Abstract
Members of the nucleotide-binding and oligomerization domain (NOD)-like receptor (NLR) family are quickly emerging as critical regulators of innate and adaptive immune responses during microbial infection and autoimmunity. The NLR family member NLRC5 was recently proposed to function as a positive and negative regulator of antiviral immune responses. NLRC5 has also been implicated in regulation of inflammasome signaling and MHC class I transcription. Some of these functions have recently been assessed in NLRC5-deficient mice and immune cells. Here, we summarize and review the newly gained knowledge on the structure, expression profile and putative functions of NLRC5 in regulating immune responses and host defense.
Keywords: NLR, Nlrc5, inflammasome, caspase-1, infection
Introduction
The innate immune system recognizes infections and cellular damage through a limited number of pattern recognition receptors (PRRs) (Kawai and Akira, 2006; Kumar et al., 2009). PRRs are thought to sense conserved microbial components that are vital for microbial survival such as flagellin and nucleic acid structures unique to bacteria and viruses (Akira et al., 2006). It is increasingly recognized that PRRs may also probe the environment for endogenous danger-associated molecular patterns (DAMPs) such as uric acid and HMGB1, which are produced or released upon tissue damage during infection or as a consequence of physicochemical stress. PRR activation triggers a number of protective responses, including the production of pro-inflammatory cytokines and chemokines that are responsible for activation of phagocytes and the recruitment of neutrophils, NK cells and lymphocytes to the site of infection. Moreover, innate immune responses contribute to adaptive immunity by instructing lymphocytes to mount T helper and humoral responses through the presentation of immunogenic peptides on MHC class I and class II receptors of professional antigen-presenting cells (Kanneganti et al., 2007).
Several PRR families can be distinguished, including Toll-like receptors (TLRs), C-type lectin receptors (CLRs), RIG-I-like receptors (RLRs), HIN-200 proteins and nucleotide binding and oligomerization domain-like receptors (NLRs) (Inohara et al., 2005; Kanneganti et al., 2007; Lamkanfi and Dixit, 2009; Meylan and Tschopp, 2006). Much research in recent years has focussed on characterizing the roles and signaling pathways of NLR family members in regulating the immune response. Bioinformatics studies revealed the existence of 22 human NLR genes and recent gene duplications gave rise to 34 mouse NLRs (Kanneganti et al., 2007). These platform proteins are characterized by the presence of a conserved nucleotide binding and oligomerization domain (referred to as NBD; NOD or NACHT domain) and located in intracellular compartments. Notably, the architecture of NLRs resembles that of a subset of plant disease-resistance (R) genes, which are involved in the hypersensitive response against virulent plant pathogens (Inohara and Nunez, 2001; Lamkanfi and Dixit, 2009). NLRs are involved in a multitude of innate immune signaling pathways ranging from the regulation of MAP kinase and NF-κB signaling pathways for Nod1 and Nod2, over modulation of MHC class II genes for CIITA, to the assembly of caspase-1-activating protein complexes named ‘inflammasomes’ for the NLR proteins NLRP1, NLRP3 and NLRC4 (Kanneganti et al., 2007; Lamkanfi and Dixit, 2009). Interestingly, the recently identified NLR family member NLRC5 (also known as NOD27, FLJ21709 and CLR16.1) has been suggested to regulate each of these signaling pathways. Moreover, NLRC5 deficient mice have been generated and used to examine the physiological role of this NLR in regulating immune responses and host defense during bacterial and viral infection in vivo. The recently gained knowledge on the expression, immune roles and signalling pathways of NLRC5 is summarized and critically reviewed in the following paragraphs.
Structure
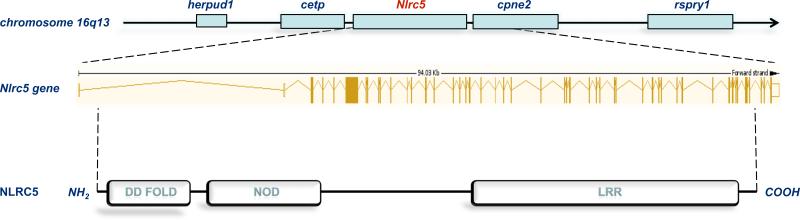
The human Nlrc5 gene is located at the locus 16q13, spanning a region of about 94 kbp. The full-length mRNA of 6822 bp is encoded by 49 exons, generating a protein that consists of 1866 amino acids (Figure 1). These features render NLRC5 the largest NLR family member. The protein has a domain architecture characteristic of all NLR members, comprising a centrally located NOD motif that is flanked at the carboxyl-terminus by an array of 20 leucine-rich repeat (LRR) motifs. Alignment of the NLRC5 LRR region suggests that the Nlrc5 gene may have arisen from an ancestral NLR gene shared with the NLRs Nod1, Nod2, NLRC3 and CIITA (Istomin and Godzik, 2009). Notable is that in addition to full-length NLRC5, five different NLRC5 splice variants were cloned from a human leukocyte cDNA library that differed in the length of the LRR motif (Neerincx et al., 2010). This suggest that NLRC5 activity may be subject to regulation by alternative splicing, but this intriguing possibility awaits confirmation that these NLRC5 variants are expressed as stable proteins. At the amino-terminus (amino acids 1-100), NLRC5 contains a death domain (DD) fold that shows little or no similarity to the caspase recruitment (CARD) and pyrin (PYD) motifs found in most NLRs (Inohara et al., 2005; Kanneganti et al., 2007). The amino-terminal (CARD and PYD) death domain folds are implicated in linking NLR family members with their downstream effector proteins through homotypic interactions involving related death domain folds found in adaptor proteins such as the bipartite inflammasome adaptor ASC (Lamkanfi and Kanneganti, 2010). However, the death domain fold of NLRC5 displays no clear homology to defined CARD, PYD and DD motifs of known adaptor and effector molecules involved in apoptosis and inflammatory signaling (Benko et al., 2010; Cui et al., 2010; Davis et al., 2011; Kuenzel et al., 2010; Meissner et al., 2010; Neerincx et al., 2010), thus offering few clues to the identity of potential NLRC5 binding partners and signaling pathways. The importance of NLRC5 signaling is reflected, however, in the fact that the protein is well-conserved throughout vertebrate evolution with orthologs identified in the chimpanzee, cow, rat and mouse genomes (Cui et al., 2010; Davis et al., 2011).
Figure 1. Structural organization of NLRC5.
The human Nlrc5 gene spans 94 kbp and is localized on chromosome 16q13 between the genes encoding plasma cholesteryl ester transfer protein (cetp) and the calcium-binding membrane protein copine II (cpne2). Nlrp3 is produced from 49 exons, of which the first two and last also encode the 5’ and 3’ untranslated regions, respectively. NLRC5 is expressed as a protein of 1866 amino acids consisting of an N-terminal death domain motif, a central NOD domain and 20 C-terminal LRR motifs. These features render NLRC5 the largest NLR family member that has been cloned.
NLRC5 expression
The NLRC5 expression profile and regulation of the Nlrc5 promoter activity by microbial components and pro-inflammatory cytokines has been examined in a variety of cells and tissues (Benko et al., 2010; Cui et al., 2010; Davis et al., 2011; Kuenzel et al., 2010; Meissner et al., 2010; Neerincx et al., 2010). Although somewhat discordant results were obtained in one study that reported a ubiquitous expression profile for NLRC5 mRNA in a variety of organs with the lowest transcript levels detected in immune-related tissues such as the spleen, lymph nodes and in immune cells (Kuenzel et al., 2010), most studies identified immune tissues including the bone marrow, lymph nodes, thymus and spleen as the preferential sources of NLRC5 mRNA expression in the human and mouse systems (Benko et al., 2010; Cui et al., 2010; Davis et al., 2011; Neerincx et al., 2010). In addition, NLRC5 transcripts were highly expressed in organs with mucosal surfaces such as the lung, small intestine, colon and uterus (Benko et al., 2010; Kuenzel et al., 2010). This suggests that NLRC5 may be involved in systemic immune signaling and regulation of host defense at mucosal interfaces. In agreement, NLRC5 mRNA and protein was detected in a variety of primary cells of myeloid and lymphoid origin as well as in the human THP-1 and murine RAW264.7 monocytic cell lines, the Jurkat T and Raji B cell lines, and the human cervic carcinoma cell line HeLa (Benko et al., 2010; Cui et al., 2010; Davis et al., 2011; Neerincx et al., 2010).
The NLRC5 promoter was shown to be highly responsive to interferon-γ (IFNγ) treatment (Kuenzel et al., 2010). In agreement, NLRC5 transcript levels were strongly induced by IFNγ in primary immune cells including macrophages, T and B lymphocytes as well as in a variety of immune and epithelial cell lines including THP-1, HeLa, CaCo2 and HT-29 (Benko et al., 2010; Kuenzel et al., 2010; Meissner et al., 2010). This suggests that the NLRC5 promoter may respond to viral infection. In agreement, NLRC5 mRNA was induced in HeLa cells, THP-1 cells and in human primary dermal fibroblasts stimulated with the double-stranded RNA mimic poly(I:C) or when infected with the single-stranded RNA Sendai virus (Neerincx et al., 2010). Notably, treatment with intracellular poly(I:C) and infection with the single-stranded RNA vesticular stomatitis virus (VSV) mostly failed to increase NLRC5 expression in RAW264.7 cells (Cui et al., 2010). However, NLRC5 mRNA expression was markedly upregulated in human foreskin fibroblasts infected with double-stranded DNA cytomegalovirus (CMV) (Kuenzel et al., 2010). NLRC5 upregulation was abrogated by a chemical inhibitor of the JAK/STAT pathway and by an IFNγ neutralizing antibody, suggesting that CMV-induced NLRC5 upregulation involved autocrine IFNγ production and IFNγ receptor-mediated activation of JAK/STAT signaling (Kuenzel et al., 2010).
Unlike the regulation of NLRC5 gene production by IFNγ and viral infection, the subcellular localization of the gene product is less clear. Immunofluorescence microscopy studies with GFP- and FLAG-tagged NLRC5 demonstrated a punctate expression profile in the cytosol of 293T, HeLa and HeLaS3 cells, but the fusion proteins were not found to be associated with the nucleus, lysosomes or mitochondria of these cells (Cui et al., 2010; Kuenzel et al., 2010; Neerincx et al., 2010). Restriction of endogenous NLRC5 expression to the cytosol was confirmed by immunofluorescence studies in human HeLaS3 cells using a rabbit antibody raised against an NLRC5-specific peptide (Kuenzel et al., 2010). In contrast to the studies above, two reports observed expression of Flag-tagged NLRC5 in both the nuclear and cytosolic compartments of overexpressing 293T and HeLa cells (Benko et al., 2010; Meissner et al., 2010). Leptomycin B, an inhibitor of CrmA-dependent nuclear export, increased the nuclear pool of overexpressed NLRC5 in 293T and HeLa cells, suggesting that the protein may shuttle between the nucleus and cytosol in a CrmA-dependent manner (Benko et al., 2010; Meissner et al., 2010). However, these results should be treated with caution since DD fold proteins are well-known to form intracellular clusters and filaments when overexpressed (Baliga et al., 2003). Thus, additional immunofluorescence microscopy and subcellular fraction studies would be helpful to resolve the current discrepancy regarding the localization of endogenous NLRC5. Moreover, such studies may reveal whether NLRC5 traffics to other compartments in response to inflammatory stimuli and during infection.
Biological functions
Despite progress in understanding the structure and the expression and induction profiles of NLRC5, its role(s) in regulating innate and adaptive immune responses and host defense remains controversial. One set of studies relied on overexpression and short hairpin (sh)RNA-mediated knockdown of NLRC5 to show a role for this NLR protein in dampening the production of pro-inflammatory cytokines in cells stimulated with LPS and poly(I:C) and in response to viral infection (Benko et al., 2010; Cui et al., 2010). This was proposed to occur through direct binding and regulation of the NF-κB regulators IKKα/IKKβ, thereby preventing recruitment of IKKγ/NEMO and nuclear translocation of NF-κB (Benko et al., 2010; Cui et al., 2010). In addition, NLRC5 may regulate this pathway by modulating the transactivation potential of nuclear NF-κB and/or by amplifying the production of the anti-inflammatory cytokine IL-10 (Benko et al., 2010). Moreover, NLRC5 was shown to negatively regulate antiviral signaling and type I IFN production through its association to the intracellular RNA virus sensors RIG-I and MDA5 (Cui et al., 2010). However, contradictory results were reported in two other studies that suggested a role for NLRC5 in inducing pro-inflammatory and IFN-dependent antiviral responses (Kuenzel et al., 2010; Neerincx et al., 2010). SiRNA-mediated knockdown of NLRC5 resulted in decreased production of type I IFN and pro-inflammatory cytokines in fibroblasts that have been infected with CMV or stimulated with poly(I:C) (Kuenzel et al., 2010). Similarly, NLRC5 downregulation in PMA-differentiated THP-1 cells and in primary human dermal fibroblasts reduced secretion of IFNβ, CXCL10, RANTES and MIP1α following Sendai virus infection and in response to poly(I:C) stimulation (Neerincx et al., 2010). However, neither a positive nor a negative role for NLRC5 in controlling antiviral immunity could be confirmed in NLRC5-deficient mice and isolated immune cells thereof (Kumar et al., 2011). Indeed, the production of IFNβ, CXCL10, RANTES, IL-6 and TNF-α were all normal following LPS, CpG DNA and poly(I:C) stimulation of NLRC5-deficient macrophages and dendritic cells (Kumar et al., 2011). Similarly, cytokine and type I IFN production NLRC5-negative dendritic cells was not affected upon infection of NLRC5-negative dendritic cells with the single-stranded RNA Newcastle disease virus or the dsDNA herpes simplex virus 1, and following stimulation with poly(dA:dT) DNA (Kumar et al., 2011). Thus, a general role for NLRC5 in regulating antibacterial and antiviral responses does not appear warranted, although it may be important for regulating particular cytokine and host response pathways or for guarding a specific set of pathogens. Such a role has recently been proposed for NLRC5 in regulating activation of the NLRP3 inflammasome and the subsequent production of IL-1β (Davis et al., 2011). However, this function was not upheld by experiments in NLRC5-deficient macrophages, which displayed normal levels of caspase-1 activation and IL-1β secretion in response to a variety of stimuli known to induce activation of the NLRP3, NLRC4 and AIM2 inflammasomes, respectively (Kumar et al., 2011). Finally, NLRC5 was proposed to regulate MHC class I transcription (Meissner et al., 2010), similar to the role of the NLR protein CIITA in inducing MHC class II gene expression (Kanneganti et al., 2007). NLRC5 overexpression in 293T cells resulted in activation of MHC class I reporter genes and enhanced MHC class I expression in lymphoid and epithelial cell lines (Meissner et al., 2010). Moreover, chromatin immunoprecipitation revealed NLRC5 occupancy at the HLA-A and HLA-B promoters and NLRC5 knockdown in HeLa cells specifically impaired expression of MHC class I and MHC class I–associated genes involved in antigen processing and presentation (Meissner et al., 2010). These results suggest NLRC5 as a transcriptional regulator of the MHC class I pathway. Surprisingly, however, shRNA-mediated knockdown of NlrC5 in RAW264.7 cells was shown to result in increased, rather than decreased, MHC class I surface expression (Benko et al., 2010). Analysis of MHC class I surface expression and gene regulation in NLRC5-deficient antigen-presenting cells may help to resolve the precise contribution of this NLR to CD8+ T cell activation.
Concluding remarks
Recent studies have identified NLRC5 as the largest and a well-conserved NLR family members that contains a rather atypical death domain fold at its amino- terminus and and elongated array of more than 20 leucine-rich repeat (LRR) motifs at its carboxyl-terminus. Moreover, consensus is emerging that the NLRC5 promoter and the gene product is regulated by IFNγ, the viral dsRNA mimic poly(I:C) and possibly by other cytokines and microbial components. However, the role(s) of NLRC5 in host defense and its role in regulating immune signaling pathways remain controversial. NLRC5 was proposed to function as a positive (Kuenzel et al., 2010; Neerincx et al., 2010) and as a negative regulator (Benko et al., 2010; Cui et al., 2010) of IFN, NF-κB, AP-1 signaling and antiviral immunity. NLRC5 has also been implicated in regulation of inflammasome signaling (Davis et al., 2011). However, none of these putative roles for NLRC5 was confirmed in NLRC5 deficient cells and mice infected with bacterial and viral pathogens or stimulated with microbial ligands (Kumar et al., 2011). Moreover, NLRC5 was proposed to regulate MHC class I transcription (Meissner et al., 2010), but this has been contested by others (Benko et al., 2010). Thus, the precise role of NLRC5 in regulating immune signaling remains unclear and further investigation is needed to resolve how it affects host defense and immune signaling.
Acknowledgements
This work was supported by National Institute of Health Grants R01AR056296, AR056296 supplements and R21AI088177 and the American Lebanese Syrian Associated Charities (ALSAC) to T-D.K. ML is supported by the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen and by the European Union Framework Program 7 Marie-Curie grant 256432.
Abbreviations
- ASC
apoptosis-associated speck-like protein containing a CARD
- BIR
baculovirus IAP repeat
- CARD
caspase recruitment domain
- CLR
C-type lectin receptor
- Ig
immunoglobulin
- IL
interleukin
- LPS
lipopolysaccharide
- LRR
leucine-rich repeat
- NLR
NOD-like receptor
- NOD
nucleotide-binding and oligomerization domain
- PRR
pattern recognition receptor
- TLR
Toll-like receptor
- RLR
RIG-I-like receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Baliga BC, Colussi PA, Read SH, Dias MM, Jans DA, Kumar S. Role of prodomain in importin-mediated nuclear localization and activation of caspase-2. J. Biol. Chem. 2003;278:4899–4905. doi: 10.1074/jbc.M211512200. [DOI] [PubMed] [Google Scholar]
- Benko S, Magalhaes JG, Philpott DJ, Girardin SE. NLRC5 limits the activation of inflammatory pathways. J. Immunol. 2010;185:1681–1691. doi: 10.4049/jimmunol.0903900. [DOI] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, Wang RF. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BK, Roberts RA, Huang MT, Willingham SB, Conti BJ, Brickey WJ, Barker BR, Kwan M, Taxman DJ, Accavitti-Loper MA, et al. Cutting Edge: NLRC5-Dependent Activation of the Inflammasome. J. Immunol. 2011;186:1333–1337. doi: 10.4049/jimmunol.1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara, Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Inohara N, Nunez G. The NOD: a signaling module that regulates apoptosis and host defense against pathogens. Oncogene. 2001;20:6473–6481. doi: 10.1038/sj.onc.1204787. [DOI] [PubMed] [Google Scholar]
- Istomin AY, Godzik A. Understanding diversity of human innate immunity receptors: analysis of surface features of leucine-rich repeat domains in NLRs and TLRs. BMC Immunol. 2009;10:48. doi: 10.1186/1471-2172-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- Kuenzel S, Till A, Winkler M, Hasler R, Lipinski S, Jung S, Grotzinger J, Fickenscher H, Schreiber S, Rosenstiel P. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J. Immunol. 2010;184:1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- Kumar H, Pandey S, Zou J, Kumagai Y, Takahashi K, Akira S, Kawai T. NLRC5 deficiency does not influence cytokine induction by virus and bacteria infections. J. Immunol. 2011;186:994–1000. doi: 10.4049/jimmunol.1002094. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Inflammasomes: guardians of cytosolic sanctity. Immunol. Rev. 2009;227:95–105. doi: 10.1111/j.1600-065X.2008.00730.x. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Kanneganti TD. Nlrp3: an immune sensor of cellular stress and infection. Int J Biochem Cell Biol. 2010;42:792–795. doi: 10.1016/j.biocel.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner TB, Li A, Biswas A, Lee KH, Liu YJ, Bayir E, Iliopoulos D, van den Elsen PJ, Kobayashi KS. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:13794–13799. doi: 10.1073/pnas.1008684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Tschopp J. Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol Cell. 2006;22:561–569. doi: 10.1016/j.molcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Neerincx A, Lautz K, Menning M, Kremmer E, Zigrino P, Hosel M, Buning H, Schwarzenbacher R, Kufer TA. A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. J. Biol. Chem. 2010;285:26223–26232. doi: 10.1074/jbc.M110.109736. [DOI] [PMC free article] [PubMed] [Google Scholar]