1. Structure
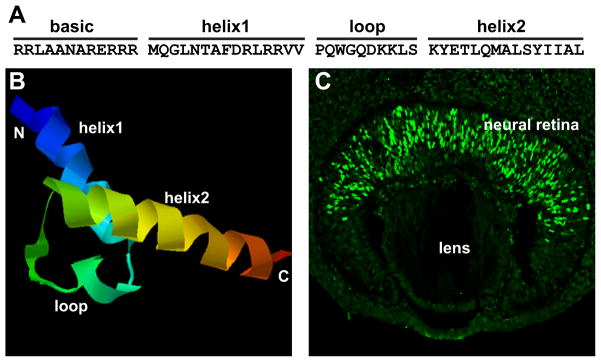
Math5 (also known as Atoh7) was first identified as a homologue of the Drosophila factor atonal. It is an evolutionarily conserved proneural transcription factor, known as Ath5 in Xenopus, zebrafish, and chick. Encoded by a single-exon gene, mouse Math5 is a 149 amino acid protein with a basic helix-loop-helix (bHLH) domain (NCBI Accession No. 016864.1) (Figure 1A). A predicted structure based on its amino acid sequence and other known bHLH structures is shown in Figure 1B. As with other bHLH transcription factors, the first helix of Math5 contains a basic region that mediates interaction with DNA. The second helix along with the flexible loop connecting the two helices mediates dimerization. As a transcription factor, Math5 must regulate genes by interacting with cis elements, but currently no definitive cis element has been identified. However, based on what is known about its homologue Atonal and other bHLH transcription factors, Math5 is likely to interact with an E-box sequence in the genome as either a homodimer or heterodimer.
Figure 1.
Structure and expression of Math1. A. Amino acid sequence of the bHLH region of mouse Math5. B. Predicted 3D structure of Math5 using PHYRE (Protein Homology/analogY Recognition Engine) at http://www.sbg.bio.ic.ac.uk/~phyre/. C. Expression of Math5 in the developing (E12.5) mouse retina. Math5 is visualized by immunofluorescence with a HA-tagged Math5 allele and anti-HA and can be seen to be expressed in a subset of retinal progenitor cells.
2. Function
The function of Math5 is best studied in the context of retinal ganglion cell (RGC) formation in retinal development, mostly in the mouse, but also in chicken, Xenopus and zebrafish. RGCs are one of the seven major retinal cell types; they are the only projection neurons in the retina, whose axons form the optic nerve and project to the brain. During development, a subset of proliferating retinal progenitor cells (RPCs) expresses Math5 (Figure 1C) and a fraction of these Math5-expressing RPCs give rise to RGCs. Once born, however, RGCs no longer express Math5, indicating its transient expression and quick action in the progenitor cells. The developmental period during which Math5 is expressed, embryonic day (E)11.5 to postnatal day (P)0, coincides closely with the beginning and end of the genesis of RGCs. Consistent with this expression pattern, Math5 loss-of-function mutants show a selective and an almost complete (95%) elimination of RGCs and optic nerves (Brown et al., 2001; Wang et al., 2001) . Such mice also have secondary abnormalities due to RGC loss, particularly in retinal vasculogenesis and circadian photoentrainment. Despite that fact that Math5-expressing cells can give rise to retinal cell types other than RGCs, generation of these other retinal cell types is not affected, except that there is an overproduction of amacrine and cone photoreceptor cells at the expense of RGCs (Brown et al., 2001; Wang et al., 2001). Therefore, Math5 is thought to confer to RPCs the competence to generate RGCs.
Math5 functions, at least in part, by regulating a number of potential genes, as shown by microarray analysis (Mu et al., 2005). Two of such genes are Pou4f2 and Isl1, which are positively regulated by Math5, and encode the POU-domain factor Pou4f2 (also known as Brn3b) and the LIM-homeodomain factor Isl1, respectively. In the absence of Pou4f2 or Isl1, RGCs differentiate in the ventricular side and migrate to the vitreal side of the retina normally but soon undergo apoptosis, indicating the indispensability of these factors for the full differentiation and/or maintenance of RGCs. In the developing retina, Math5 itself is regulated positively by Pax6 and negatively by the Notch pathway. Based on these findings, a gene regulatory network involved in RGC development has been constructed in which Math5 occupies a key position, downstream of Pax6 and upstream of Pou4f2 and Isl1.
Apart from being a pro-RGC factor, Math5 carries out a number of other functions (Mu and Klein, 2008). It inhibits such bHLH factors as Ngn2, NeuroD1, Math3 and Bhlhb5, either cell autonomously or cell non-autonomously, thereby suppressing the formation of photoreceptor, amacrine and bipolar cells. Also, it appears to promote the cell cycle exit of RPCs, as evidenced by their improper expression of p27/Kip1 and inability to become fully post-mitotic in Math5−/− retina. Finally, besides the retina, Math5 expression domains are also found in the developing cerebral cortex, cerebellar cortex and hippocampus although if and how Math5 participates in the development of these brain tissues is not yet known.
3. Disease involvement
bHLH transcription factors are involved in several aspects of vertebrate neurogenesis, including the differentiation of retinal neurons, and their mutations can lead to various neurological malformations. Since mutations in Math5 in the mouse and ath5 in zebrafish result in RGC and optic nerve agenesis and the human ATOH7 shows a very high homology in amino acid sequence within bHLH domain, mutated human ATOH7 might be implicated in RGC and optic nerve defects. In fact, nonsyndromic congenital retinal nonattachment, a form of blindness involving the detachment of retina from the posterior globe and absence of optic nerves, has recently been found to be caused by the deletion of a ‘shadow’ enhancer, which is secondary to the primary enhancer but can yield the same expression patterns, of ATOH7. In addition, recently several studies have suggested that single nucleotide polymorphisms (SNPs) associated with ATOH7 are risk factors in open angle glaucoma. Further study of the biological significance of these SNPs could provide insight in the development of glaucoma. In the future, it is possible that more ATOH7 mutations/variations will be found to be implicated in other types of eye diseases.
4. Future studies
While the role of Math5 in RGC differentiation has been firmly established, several issues with regard to the mechanism of Math5 function remain unresolved. First, and probably the most important, is identification of the factor(s) that makes certain Math5- expressing RPCs commit to RGCs. This is because lineage tracing experiments using the Cre-loxP recombination system have shown that Math5-expressing RPCs can differentiate into horizontal, amacrine and photoreceptor cells in addition to RGCs. Intrinsic factor and/or extracellular signal could be acting on Math5-positive progenitors to specify RGC fate. The second issue is the molecular mechanism by which Math5 makes RPCs competent to produce RGCs. This would certainly entail the identification of minimal set of downstream factors required for the proper differentiation of RGCs and the mechanism of regulation of these factors by Math5. Investigating the role of Math5 in non-RGC destined RPCs and the extracellular signals acting on Math5-positive progenitors would make the answer to this question more comprehensive. Third, there seem be conflicting data on when during the cell cycle Math5 is expressed and RGC fate decision is made within RPCs, but recent experiments have firmly established that Math5 is expressed in proliferating RPCs. However, it is not completely clear how many times a Math5-expressing RPC will further divide and what progenies it can produce. Since Math5 is an essential regulator for RGC formation, understanding the mechanisms underlying its function will also pave the way for in vivo generation of RGCs to be used in cell replacement therapies in glaucoma and other retinal diseases involved in RGC loss.
Acknowledgments
Our research is founded by The Whitehall Foundation; the National Eye Institute (EY020545); and an unrestricted grant from RPB to the Department of Ophthalmology of University at Buffalo.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Beremand PD, Thomas TL, Klein WH. A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev Biol. 2005;280:467–481. doi: 10.1016/j.ydbio.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Mu X, Klein WH. Gene regulatory networks and retinal ganglion cell development. In: Chalupa LM, Williams RW, editors. Eye, retina, and visual system of the mouse. MIT Press; Cambridge: 2008. pp. 321–332. [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]