clearance of apoptotic cells, such as DNAse 1, serum amyloid P/C-reactive protein, C1q, IgM, and the Mer receptor kinase, develop anti-nucleosome antibodies and glomerulonephritis [19, 55]. Lupus-derived autoantibodies strongly react with histones modified during apoptosis [19]. These apoptosis-induced histone modifications include histone acetylation, de-ubiquitination of H2A, transglutamation of H2B, and phosphorylation of H2A, H2A.X, H2B, and H3 [57, 58]. Because peptides that carry apoptosis-induced acetylation motifs can accelerate both disease onset and severity in lupus-prone mice, it appears possible that apoptosis-induced chromatin changes can disrupt immune tolerance and result in autoimmune diseases [4, 19, 57]. More than 200 non-histone proteins have also been identified as HDAC substrates. Given the results discussed above, some of these proteins are likely targets in SLE and other autoimmune diseases [28].
These findings suggest that the presence of extracellular demethylated DNA and specific histone modifications, particularly those associated with apoptosis, play a central role in the pathogenesis of SLE and other autoimmune diseases (Box 2). Because a number of the aforementioned modifications to CpG-DNA sequences, histones or other nuclear proteins are disease specific, they may prove useful as disease biomarkers [41, 59].
Box 2. Epigenetic mechanisms in autoimmune diseases.
In rheumatoid arthritis, various disease-specific epigenetic patterns have been reported. Rheumatoid arthritis synovial fibroblasts (RASF) play a central role in disease onset and progression [61]. Decreased global (RASF) and local (LINE-1 promoter) DNA-metylation has been reported [61–63]. Demethylated CpG elements within the IL6 promoter of monocytes are associated with monocyte activation and inflammation [61, 64]. In accordance with these finding, demethylation of normal fibroblasts with 5-acazytidine results in RA-like phenotypes [61]. Interestingly, due to increased DNA methylation, death-receptor-3 (DR-3) is downregulated in rheumatoid arthritis patients’ monocytes. This results in resistance to apoptosis and may account for extended pro-inflammatory responses [13, 65]. Furthermore, the balance between histone acetylation and deacetylation through HDACs is disrupted in rheumatoid arthritis, resulting in a hyperacetylated genome [61, 66]. HDAC-inhibitors on the other hand improve symptoms in murine rheumatoid arthritis models and reduce the expression of vascular endothelial growth factor in-vivo [54]. This results in reduced angiogenesis in synovial tissue in collagen-antibody induced arthritis [66].
Scleroderma is characterized by tissue and organ fibrosis. Cultured scleroderma fibroblasts exhibit disease-specific cytokine-profiles, including over-expression of TNF-α. Fibroblasts maintain a profibrotic phenotype when transferred to in-vitro settings [67]. An involvement of aberrant DNA methylation and histone acetylation has been suggested [4, 13, 67], but direct evidence remains to be provided. Co-culture of fibroblasts with HDAC-inhibitors results in increased expression of the transcription factor FL1, resulting in increased collagen-production in fibroblasts [67].
In type 1 diabetes, susceptibility genes have been reported, but additional factors are involved in the pathogenesis [13, 28]. Epigenetic analysis of concordant twins demonstrated significantly increased DNA methylation, when compared to healthy individuals. Epigenetic variation may result in impaired homocysteine metabolism and subsequent tissue damage, impaired lymphocyte function and pancreatic islet-cell repair mechanisms [13].
In multiple sclerosis patients, a 30% reduction of DNA methylation in white matter lesions has been reported. Furthermore, hypomethylation of the promoter region of the peptidyl-arginine-deaminase II (PAD2) gene has been demonstrated [13]. PAD2 is involved in the citrullination of myelin-basic-protein (MBP) and overexpressed in multiple sclerosis patients. Citrullination of MBP may play an important role in protein auto-cleavage [13]. Thus, proteolytic digestion and myelin instability may result in enhanced T lymphocyte responses and inflammation [13]. The clinical improvement of mice that were induced to develop multiple sclerosis in response to HDAC-inhibitor treatment suggests an involvement of histone modifications [68]. Still, findings are somewhat preliminary and genome-wide studies on epigenetic mechanisms, including DNA methylation and histone modifications are in progress.
miRNAs
Mi-RNAs are 21 to 23 base pair RNAs and function as post-transcriptional and post-translational regulators of gene expression. MiRNAs are transcribed from genomic, intergenic DNA by RNA polymerase II or III as preliminary transcripts (pri-miRNAs) (Figure 3) that are then cleaved by the nuclear ribonuclease Drosha [68–70] and exported to the cytoplasm. The cytoplasmic enzyme Dicer further processes the transcripts into mature miRNAs [71, 72]. One or both strands form complexes with ribonucleoproteins (RNPs) and express regulatory effects [28, 73, 74]. MiRNA function can be executed by duplex formation with target genes in the 3′ untranslated region (3′UTR) of messenger RNA (mRNA). This interaction can lead to downregulation of gene expression by translational repression, mRNA cleavage, or translational arrest [75, 76]. Up to 1000 miRNAs are suspected to control 1/3 of the human transcriptome and are involved in the regulation of cell-differentiation, cell-cycle programming, apoptosis, and immune-regulation [28, 76–78]. Several authors categorize miRNAs, together with DNA methylation and histone acetylation, as one of the three central epigenetic mechanisms [4, 28]. This is supported by the discussion that miRNAs originate from larger intergenic transcripts, and that intergenic transcription was suggested to be at the interface between chromatin remodeling and transcription of adjacent genes [3]. There is a tight correlation between chromatin structure and the presence of intergenic transcripts which seem to permit interactions between distal regulatory regions and core promoters [3]. Other groups discuss miRNAs as a separate regulatory mechanism. Still, miRNAs are characterized by a close relationship and complex interplay with DNA methylation and histone modifications [79]. Using mouse models, several authors addressed the question of whether miRNAs are contributing to immune-regulation. MiR-181a has been shown to be involved in T cell development by modulating TCR signaling-related phosphatases [80]. Several groups have demonstrated an important role of miR-150 in B lymphocyte development [70, 81, 82] by regulating the expression of c-Myb. MiR-150−/− mice presented with dysregulation of B lymphocyte development and displayed steady state levels of serum immunglobulins, as well as enhanced T-lymphocyte dependent immune responses, presumably secondary to increased c-Myb levels [70].
Figure 3. Schematic display of gene regulation by miRNAs.
MicroRNAs (miRNAs) are derived from primary genomic DNA transcripts (pri-miRNAs). Pri-miRNAs are processed by the ribonuclease Drosha to pre-miRNAs. These are then transported to the cytoplasm and subsequently processed into mature 21 to 23 nucleotide miRNAs by Dicer. MiRNAs are incorporated into a RNP complex that exerts regulatory miRNA functions.
MiRNA regulatory functions are transcript degradation and translational arrest.
The close involvement of miRNAs in multiple regulatory mechanisms of cell-differentiation and immune-regulation comprises a huge potential for cell dysfunction and expression of disease pathology. Still, the number of miRNAs that seem to be associated in the pathogenesis of patients with SLE and lupus-prone mice remains limited. The groups of miRNAs identified so far are involved in 1) the regulation of innate immunity (miR-146a) by abnormal activation of the type I interferon pathway [83], 2) inflammatory responses (miR-125a) by suppressing KLF13 and RANTES [84], and 3) DNA methylation (miR21 and miR-148a) [70, 85]. To date, miR-17-92 mice, which over-express miR-17-92 in B and T lymphocytes, are the only available murine miRNA model system that displays SLE-like manifestations [86]. Mice that ectopically express the human miR-17-92 cluster develop generalized lymphoproliferative disease with increased numbers of activated CD4+ T lymphocytes, antigen-experienced CD4+ and CD8+ T lymphocytes, B1a, and germinal-center B lymphocytes. Animals develop autoimmune phenomena with elevated anti-DNA antibody titers. Pro-apoptotic-factors Pten and Bim are predicted targets of miR-17-92. Pten and Bim levels were reduced in CD4+ T lymphocytes of miR-17-92 mice, suggesting that lymphocyte expansion may be caused by downregulation of these regulatory factors [86].
There seems to be a strong interplay between miRNAs and other epigenetic control mechanisms. Among the known genes regulated by miRNAs, there are several which are involved in the epigenetic regulation of cellular functions, including DNA methylation and histone modifications. MiR-29 and miR-143 directly influence DNA methylation by regulating DNMT3a and DNMT3b-expression [87–89]. A recent study demonstrated the involvement of miR-126 in the regulation of DNA methylation in SLE CD4+ T cells by interaction with the 3′UTR of DNMT1. Over-expression of miR-126 in CD4+ T cells resulted in demethylation of CD11A and CD70, causing T and B cell hyperactivity [90].
Recently, downregulation of miR-181-a in children with SLE was demonstrated, resulting in upregulation of the miR-181-a target gene P300/CBP-associated factor (PCAF). It was proposed that upregulation of PCAF may impact ubiquitination levels of Hdm2, a negative regulator of tumor suppressor protein p53, resulting in the induction of apoptosis in children with SLE [91]. MiRNA expression itself can be regulated by DNA methylation and histone modifications. DNMT-inhibitors and HDAC-inhibitors have been shown to result in an upregulation of miRNA expression [28, 92, 93].
Several of the aforementioned miRNAs and others are involved in other autoimmune disorders and discussed in Box 3.
Box 3. MiRNAs in autoimmune diseases.
A growing number of reports hint to the involvement of miRNAs in autoimmune diseases other than SLE.
In rheumatoid arthritis animal models, Rheumatoid arthritis synovial fibroblasts (RASFs) are characterized by the expression of a number of distinct miRNAs, including the aforementioned miR-146a and miR-155 [94]. TNF-α, and IL-1β enhance miR-155, which suppresses metalloproteinases in RASFs. MiR-203 was up-regulated in RASF in a DNA methylation-dependent manner. MiR-203-over-expression results in a pro-inflammatory phenotype with upregulation of IL-6 and matrix-metalloprotease-1 [95]. HDACs are regulated by miR-140 (HDAC4 in a mouse cartilage cell line), miR-2861 (HDAC5 in osteoblasts), and miR-449a (HDAC1 in a prostate cancer cell line) [96–98]. MiR-146 is up-regulated by pro-inflammatory cytokines and exhibits negative regulatory functions on NFκB pathways in monocytes from rheumatoid arthritis patients [14].
The same miR-146, and two further miRNAs, miR-574-3p and miR-768-3p, were confirmed to be up-regulated in the salivary glands and peripheral tissues of Sjogren’s syndrome patients [99, 100]. Diabetic mice with associated Sjogren’s syndrome exhibit upregulation of miR-146 and miR-150 in salivary glands and peripheral lymphocytes [100].
Concluding remarks
A growing body of evidence implicating the involvement of epigenetic mechanisms in immune programming and the development of autoimmune diseases has accumulated over recent years. Reduced DNA methylation, histone hypoacetylation and hyperacetylation, and the overexpression of certain miRNAs, resulting in immune imbalance, have been shown to be associated with the onset and progression of autoimmune diseases (Table 1). Nevertheless, recent studies provide only the basis for understanding the impact of epigenetic modifications in the expression of autoimmune diseases. Current research aims to address the interconnections between chromatin state, transcription factor expression and DNA-accessibility, resulting cytokine dysregulation, and clinical phenotypes. Further understanding of molecular mechanisms that cause and result from disease-associated epigenetic patterns is required in order to understand the pathophysiology of autoimmune diseases and to be able to introduce preventive measures for individuals with genetic predisposition and more rational therapeutic modalities. Understanding the pathophysiology of SLE, and other autoimmune diseases with overlapping and variable features, will help to establish more individually optimized treatment options and provide the option to reduce disease and treatment associated morbidity and mortality.
Table 1.
Epigenetic mechanisms in autoimmune diseases.
| Epigenetic mechanism | Disease | Refs. |
|---|---|---|
| DNA de-methylation | ||
|
| ||
| Global hypomethylation of B and T lymphocytes | SLE | [4, 19, 25] |
|
| ||
| Altered DNMT expression in SLEa CD4+ T cells | ||
| - Negative correlation of DNMT1b and DNMT3a expression with disease activity | SLE | [4, 19, 38] |
| - DNMT, DNMT3a and DNMT3b: no correlation with disease activity | SLE | [40] |
|
| ||
| MAPK/ERKc over-expression in SLE results in impaired DNMT1 expression | SLE | [4, 19] |
| GADD45ad is associated with DNA de-methylation and GADD45a is overexpressed in SLE | SLE | [34, 35–37] |
|
| ||
| Cytokine genes: de-methylation | ||
| - IL4, IL6, IL10, IL13 | SLE/RAe | [20–22] |
| - IL6, | SLE | [30, 41, 62] |
|
| ||
| De-methylation of co-stimulatory molecules | SLE | [23–26, 32] |
| - CD6, CD11A, CD40L, CD70, CD5 | ||
|
| ||
| De-methylation of PRF1f in SLE CD4+ T lymphocytes results in monocyte lysis | SLE | [27] |
|
| ||
| De-methylation of the PP2Ag promoter results in increased expression of PP2Acα | SLE | [29, 30] |
|
| ||
| In-vivo and in-vitro de-methylation results in lupus-like symptoms | SLE | [4, 19] |
|
| ||
| In-vitro de-methylation of fibroblasts results in RA-like phenotype | RA | [41] |
|
| ||
| De-methylated DNA-fragments induce anti-DNA-antibody production | SLE | [4, 19, 55] |
|
| ||
| De-methylation of pro-inflammatory genes (IFGNR2 – Interferon-gamma-receptor 2, MMP14 - Matrix metalloproteinase-14, LCN2 - Lipocalin-2, CSF3R – colony-stimulating- factor 3-receptor, and AIM2 - Interferon-inducible-protein/absent in melanoma 2) is associated with increased disease activity in SLE | SLE | [4, 19] |
|
| ||
| De-methylation of HERVh element LINE-1 in CD4+, CD8+ T cells, and B lymphocytes | RA/SLE | [31, 41, 60, 62] |
|
| ||
| Increased methylation of the death-receptor-3 (DR-3) gene | RA | [14, 63] |
|
| ||
| Aberrant CpG-DNA methylation of fibroblasts results in pro-inflammatory and pro-fibrotic phenotype | SSc | [14, 64, 66] |
|
| ||
| Increased CpG-DNA methylation results in: | T1Di | [14] |
| - Impaired homocysteine metabolism and tissue damage | ||
| - Impaired lymphocyte function and inflammation | ||
|
| ||
| Global de-methylation of CpG-DNA in inflammatory white matter lesions | MSj | [14] |
|
| ||
| De-methylation of the PAD2k promoter | MS | [14] |
| - Resulting in impaired proteolytic digestion of myelin, myelin instability | ||
| - Enhanced T lymphocyte responses | ||
|
| ||
| Histone modifications | ||
|
| ||
| Evidence in support of increased acetylation: | ||
| - Histone acetylation around the TNF promoter and increased monocyte maturation and TNF-α expression | SLE | [51] |
| - HDIl suppress IL-2 expression, causing SLE-like signaling aberrations | SLE | [47] |
| - Global hyper-acetylation | RA | [41, 57] |
| - HDI mediate increased FL1 expression and pro-fibrotic phenotypes in fibroblasts | SScm | [64] |
|
| ||
| Evidence in support of reduced acetylation: | ||
|
| ||
| - HDIs down-regulate IL-12, IFN-γ, IL-6, and IL-10 in MRL/lpr mice | SLE | [46] |
| - HDACn recruitment to the IL2 promoter by CREMo: | ||
| => IL-2 expression is down-regulated in SLE T cells | SLE | [50] |
| => HDAC recruitment as a putative mechanism of IL2 gene silencing | ||
| - HDAC Sirtuin-1 (Sirt1) is overexpressed in MRL/lpr CD4+ T lymphocytes | SLE | [51] |
| - Suppression of Sirt1 activity by anti-Sirt1 miRNA results in: | SLE | [51] |
| => elevation of H3 and H4p acetylation, | ||
| => reduced anti-DNA titers, renal IgG deposition, and renal pathology scores | ||
| - Reduced expression of histone acetyltransferases: E1A binding protein p300, p300/CBP-associated factor and HDAC7 in lupus-prone mice | SLE | [51] |
| - HDIs improve symptoms in murine RA models | RA | [65] |
|
| ||
| Histone-modifications and auto-antibody production: | ||
|
| ||
| - SLE autoantibodies react with apoptosis-induced histone-modifications | SLE | [4, 19, 52] |
| - Apoptosis-induced acetylation-motifs accelerate disease onset and severity in lupus-prone mice | SLE | [52] |
|
| ||
| miRNAs | ||
|
| ||
| Over-expression of miRNAs: | ||
|
| ||
| - miR-146a: abnormal activation of type I IFN/NFκBq pathways | SLE/SjSr/RA | [83] |
| - miR-125a: inflammatory responses through suppression of KLF13s and RANTESt | SLE | [84] |
| - miR-148a and miR-21: direct and indirect targeting of DNMT1 | SLE | [85] |
- miR-17~92 mouse model exhibits lupus-like symptoms:
|
SLE | [86] |
| - MiR-203 is up-regulated in RASF in a DNA methylation-dependent manner | RA | |
| - MiR-203-over-expression: upregulation of IL-6 and matrix-metalloprotease-1 | RA | [95] |
| - miR-155: suppression of metalloproteases in RASF | SjS | [94] |
| - miR-574-3p and miR-768-3p: miRNA over-expression in salivary glands | SjS | [99, 100] |
- miR-150: up-regulated in salivary glands of diabetic mice with SjS:
|
[100] | |
|
| ||
| Downregulation of miRNAs in AID: | ||
|
| ||
| - miR-181-a in pediatric SLE patients | SLE | [91] |
| - miR-181-a regulates P300/CBP-associated factor (PCAF) | SLE | [91] |
| - upregulation of PCAF: impaired ubiquitination of Hdm2, and apoptosis induction | SLE | [91] |
|
| ||
| miRNAs are involved in DNA methylation: | ||
|
| ||
| - miR-126 regulates DNA methylation in SLE T cells (interaction with DNMT1 3′UTR) | SLE | [90] |
| - overexpression of miR-126 in CD4+ T cells: CD11A and CD70 demethylation | SLE | [90] |
| - miR-140: regulates HDAC4 in mouse cartilage cells | RA | [97] |
| - miR-2861: regulates HDAC5 in osteoblasts | RA | [96–98] |
| - miR-449a: regulates HDAC1 in prostate cancer cells | RA | [98] |
|
| ||
| Epigenetic modifications influence miRNA expression: | ||
|
| ||
| - HDIs and DNMT inhibitors mediate upregulation of miRNA expression | RA | [28, 94, 95] |
SLE: Systemic lupus erythematosus;
DNMT: DNA methyltransferase;
MAPK/ERK: mitogen-activated protein kinase/extracellular-signal regulated kinase;
GADD45a: growth arrest and DNA damage-inducible protein 45 alpha;
RA: Rheumatoid arthritis;
PRF1:Perforin-1; RASF: RA synovial fibroblasts;
PP2A: serine/threonine protein-phosphatase 2A;
HERV: human endogenous retroviral elements;
T1D: type 1 diabetes;
MS: Multiple sclerosis;
PAD2: peptidyl-arginine-deaminase II;
HDI: histone deacetylase inhibitor;
SSc: systemic scleroderma;
HDAC: histone deacetylase;
CREM: cAMP response element (CRE) modulator;
H3/H4: histone H3/H4;
IFN: interferon/nuclear factor κB (NFκB);
SjS: Sjogren’s syndrome;
KLF13: Kruppel-like factor 13;
RANTES: Regulated upon Activation, Normal T-cell Expressed, and Secreted, also CCL5.
Acknowledgments
The authors thank Christine Hendrix for help with the preparation of the manuscript. Work performed in the authors’ lab was supported by grants from National Institute of Allergy and Infectious Diseases and the National Institutes of Health.
Glossary
- Cis-DNA sequences
elements that regulate gene expression within the same molecule (usually chromosomes). Cis elements summarize CpG DNA, promoter and enhancer regions, locus control regions, and matrix attachment sites. In contrast to cis regulatory elements, trans elements can regulate the expression of distant genes. Examples for trans regulatory elements are transcription factors, RNA polymerases, chromatin remodeling complexes, and histone modifying enzymes (such as de-acetylases, methyltransferases, etc.)
- CpG sites/CpG DNA
sequences within double-stranded (genomic) DNA in which a cytosine nucleotide is followed by a guanine nucleotide. In mammalian DNA, cytosine and guanine are separated by a phosphate group that links nucleotides together. Thus, the term “CpG” is used to distinguish between linear DNA sequences from other “CG” base-pairing. CG-rich regions are usually located within regulatory regions of promoters and are frequently referred to as CpG islands. In mammalian genomes, CpG islands are usually 300 to 3,000 base pairs long and appear in up to 50% of all human promoter regions
- Epigenetics
group of regulatory mechanisms that modify gene expression, without changing the underlying genomic sequence. This is accomplished by the reorganization of nucleosomes, resulting in variable transcription factor and RNA polymerase binding to DNA. Examples for epigenetic modifications are DNA methylation, and histone modifications, which will be explained in more detail in the article. Epigenetic modifications are generally dynamic, but can also remain through cell division and be passed on from generation to generation. Epigenetic modifications have been shown to be involved in the pathophysiology of multiple disorders, including autoimmune disease
- Glomerulonephritis
renal disorder that is characterized by inflammation of glomerula (the filtration organelles of the kidney) and/or small blood vessels. It is frequently associated with systemic autoimmune disorders, including SLE. Glomerulonephritis is one of the main contributors to morbidity and mortality in SLE. Diagnosing microscopical pathological patterns can be critical for the diagnosis of autoimmune diseases and optimized, subtype specific treatment
- Human endogenous retroviral elements (HERV)
sequences in the human genome that are derived from ancient viral infections. HERVs are suspected to be of retroviral origin and became integrated into the human genome. They are usually silenced by DNA methylation. Activation of HERVs has been suggested to result in a general disturbance of gene expression, in particular of genes that may contribute to the development of autoimmune diseases
- Proteinuria
presence of (excess) serum protein in the urine. In clinical settings, especially during the diagnosis and follow-up of patients with glomerulonephritis, proteinuria is used as a measure for disease activity and severity
- Transcription factors
proteins that specifically bind to DNA sequences within regulatory elements. Transcription factors are essential for gene regulation, by controlling the transcription of RNA from coding regions of genes. The process of gene activation through transcription factor binding is called “transactivation”. Impaired function or expression of various transcription factors has been reported to be responsible for a number of autoimmune disorders, including SLE
Footnotes
Conflict of interest statement:
The authors have no conflicts of interest.
References
- 1.Altshuler D, et al. Genetic mapping in human disease. Science. 2008;322(5903):881–8. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hedrich CM. Genetic Variation and Epigenetic Patterns in Autoimmunity. J Genet Syndr Gene Ther. 2011 doi: 10.4172/2157-7412.10000e2. ( http://omicsonline.org) [DOI]
- 3.Hedrich CM, Bream JH. Cell type-specific regulation of IL-10 expression in inflammation and disease. Immunol Res. 2010;47(1–3):185–206. doi: 10.1007/s12026-009-8150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renaudineau Y, Youinou P. Epigenetics and autoimmunity, with special emphasis on methylation. Keio J Med. 2011;60(1):10–6. doi: 10.2302/kjm.60.10. [DOI] [PubMed] [Google Scholar]
- 5.Gualtierotti R, et al. Updating on the pathogenesis of systemic lupus erythematosus. Autoimmun Rev. 2010;10(1):3–7. doi: 10.1016/j.autrev.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, et al. B cells contribute to ischemia/reperfusion-mediated tissue injury. J Autoimmun. 2009;32(3–4):195–200. doi: 10.1016/j.jaut.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crispín JC, Tsokos GC. Interleukin-17-producing T cells in lupus. Curr Opin Rheumatol. 2010;22(5):499–503. doi: 10.1097/BOR.0b013e32833c62b0. [DOI] [PubMed] [Google Scholar]
- 8.Meyer O, Hauptmann, et al. Genetic deficiency of C4, C2 or C1q and lupus syndromes. Association with anti-Ro (SS-A) antibodies. Clin Exp Immunol. 1985;62(3):678–84. [PMC free article] [PubMed] [Google Scholar]
- 9.Lee-Kirsch MA, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J Mol Med. 2007;85(5):531–7. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 10.Suggs MJ, et al. HLA DRB*1503 allelic haplotype predominance and associated immunodysregulation in systemic lupus erythematosus. Exp Mol Pathol. 2011 doi: 10.1016/j.yexmp.2011.03.006. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Moser KL, et al. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10(5):373–9. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karouzakis E, et al. Epigenetic control in rheumatoid arthritis synovial fibroblasts. Nat Rev Rheumatol. 2009;5(5):266–72. doi: 10.1038/nrrheum.2009.55. [DOI] [PubMed] [Google Scholar]
- 13.Meda F, et al. The epigenetics of autoimmunity. Cell Mol Immunol. 2011 doi: 10.1038/cmi.2010.78. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dieudé P. Rheumatic diseases: environment and genetics. Joint Bone Spine. 2009;76(6):602–7. doi: 10.1016/j.jbspin.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Zuvich RL, et al. Genetics and pathogenesis of multiple sclerosis. Semin Immunol. 2009;21(6):328–33. doi: 10.1016/j.smim.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao MJ, et al. Epigenetics in multiple sclerosis susceptibility: difference in transgenerational risk localizes to the major histocompatibility complex. Hum Mol Genet. 2009;18(2):261–6. doi: 10.1093/hmg/ddn353. [DOI] [PubMed] [Google Scholar]
- 17.Mastronardi FG, et al. Peptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. J Neurosci Res. 2007;85(9):2006–16. doi: 10.1002/jnr.21329. [DOI] [PubMed] [Google Scholar]
- 18.Javierre BM, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20(2):170–9. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zouali M. Epigenetics in lupus. Ann N Y Acad Sci. 2011;1217:154–65. doi: 10.1111/j.1749-6632.2010.05831.x. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, et al. Decreased DNA methyltransferase levels contribute to abnormal gene expression in “senescent” CD4(+)CD28(-) T cells. Clin Immunol. 2009;132(2):257–65. doi: 10.1016/j.clim.2009.03.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lal G, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182(1):259–73. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao M, et al. Hypomethylation of IL10 and IL13 promoters in CD4+ T cells of patients with systemic lupus erythematosus. J Biomed Biotechnol. 2010;2010:931018. doi: 10.1155/2010/931018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Q, et al. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol. 2005;174(10):6212–9. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 24.Singer NG, et al. Role of the CD6 glycoprotein in antigen-specific and autoreactive responses of cloned human T lymphocytes. Immunology. 1996;88(4):537–43. [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Q, et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 2002;46(5):1282–91. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- 26.Lu Q, et al. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179(9):6352–8. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan MJ, et al. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J Immunol. 2004;172(6):3652–61. doi: 10.4049/jimmunol.172.6.3652. [DOI] [PubMed] [Google Scholar]
- 28.Brooks WH, et al. Epigenetics and autoimmunity. J Autoimmun. 2010;34(3):J207–19. doi: 10.1016/j.jaut.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Basu D, et al. Stimulatory and inhibitory killer Ig-like receptor molecules are expressed and functional on lupus T cells. J Immunol. 2009;183(5):3481–7. doi: 10.4049/jimmunol.0900034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunahori K, et al. Methylation status of CpG islands flanking a cAMP response element motif on the protein phosphatase 2Ac alpha promoter determines CREB binding and activity. J Immunol. 2009;182(3):1500–8. doi: 10.4049/jimmunol.182.3.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunahori K, et al. Promoter Hypomethylation Results in Increased Expression of Protein Phosphatase 2A in T Cells from Patients with Systemic Lupus Erythematosus. J Immunol. 2011;186(7):4508–17. doi: 10.4049/jimmunol.1000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garaud S, et al. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J Immunol. 2009;182(9):5623–32. doi: 10.4049/jimmunol.0802412. [DOI] [PubMed] [Google Scholar]
- 33.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21(3):359–66. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 34.Nakkuntod J, et al. Hypomethylation of LINE-1 but not Alu in lymphocyte subsets of systemic lupus erythematosus patients. Clin Chim Acta. 2011 doi: 10.1016/j.cca.2011.04.002. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 35.Gorelik G, et al. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol. 2007;179(8):5553–63. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 36.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–12. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, et al. Overexpression of the growth arrest and DNA damage-induced 45alpha gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis Rheum. 2010;62(5):1438–47. doi: 10.1002/art.27363. [DOI] [PubMed] [Google Scholar]
- 38.Lei W, et al. Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand J Rheumatol. 2009;38(5):369–74. doi: 10.1080/03009740902758875. [DOI] [PubMed] [Google Scholar]
- 39.Januchowski R, et al. Prevalence of ZAP-70, LAT, SLP-76, and DNA methyltransferase 1 expression in CD4+ T cells of patients with systemic lupus erythematosus. Clin Rheumatol. 2008;27(1):21–7. doi: 10.1007/s10067-007-0644-8. [DOI] [PubMed] [Google Scholar]
- 40.Wang GS, et al. Ultraviolet B exposure of peripheral blood mononuclear cells of patients with systemic lupus erythematosus inhibits DNA methylation. Lupus. 2009;18(12):1037–44. doi: 10.1177/0961203309106181. [DOI] [PubMed] [Google Scholar]
- 41.Balada E, et al. Transcript levels of DNA methyltransferases DNMT1, DNMT3A and DNMT3B in CD4+ T cells from patients with systemic lupus erythematosus. Immunology. 2008;124(3):339–47. doi: 10.1111/j.1365-2567.2007.02771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dieker J, Muller S. Epigenetic histone code and autoimmunity. Clin Rev Allergy Immunol. 2010;39(1):78–84. doi: 10.1007/s12016-009-8173-7. [DOI] [PubMed] [Google Scholar]
- 43.Wilson CB, et al. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9(2):91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 44.Brenner C, Fuks F. A methylation rendezvous: reader meets writers. Dev Cell. 2007;12(6):843–4. doi: 10.1016/j.devcel.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 45.Ng HH, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23(1):58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38(13):4246–53. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra N, et al. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J Clin Invest. 2003;111(4):539–52. doi: 10.1172/JCI16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nambiar MP, et al. Effect of trichostatin A on human T cells resembles signaling abnormalities in T cells of patients with systemic lupus erythematosus: a new mechanism for TCR zeta chain deficiency and abnormal signaling. J Cell Biochem. 2002;85(3):459–69. doi: 10.1002/jcb.10160. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan KE, et al. The TNFalpha locus is altered in monocytes from patients with systemic lupus erythematosus. Clin Immunol. 2007;123(1):74–81. doi: 10.1016/j.clim.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crispín JC, Tsokos GC. Transcriptional regulation of IL-2 in health and autoimmunity. Autoimmun Rev. 2009;8(3):190–5. doi: 10.1016/j.autrev.2008.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tenbrock K, et al. The transcriptional repressor cAMP response element modulator alpha interacts with histone deacetylase 1 to repress promoter activity. J Immunol. 2006;177(9):6159–64. doi: 10.4049/jimmunol.177.9.6159. [DOI] [PubMed] [Google Scholar]
- 52.Hu N, et al. Aberrant expression pattern of histone acetylation modifiers and mitigation of lupus by SIRT1-siRNA in MRL/lpr mice. Scand J Rheumatol. 2009;38(6):464–71. doi: 10.3109/03009740902895750. [DOI] [PubMed] [Google Scholar]
- 53.Nishida K, et al. Histone deacetylase inhibitor suppression of autoantibody-mediated arthritis in mice via regulation of p16INK4a and p21(WAF1/Cip1) expression. Arthritis Rheum. 2004;50(10):3365–76. doi: 10.1002/art.20709. [DOI] [PubMed] [Google Scholar]
- 54.Arbuckle MR, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;16;349(16):1526–33. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 55.Savill J, et al. A blast from the past: clearance of apoptotic cells regulates immune responses. Nat Rev Immunol. 2002;2(12):965–75. doi: 10.1038/nri957. [DOI] [PubMed] [Google Scholar]
- 56.Herrmann M, et al. Impaired phagocytosis of apoptotic cell material by monocyte-derived macrophages from patients with systemic lupus erythematosus. Arthritis Rheum. 1998;41(7):1241–50. doi: 10.1002/1529-0131(199807)41:7<1241::AID-ART15>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 57.Dieker JW, et al. Apoptosis-induced acetylation of histones is pathogenic in systemic lupus erythematosus. 2007;56(6):1921–33. doi: 10.1002/art.22646. [DOI] [PubMed] [Google Scholar]
- 58.Godde JS, Ura K. Cracking the enigmatic linker histone code. J Biochem. 2008;143(3):287–93. doi: 10.1093/jb/mvn013. [DOI] [PubMed] [Google Scholar]
- 59.Muñoz LE, et al. The role of defective clearance of apoptotic cells in systemic autoimmunity. Nat Rev Rheumatol. 2010;6(5):280–9. doi: 10.1038/nrrheum.2010.46. [DOI] [PubMed] [Google Scholar]
- 60.Neumann E, et al. Rheumatoid arthritis progression mediated by activated synovial fibroblasts. Trends Mol Med. 2010;16(10):458–68. doi: 10.1016/j.molmed.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 61.Karouzakis E, et al. DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2009;60(12):3613–22. doi: 10.1002/art.25018. [DOI] [PubMed] [Google Scholar]
- 62.Neidhart M, et al. Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis Rheum. 2000;43(12):2634–47. doi: 10.1002/1529-0131(200012)43:12<2634::AID-ANR3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 63.Nile CJ, et al. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58(9):2686–93. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- 64.Takami N, et al. Hypermethylated promoter region of DR3, the death receptor 3 gene, in rheumatoid arthritis synovial cells. Arthritis Rheum. 2006;54(3):779–87. doi: 10.1002/art.21637. [DOI] [PubMed] [Google Scholar]
- 65.Huber LC, et al. Histone deacetylase/acetylase activity in total synovial tissue derived from rheumatoid arthritis and osteoarthritis patients. Arthritis Rheum. 2007;56(4):1087–93. doi: 10.1002/art.22512. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, et al. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006;54(7):2271–9. doi: 10.1002/art.21948. [DOI] [PubMed] [Google Scholar]
- 67.Camelo S, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;164(1–2):10–21. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 68.Denli AM, et al. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 69.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 70.Thai TH, et al. Is there a link between dysregulated miRNA expression and disease? Discov Med. 2010;10(52):184–94. [PubMed] [Google Scholar]
- 71.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106(1):23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 72.Hutvágner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 73.Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 431(7006):338–42. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 74.Pillai RS, et al. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17(3):118–26. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 75.Fabian MR, et al. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 76.Chen CZ, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 77.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29(7):343–51. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 78.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 79.Chuang JC, et al. Epigenetics and microRNAs. Pediatr Res. 2007;61(5 Pt 2):24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 80.Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–61. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 81.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–59. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 82.Zhou B, et al. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104(17):7080–5. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tang Y, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60(4):1065–75. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 84.Zhao X, et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010;62(11):3425–35. doi: 10.1002/art.27632. [DOI] [PubMed] [Google Scholar]
- 85.Pan W, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184(12):6773–81. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 86.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104(40):15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garzon R, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113(25):6411–8. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ng EK, et al. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer. 2009;101(4):699–706. doi: 10.1038/sj.bjc.6605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao S, et al. MicroRNA-126 regulates DNA methylation in CD4(+) T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2010 doi: 10.1002/art.30196. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 91.Lashine YA, et al. Expression signature of microRNA-181-a reveals its crucial role in the pathogenesis of paediatric systemic lupus erythematosus. Clin Exp Rheumatol. 2011;29(2):351–7. [PubMed] [Google Scholar]
- 92.Han L, et al. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6(8):1284–8. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- 93.Scott GK, et al. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66(3):1277–81. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 94.Stanczyk J, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58(4):1001–9. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 95.Stanczyk J, et al. Altered expression of miR-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 2010 doi: 10.1002/art.30115. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li H, et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest. 2009;119(12):3666–77. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tuddenham L, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580(17):4214–7. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 98.Noonan EJ, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28(14):1714–24. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 99.Alevizos I, et al. MicroRNA expression profiles as biomarkers of minor salivary gland inflammation and dysfunction in Sjögren’s syndrome. Arthritis Rheum. 2011;63(2):535–44. doi: 10.1002/art.30131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bulosan M, et al. Inflammatory caspases are critical for enhanced cell death in the target tissue of Sjögren’s syndrome before disease onset. Immunol Cell Biol. 2009;87(1):81–90. doi: 10.1038/icb.2008.70. [DOI] [PMC free article] [PubMed] [Google Scholar]