Abstract
Receptor-regulated cellular signaling often is mediated by formation of transient, heterogeneous protein complexes of undefined structure. We used single and two-color photoactivated-localization microscopy (PALM) to study complexes downstream of the T cell antigen receptor (TCR) in single molecule detail at the plasma membrane of intact T cells. The kinase ZAP-70 distributed completely with the TCRζ chain and both partially mixed with the adapter LAT in activated cells thus showing localized activation of LAT by TCR-coupled ZAP-70. In resting and activated cells LAT primarily resided in nanoscale clusters as small as dimers whose formation depended on protein-protein and protein-lipid interactions. Surprisingly, the adapter SLP-76 localized to the periphery of LAT clusters. This nanoscale structure depended on polymerized actin and its disruption affected TCR-dependent cell function. These results extend our understanding of the mechanism of T cell activation and the formation and organization of TCR-mediated signaling complexes, findings also relevant to other receptor systems.
Introduction
Signaling complexes are heterogeneous, dynamic multi-molecular structures induced following receptor binding and which mediate signal transduction. They are nucleated by activated receptors and membrane-bound scaffold proteins and are often found in microclusters (Cebecauer et al., 2010; Harding and Hancock, 2008; Schlessinger, 2000). Using diffraction-limited light microscopy, studies of T cell activation have demonstrated that microclusters form at the plasma membrane (PM) of T cells upon engagement of the antigen specific receptor (TCR) (Campi et al., 2005; Yokosuka et al., 2005). These microclusters contain molecular complexes comprised of the TCR, signaling enzymes such as ZAP-70 and phospholipase C-γ1 (PLC-γ1) and several adapter proteins including LAT, SLP-76, Grb2 and Gads (Bunnell et al., 2002). Similar structures have been seen in B cells and mast cells following engagement of the immunoreceptors in these cells (Harwood and Batista, 2010b; Wilson et al., 2001).
Although signaling complexes and microclusters have been studied extensively by various biochemical and imaging techniques, the spatial organization of individual molecules within signaling complexes or of complexes within microclusters has not been addressed. Much remains to be learned about complexes and microclusters including, their full size distribution, their potential heterogeneity, their arrangement in the PM and the molecular requirements for their formation. Imaging the spatial relationship of molecules at the single molecule level might resolve these questions. Toward this goal, we studied TCR interactions with signaling molecules and signaling complexes downstream of the TCR. We found a nanoscale organization to these interactions and structures with unexpected properties relevant for understanding T cell activation.
Results
Most LAT molecules are not within microclusters
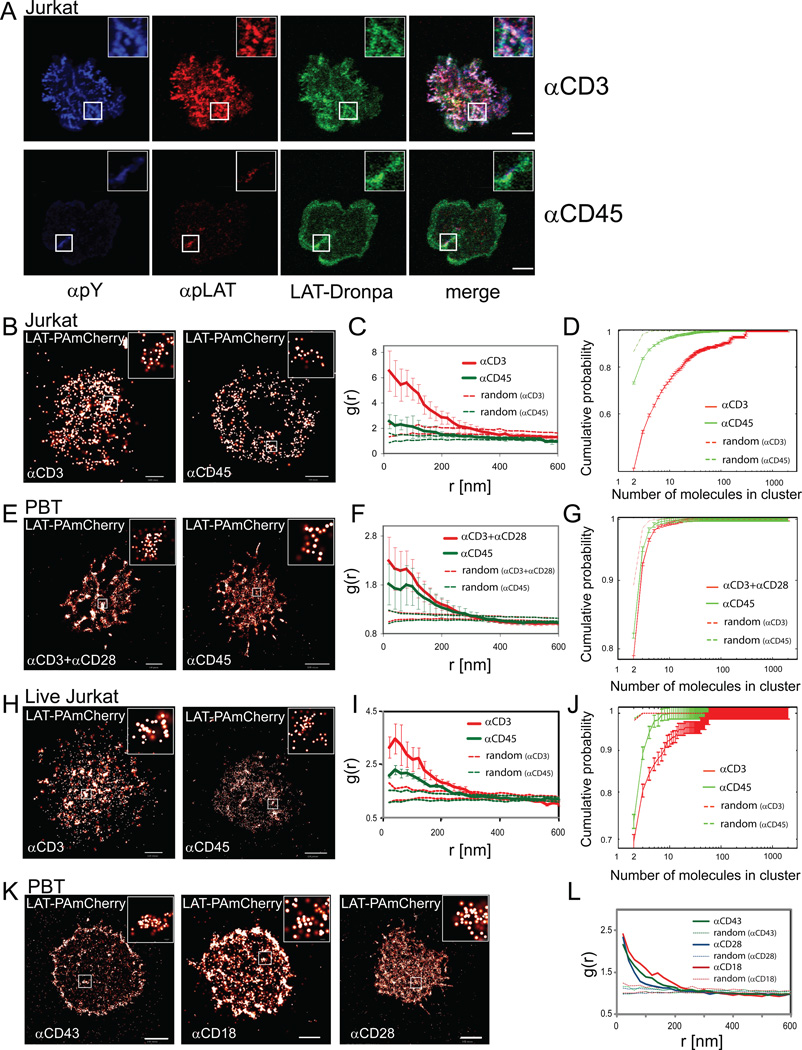
We began by studying the organization of LAT at the PM by diffraction-limited microscopy. Cells stably expressing LAT-Dronpa [(Ando et al., 2004); Figure S1A,B] were dropped and imaged on antibody coated coverslips (Bunnell et al., 2003) that either stimulate the TCR (αCD3) or avoid TCR stimulation (αCD45). Using confocal microscopy, LAT molecules showed pronounced signaling microclusters in both Jurkat T cells (Figure 1A, αCD3) and human peripheral blood T cells (PBTs) upon activation (Figure S1C, αCD3+αCD28). On control, non-activating surfaces, there were fewer cells with LAT clusters as detected by intensity thresholding (Figure 1A, αCD45, Figure S1D,F, and see SI for further details). The extent of spreading under both conditions was similar (Figure S1E,G), but there was a far lower extent of LAT or phosphorylated proteins in clusters under non-stimulating conditions (Figure S1H). Importantly, we note that LAT microclusters accounted for only a small part of total LAT molecules at the cell surface as measured by total fluorescence using either confocal (~24% and ~8% for stimulating and non-stimulating conditions; see Figure S1I) or total internal reflection (TIRF) microscopy (~20% and 12% for stimulating and non-stimulating conditions; see Figure S1J). These observations emphasize the need to characterize the role and organization of all of the LAT molecules at the PM including those that are not in microclusters (Lillemeier et al., 2010; Lillemeier et al., 2006; Wilson et al., 2001).
Figure 1. LAT clusters at the PM are mainly in small nanoclusters.
(A) Confocal images of Jurkat E6.1 T cells on αCD3- or αCD45-coated coverslips. Cells were stained for pY (blue), pLAT(pY191, red) and stably expressed LAT-Dronpa (green). Bar - 10 µm. Insets show a magnified view of a microcluster.
(B) PALM images of Jurkat T cells expressing LAT-PAmCherry spread on αCD3 or spread on αCD45. Insets show nano-scale organization of individual molecules within apparent microclusters. Color codes (heat map, white is highest) for overlapping probability density functions of individual molecules, with a maximal value of 410 molecules/µm2 on αCD3 and 360 molecules/µm2 on αCD45. Bars – 0.5 µm and 1 µm, respectively. (C) Pair correlation function (PCF) of LAT-PAmCherry molecules (n=7 cells for stimulating and non-stimulating conditions; dashed lines indicate upper and lower 95% confidence levels of a Heterogeneous Poisson process; see Analyses section in SI for further details). (D) Results of a clustering algorithm to resolve individual clusters and generate cumulative size-distribution curves of the LAT clusters seen under stimulating and non-stimulating conditions. (n=12 cells for stimulating and 10 for non-stimulating conditions; dashed lines represent clustering of random sets; the two dashed lines overlap; see SI for further details on statistical analyses). (E) PALM images of peripheral blood lymphocytes (PBLs) expressing LAT-PAmCherry spread on either αCD3+αCD28- or on αCD45-coated coverslips. Insets show nano-scale organization of individual molecules within apparent microclusters. Maximal density values are 740 molecules/µm2 and 490 molecules/µm2, respectively. Bars – 1 µm (left) and 2 µm (right). (F) The PCF of LAT-PAmCherry molecules (n=8 cells for stimulating and non-stimulating conditions; dashed lines defined as in panel C). (G) The cluster analyses showing cumulative size-distribution curves. (n=12 cells for stimulating and n=10 for non-stimulating conditions; dashed lines represent clustering of random sets; the two dashed lines overlap; see SI for further details on statistical analyses). (H) PALM image of a live Jurkat T cell expressing LAT-PAmCherry spread on αCD3- or on αCD45-coated surface. Maximal density value 50 molecules/µm2 for both panels. Bars - 2 µm (I) PCF of LAT-PAmCherry molecules (n=3 cells for stimulating and non-stimulating conditions; dashed lines defined as in panel C). (J) Clustering analyses (n=3 cells for stimulating and non-stimulating conditions). See Materials and Methods section in SI and Figure S1K for further details regarding live cell imaging with PALM. (K) Peripheral blood T (PBT) cells expressing LAT-PAmCherry plated onto αCD43-, αCD18- or αCD28-coated coverslips. Bars - 2µm. (L) PCFs of LAT-PAmCherry molecules (dashed lines defined as in panel C).
LAT resides in pre-existing nanoclusters with a shift towards larger clusters upon stimulation
To better characterize PM LAT clustering with single molecule resolution, we used photoactivated-localization microscopy (PALM) imaging to observe individual LAT molecules, tagged with photoactivatable fluorescent proteins [LAT-Dronpa (Ando et al., 2004) or LAT-PAmCherry (Subach et al., 2009)]. We chose stimulation conditions that both maximized cell spreading and the presence of LAT in the narrow optical section detected by PALM in TIRF mode (Figure S2A,B). PALM images from Jurkat T cells (Figure 1B,H) and normal human peripheral blood T cells (PBTs) (Figure 1E) were analyzed under activating and non-activating conditions. We measured the probability density of locating individual LAT-PAmCherry molecules with a precision of ~20nm (Figure S2C). We next analyzed the scale of LAT clustering using pair-correlation functions (PCFs or g(r); Figure 1C,F,I; Figure S1L; see also Analyses section in SI). In addition, using a cluster analysis algorithm and a careful choice of the distance threshold for clustering (see Analyses section in SI and Figure S1M,N), we were able to identify individual LAT clusters in the PM and describe their size distribution in detail (Figure 1D,G,J). We found that LAT was always significantly pre-clustered under non-stimulating conditions (i.e., αCD45-coated coverslips) as well as activating conditions on the PM of both Jurkat (Figure 1B-D,H-J) and PBTs (Figure 1E-G). Clustering can be observed in univariate PCF data (Figure 1C,F,I) when the value of g(r) (bold lines) exceeds the 95% confidence interval due to calculated random molecular distribution (dashed lines; refer to the Heterogeneous Poisson null model in the Analyses section in SI). PCFs also revealed that molecules in defined clusters were separated by as little as 20 nm. The majority of LAT molecules resided in very small nanoclusters containing only two, three or a few detectable molecules (Figure 1D,G,J). Though small nanoclusters predominated, there was a wide size distribution as indicated by the cluster size statistics (Figure S1L).
We were struck by the presence of LAT clustering as seen by confocal (Figure S1D) and PALM imaging (Figure 1B,E,H) in cells spread on the coverslip in response to non-activating αCD45 antibodies. Therefore, we tested coverslips coated with antibodies against other surface molecules including CD28, CD43 or CD18, which also lead to cell adherence and spreading without activating the TCR (Figure 1K; confocal imaging results not shown). Regardless of which control antibodies we used or their effect on cell or microcluster appearance, we found that LAT was distributed in nanoclusters (Figure 1L). We conclude that in the basal state LAT is organized in nanoclusters at the PM in Jurkat and PBT cells.
Upon stimulation with αCD3, we observed an increase in the extent of LAT clustering (Figure 1C,F,I) and a modest shift towards larger clusters (Figure 1D,G,J) in the size distribution curves, without a change in the overall shape of the distributions (Figure 1D,G,J). These changes were more pronounced in Jurkat T cells than in PBT cells. Differences between cell types and cell-to-cell variability in the pattern of cellular spreading and the extent of protein expression contributed to the differences in the amount of clustering observed in our analyses (Figure 1C,D,F,G,I,J). A similar size distribution was determined for LAT clusters at the PM of both fixed and live cells (Figure 1H-J and Figure S1K) imaged under stimulating and non-stimulating conditions. The live cell PALM result confirms that the nanocluster distribution we observed was not an artifact of fixation.
Two factors might contribute to the dominance of small nanoclusters in the size-distribution and PCFs derived from unstimulated and stimulated cells. First, most of the LAT molecules at the PM do not reside in microclusters (Figure S1I,J). Second, large aggregates such as microclusters are broken down by our analyses into their smallest detectable constituents, such that a single microcluster viewed by diffraction-limited microscopy is revealed to be a collection of much smaller nanoclusters.
LAT clusters are recruited to early contact areas within lamellae
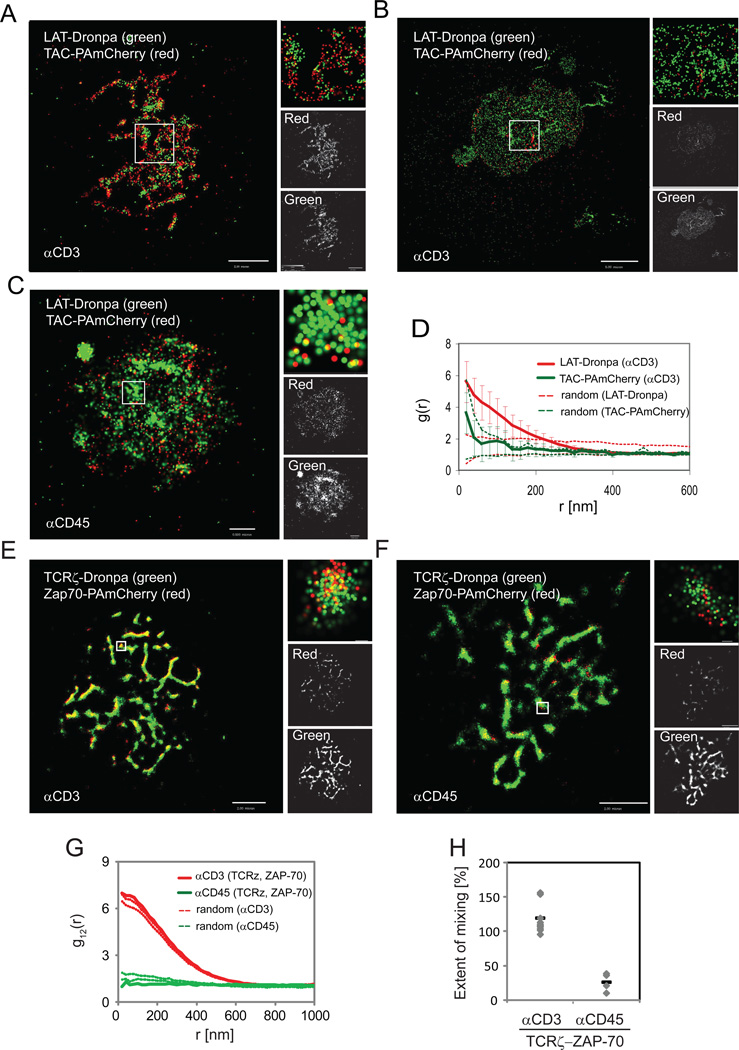
To further understand the significance of the irregular distribution of LAT on the coverslip and LAT organization in basal nanoclusters, we conducted two-color PALM imaging of cells expressing the fluorescent conjugates LAT-Dronpa and a PM marker, TAC-PAmCherry (Subach et al., 2009) (TAC is CD25, the IL-2 receptor alpha subunit; see Materials and Methods section in SI and Figure S2D,E regarding two-color PALM imaging). In this experiment LAT clusters were found at the sites of contact where PM lamellae containing the randomly distributed TAC-PAmCherry first touched the stimulating surface (Figure 2A). As the cells spread over time, more of the PM made contact with the surface and only LAT clusters were retained with the irregular lamellar pattern at sites of initial cellular contact (Figure 2B; see also Figure S2A). An irregular distribution of PM at the coverslip was also observed using non-stimulating conditions (Figure 2C). We conclude that these results explain the first-order heterogeneity of LAT distribution, which we define as the irregular distribution of micron-sized LAT clusters at the PM. That is, LAT microclusters first appear and persist in areas of membrane contact with the activating surface, resulting in micron-sized features that reflect these initial contacts. Importantly this experiment also shows that second-order molecular heterogeneity, the nanoscale organization of LAT microclusters, is not a feature of all membrane proteins. The PCF of TAC showed that it is randomly distributed (the TAC PCF curve falls within the 95% confidence interval of the heterogeneous Poisson null model) and not clustered within the PM (Figure 2D; for data of unstimulated cells, see Figure S2F,G).
Figure 2. Two-color PALM captures the mutual organization and interactions of signaling molecules at the PM.
(A,B,C) LAT is recruited to early contact areas within lamellae. Two-color PALM images of Jurkat cells expressing LAT-Dronpa (green) and TAC-PAmCherry (red) under stimulating conditions, showing (A) partial or (B) complete spreading, or (C) cells under non-stimulating conditions showing partial spreading. Right panels show a zoomed image of the region of interest, a single color rendering of TAC-PAmcherry (labelled red) and LAT-dronpa (labelled green). Maximal probability density values (as detailed in Figure 1B) for LAT-Dronpa and TAC-PAmCherry – (A) 270 and 1340 molecules/µm2, respectively; (B) 190 and 10 molecules/µm2; (C) 126 and 59 molecules/µm2. Bars – (A) 2µm, (B) 5µm, (C) 0.5µm. (D) The PCF of LAT-Dronpa (red line) and TAC-PAmCherry (green line) under stimulating conditions (n=5 cells; Dashed lines indicate upper and lower 95% confidence levels of a Heterogeneous Poisson process). See Figure S2F,G for the related PCFs for non-stimulating conditions.
(E,F,G,H) ZAP-70 is well co-localized with TCRζ in activated T cells. Two-color PALM images of Jurkat T cells expressing TCRζ-Dronpa and ZAP-70-PAmCherry on (E) αCD3- and (F) αCD45-coated coverslips. Insets show zoomed images of individual TCRζ and ZAP-70 clusters (bars – 200nm). (G) Bivariate correlation curves of TCRζ and ZAP-70 for αCD3 (red lines) and αCD45 (green lines). Dashed lines indicate upper and lower 95% confidence levels of a random labeling (homogeneous mixing) model (H) The extent of mixing between TCRζ and ZAP-70 (as defined in the text) are compared between activating and non-activating conditions for multiple cells expressing TCRζ and ZAP-70 (n=13 for αCD3, and n=5 for αCD45). Maximal probability density values (as detailed in Figure 1B) for TCRζ-Dronpa and ZAP-70-PAmCherry – (E) 1580 and 1630 molecules/µm2, respectively, (F) 1070 and 1610 molecules/µm2. Bars – 2µm.
In the next section interactions between the TCR, ZAP-70 and LAT are extensively studied using two-color PALM and bivariate analyses. The distribution of these molecules individually can also be studied as above with univariate statistics (such as PCFs). The distribution of the TCR has been studied previously biochemically and by PALM (Lillemeier et al., 2010; Schamel et al., 2005). In T cells interacting with αCD45 on the coverslip we found that the TCRζ chain was found in nanoclusters with a distribution favoring small nanoclusters much like LAT (Figure S2H). Since the mechanism of activation in our studies is antibody-induced clustering of the TCR, we can not comment on the distribution of the TCR following physiologic activation.
Two-color PALM captures the efficient interaction of TCRζ and ZAP-70 in activated T cells
We used two-color PALM to study the organization of signaling molecules downstream of the TCR. We began by imaging TCRζ-Dronpa and ZAP-70-PAmCherry. Upon TCR activation, the protein tyrosine kinase ZAP-70 is recruited to the phosphorylated zeta chains of the TCR (TCRζ), becomes activated and then phosphorylates multiple tyrosines on LAT and other protein substrates (Wang et al., 2010). Phosphotyrosines on LAT then serve as docking sites for multiple adapters and enzymes (Balagopalan et al., 2010). As expected from previous imaging and biochemical studies (Bunnell et al., 2002; Wang et al., 2010), we found that ZAP-70 co-localized with TCRζ upon activation (Figure 2E) and was far less efficiently recruited to TCRζ without TCR activation (Figure 2F). We used bivariate correlation statistics to test whether the two molecules, TCRζ and ZAP-70, mixed randomly. If they did, the bivariate curve (g12(r), bold lines in Figure 2G) would fall between the two curves that define the 95% confidence interval of a random mixing model (dotted lines in Figure 2G), which serves as a null hypothesis (see Analyses section in SI). Displacement of the curve above the 95% confidence interval would indicate significant attraction between the two molecules beyond the random mixing model, while displacement below the 95% confidence interval would suggest that repulsion or segregation govern the relative positioning of the two molecules.
For the case of TCRζ and ZAP-70, the bivariate analysis demonstrated complete mixing of the two molecules following TCR activation (Figure 2G, red lines). This result shows that ZAP-70 is recruited to TCR clusters in a manner independent of TCR cluster size. In contrast, the cells that engaged the non-activating surface revealed little interaction between TCRζ and ZAP-70 consistent with the little or absent TCRζ phosphorylation found in unactivated T cells (Figure 2G, green lines). We also note systematic differences in the absolute height of the bivariate correlation curves and their 95% confidence intervals between activated and non-activated cells. To quantify the significance of these differences, we plotted the normalized level of bivariate correlation for multiple individual cells at scales below 60nm (essentially, the y-axis intercepts of the bold lines in comparison to the intercepts of the dotted lines shown in Figure 2G). The results indicate that TCRζ and ZAP-70 consistently mix in a more homogenous way (approximately 100%) under activating conditions than under non-activating conditions (far less than 50%, Figure 2H).
TCRζ and LAT exist in overlapping pools that segregate more upon T cell activation and serve as hot spots for LAT activation
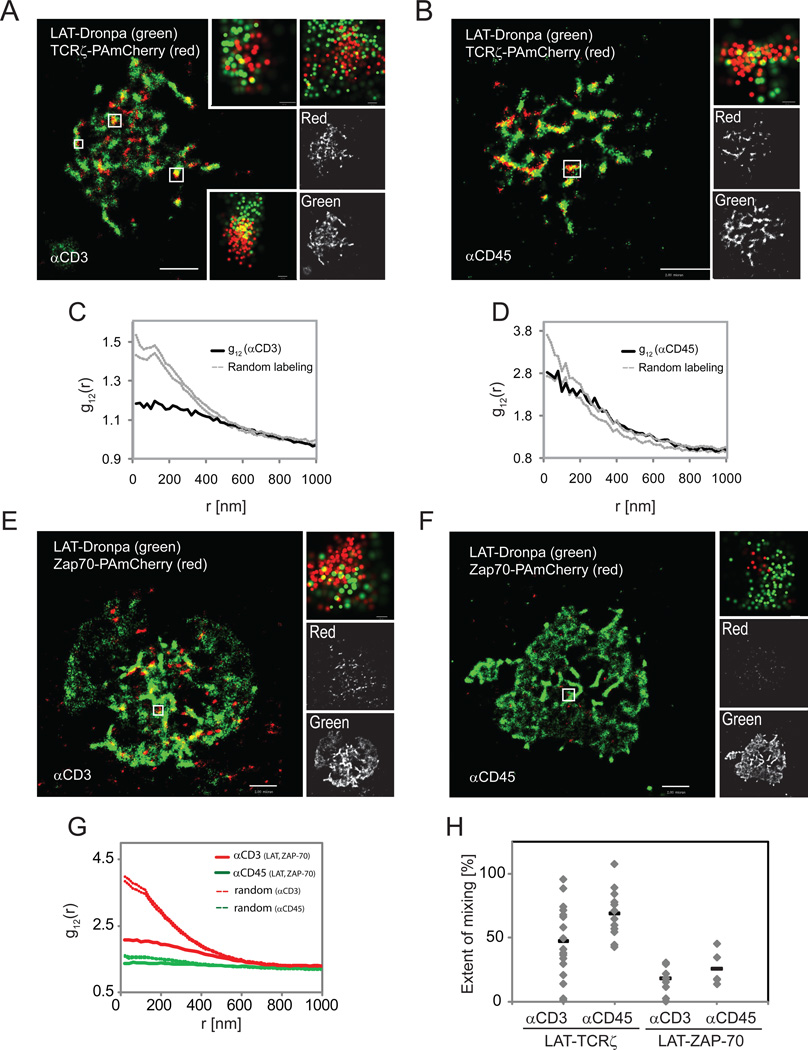
The TCR and LAT have been suggested to exist in separate domains that concatenate upon cell activation without mixing (Lillemeier et al., 2010). We used 2-color PALM to image LAT-Dronpa and TCRζ-PAmCherry expressed in Jurkat T cells on αCD3-coated or on control αCD45-coated coverslips. Two-color PALM images showed nanoscale domains of both LAT and TCRζ that significantly overlapped under both activating and non-activating conditions (Figure 3A,B).
Figure 3. LAT and TCRζ exist in overlapping pools, where nanoscale domains could function as hot spots of T cell activation.
Two-color PALM images of a LAT-Dronpa- and TCRζ-PAmCherry-expressing Jurkat T cell (A) on αCD3- or (B) αCD45-coated coverslips. Zoomed images (insets) of individual TCRζ and LAT clusters are shown (bars – 100nm). (C) Bivariate correlation curves for αCD3 (Dashed lines indicate upper and lower 95% confidence levels of a Heterogeneous Poisson process). (D) Bivariate correlation curve for non-activating conditions (Dashed lines defined as in panel C). Maximal probability density values (as detailed in Figure 1B) for LAT-Dronpa and TCRζ-PAmCherry – (A) 1380 and 1120 molecules/µm2, respectively; (D) 510 and 400 molecules/µm2. Bars – 2µm.
(E,F,G,H) ZAP-70 is poorly co-localized with LAT. Two-color PALM images of Jurkat T cells expressing LAT-Dronpa and ZAP-70-PAmCherry on (E) αCD3- or (F) αCD45-coated coverslips. Zoomed images (insets) of individual LAT and ZAP-70 clusters (bars – 200nm). (G) Bivariate correlation curves of LAT and ZAP-70 for αCD3 (red lines) and for αCD45 (green lines). Dashed lines defined as in panel C. (H) The extent of mixing between LAT and TCRζ or LAT and ZAP-70 are compared between activating and non-activating conditions for multiple cells (LAT and TCRζ - n=19 for αCD3, and n=14 for αCD45; LAT and ZAP-70 - n=13 for αCD3, and n=4 for αCD45). See definition of the mixing coefficient in the text. Maximal probability density values (as detailed in Figure 1B) for TCRζ-Dronpa and ZAP-70-PAmCherry – (A) 1580 and 1630 molecules/µm2, respectively, (B) 1070 and 1610 molecules/µm2 (C) 1500 and 1260 molecules/µm2, (D) 1400 and 440 molecules/µm2. Bars – 2µm.
In activated samples (Figure 3C) the bivariate analysis revealed homogeneous mixing of the two molecules at separations over ~450nm, but a strong deviation from this model below this length scale as indicated by the location of the curve below the zone defined for 95% confidence level of the homogeneous mixing (random labeling) model. However, under non-activating conditions the bivariate correlation between the molecules was much closer to homogeneous mixing at all length-scales (Figure 3D). Comparing the differences between the two conditions for multiple cells, we found that TCRζ and LAT tend to mix in a more homogenous way under non-activating conditions over all size ranges and then segregate more from each other upon activation (Figure 3H).
In light of the partial overlap of molecular distribution between TCRζ and LAT following activation, we were interested in the localization of ZAP-70 relative to LAT to provide information on the means by which LAT becomes phosphorylated. In contrast to its complete mixing with TCRζ, ZAP-70 showed only a partial overlap in distribution with LAT molecules upon T cell activation (Figure 3E,G,H) or under non-activating conditions (Figure 3F,G,H). This distribution was similar to the partial mixing of LAT with TCRζ (Figure 3A,C). Since we failed to detect efficient mixing of ZAP-70 with LAT with activation, we propose that ZAP-70 phosphorylates LAT when it is bound to TCRζ or in very close proximity to this TCR subunit. At any point in time there is a limited number of such “hot spots” where activated TCRs interact with and phosphorylate LAT.
Protein-protein and protein-lipid interactions are both required for intact LAT nanocluster formation
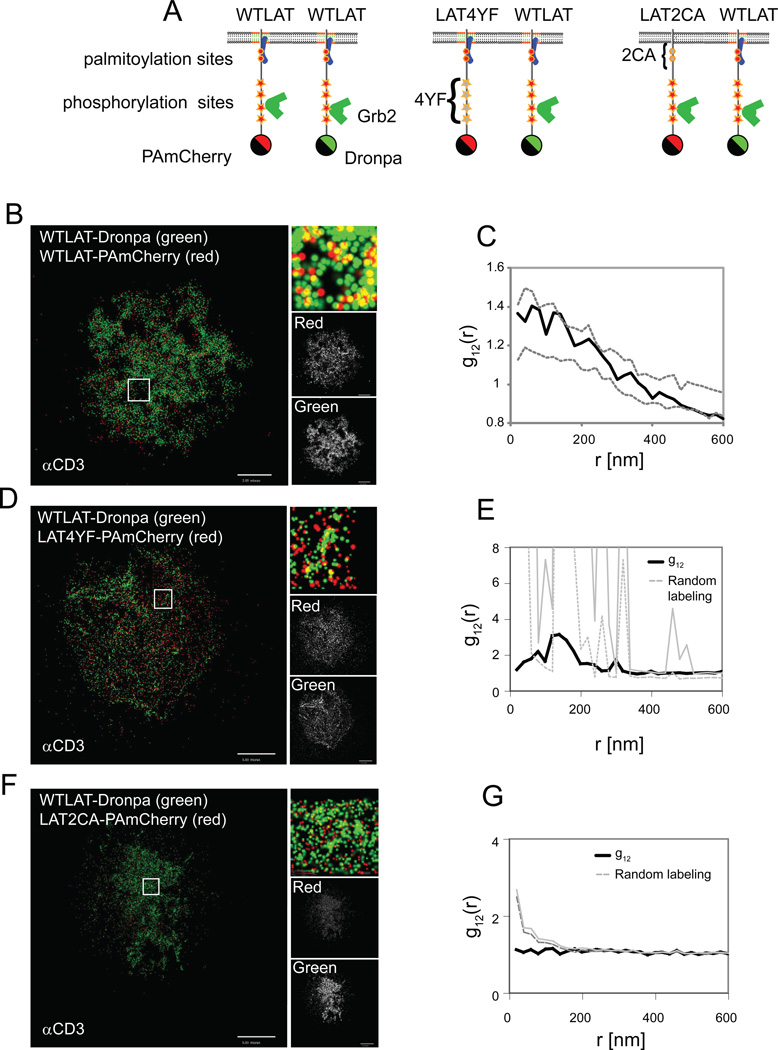
The mechanisms responsible for the formation of LAT-nucleated signaling complexes have been studied with controversial results. Early studies identified LAT as a molecule residing in lipid rafts (Wilson et al., 2004; Zhang et al., 1998) and aggregation of lipid rafts has been proposed as a mechanism for generating activation-induced signaling complexes (Harder, 2004; Zhang et al., 1998). However, other studies favored the role of an extended network of protein-protein interactions as the formation mechanism (Bunnell et al., 2002; Douglass and Vale, 2005; Houtman et al., 2006). Our discovery of a LAT nanoscale distribution led us to search for the molecular requirements for LAT nanoclustering. For this reason, we conducted two-color PALM imaging of wildtype (WT) LAT-Dronpa with various LAT constructs conjugated to PAmCherry (Figure 4A). We examined the clustering of a mutant that is incapable of phosphorylation-dependent protein-protein interactions due to the replacement of the four distal tyrosines of LAT with phenylalanines (Bunnell et al., 2006) (LAT4YF-PAmCherry) and a mutant in which the absence of two juxtamembrane cysteines blocks palmitoylation and lipid raft recruitment (Zhang et al., 1998) (LAT2CA-PAmCherry). In a cell expressing both WTLAT-Dronpa and WTLAT-PAmCherry, both molecules were incorporated into nanoclusters (Figure 4B, Figure S3A) with homogeneous mixing as determined by bivariate analysis, indicating that the two chimeras reside in the same clusters (Figure 4C, Figure S3D). In contrast, there was only a slight mixing of WTLAT and LAT4YF (Figure 4D,E, Figure S3D). Moreover, there was no interaction in samples imaged on αCD45-coated coverslips (Figure S3E,F).
Figure 4. Protein-protein and protein-lipid interactions are both required for LAT intact nanocluster formation.
(A) A cartoon showing the position of LAT mutations and their effect on LAT interactions. The 4YF mutations prevent the binding of adapter proteins such as Grb2 and the 2CA mutations prevent LAT palmitoylation and recruitment to lipid rafts. Two-color PALM images of activated Jurkat T cells expressing (B) WTLAT-Dronpa and WTLAT-PAmCherry and (C) their bivariate analyses or Jurkat cells expressing (D,E) WTLAT-Dronpa and 4YF-LAT-PAmCherry or (F,G) WTLAT-Dronpa and 2CA-LAT-PAmCherry. In each panel, the PALM images show two-color rendering of WTLAT-Dronpa (green), WT- or mutated-LAT-PAmCherry (red) and a zoomed image in the inset. The univariate PCF curves of the cells are shown in Figure S3A-C. Dashed grey lines in panels C, E and G indicate upper and lower 95% confidence levels of the random labeling model. See Analyses section in SI for further details. Color codes of PALM images (bright is highest) for individual probability density functions of single molecules, with maximal values for WTLAT-Dronpa and mutant or WTLAT-PAmCherry of – (B) 100 and 100 molecules/µm2, respectively; (D) 180 and 120 molecules/µm2; (F) 310 and 470 molecules/µm2, respectively. Bars – 5µm
Recent studies have shown that LAT palmitoylation is required for normal transport of LAT to the PM (Hundt et al., 2009). However, our TIRF-based assay allowed us to image the fraction of LAT2CA molecules that resided in the membrane. We observed complete independence (see Analyses section in SI) of WTLAT and LAT2CA (Figure 4F,G, Figure S3D). The univariate statistics also revealed interesting information. LAT4YF molecules preferred to self-associate rather than mix with WTLAT, as they showed residual nanoscale clustering by themselves under both stimulating (Figure S3B) and non-stimulating (Figure S3E) conditions. In contrast, LAT2CA molecules were randomly located at the PM with no significant self-association (Figure S3C), suggesting an essential role of the cysteine residues, perhaps by their ability to become palmitoylated, in the nucleation of LAT nanoclusters. These results together show that mutation of LAT at residues required for either protein-protein interactions or protein-lipid interactions blocked full integration into intact nanoclusters.
LAT clusters of all sizes can recruit signaling molecules and can be phosphorylated upon TCR stimulation
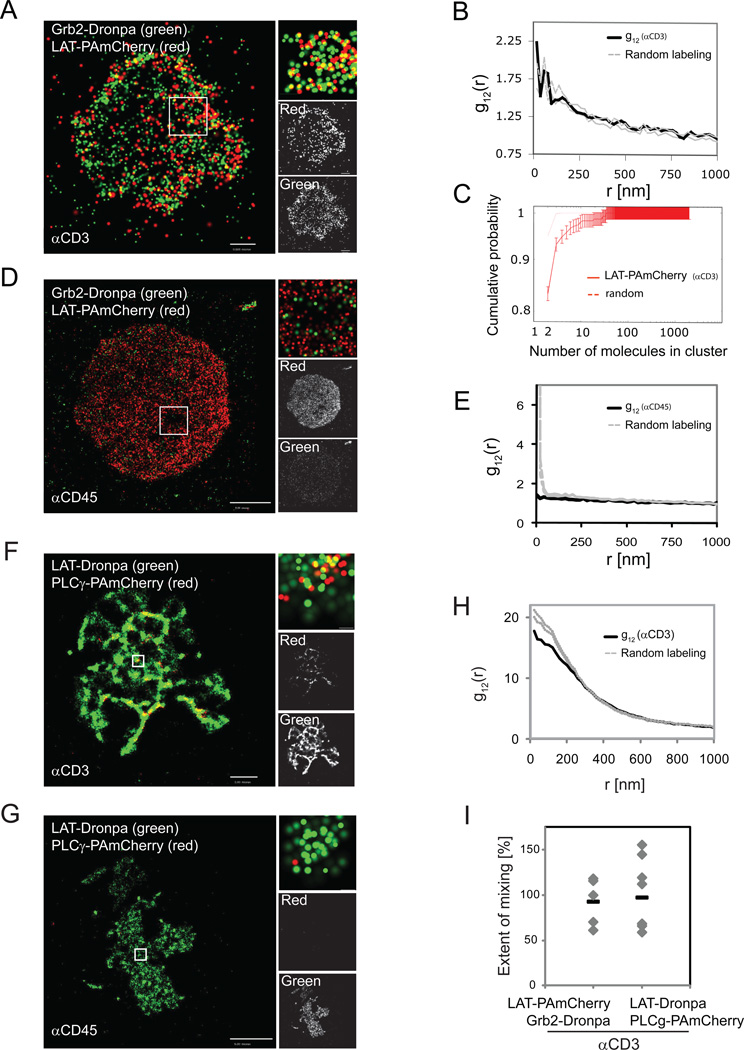
LAT nanoclusters ranged in size from a few, predominantly, to hundreds of detected molecules. The question arose whether signaling occurs only in large LAT aggregates or whether the entire distribution of LAT nanoclusters is capable of signal transduction upon TCR stimulation. To this end, we conducted two-color PALM imaging of LAT clusters and Grb2. Grb2 is a key adapter molecule that binds to any of the three LAT distal phosphotyrosines upon cell activation, and in turn recruits molecules such as the guanine nucleotide exchange factor SOS1, resulting in activation of the Ras pathway (Houtman et al., 2006) or the ubiquitin ligase Cbl, resulting in protein recycling and degradation (Balagopalan et al., 2007). In addition, the Grb2-SOS1 complex has been proposed as a mediator of LAT clustering (Houtman et al., 2006). PALM images of LAT and Grb2 showed no co-localization under non-stimulating conditions (Figure 5D,E). Grb2-Dronpa was barely seen at the PM under these conditions although it was well expressed in the cytosol, as observed by epifluorescence. However, following TCR stimulation and LAT phosphorylation , LAT and Grb2 showed high colocalization that was homogeneous with LAT cluster size (Figure 5A,B). The extent of mixing between LAT and Grb2 averaged 93±12% (Figure 5I). Because LAT cluster size distribution is dominated by very small clusters (Figure 5C), we conclude that LAT phosphorylation and downstream signaling do not require the formation of large assemblies of LAT molecules.
Figure 5. LAT clusters of all sizes can recruit Grb2 upon TCR stimulation, while PLC-γ1 is most efficiently recruited in isolated clusters.
(A) PALM of Jurkat cells expressing Grb2-Dronpa and WTLAT-PAmCherry on αCD3-coated coverslips. (B) Bivariate PCFs indicate colocalization of Grb2 and LAT on αCD3-coated coverslips (in black; random labeling model in grey, upper and lower confidence levels of 95%). (C) Cumulative probability function (n=7 cells, as detailed in Figure 1). (D) PALM of Jurkat cells expressing Grb2-Dronpa and WTLAT-PAmCherry on αCD45-coated coverslips. (E) Bivariate PCF between LAT and Grb2 on αCD45-coated coverslips (dashed grey lines defined as in panel B). (F-I) PALM analysis of Jurkat cells expressing LAT-Dronpa and PLC-γ1-PAmCherry spread on (F) αCD3- or (G) αCD45-coated coverslips. (H) Bivariate PCFs indicate colocalization of PLC-γ1 and LAT on αCD3-coated coverslips (in black; dashed grey lines defined as in panel B). (I) The extent of mixing between of LAT-PAmCherry and Grb2 Dronpa or PLC-γ1 and LAT (n=5 for LAT-PAmCherry and Grb2-Dronpa, and n=8 for LAT-Dronpa and PLCγ1-PAmCherry). Maximal probability density values (as detailed in Figure 1B) for Grb2-Dronpa and LAT-PAmCherry – (A) 640 and 260 molecules/µm2, respectively; (D) 540 and 860 molecules/µm2. Bars – (A) 0.5 µm, (D) 2 µm. Maximal probability density values (as detailed in Figure 1B) for LAT-Dronpa and PLC-γ1-PAmCherry – (F) 730 and 290 molecules/µm2, respectively; (G) 370 and 120 molecules/µm2. Bars – (F) 2 µm, (G) 5 µm.
PLC-γ1 is highly co-localized with LAT in isolated clusters
PLC-γ1 is another signaling molecule that is recruited to phosphorylated LAT upon TCR activation (Braiman et al., 2006; Sommers et al., 2002). Upon activation it cleaves phosphoinositides to products that regulate the elevation of intracellular Ca++ and activation of critical enzymes such as protein kinase C and RasGRP. As expected, PALM showed that TCR stimulation led to the recruitment of PLC-γ1 to LAT (Figure 5F) while there was no co-localization of LAT and PLC-γ1 under non-stimulating conditions (Figure 5G). Closer examination reveals that, in contrast to Grb2, PLC-γ1 was not distributed evenly with LAT, but was located in isolated clusters, within LAT-rich lamellae and microclusters (Figure 5F, inset).
LAT and PLC-γ1 maintained a high extent of co-localization, as revealed by the proximity of their bivariate correlation function to the model of random labeling (Figure 5H). The extent of mixing between the two molecules averaged 87 ±10 %. Nevertheless, several cells (4 out of 8) demonstrated bivariate correlation curves above the 95% confidence levels of the random-labeling model (Figure 5I), suggesting that PLC-γ1 molecules can preferentially become closer to LAT than expected by the model. On the other hand, the fact that some cells demonstrate an extent of mixing significantly below 100% indicates that occasionally PLC-γ1 can be recruited to the PM at sites without LAT or that its position relative to LAT is also controlled by other molecules.
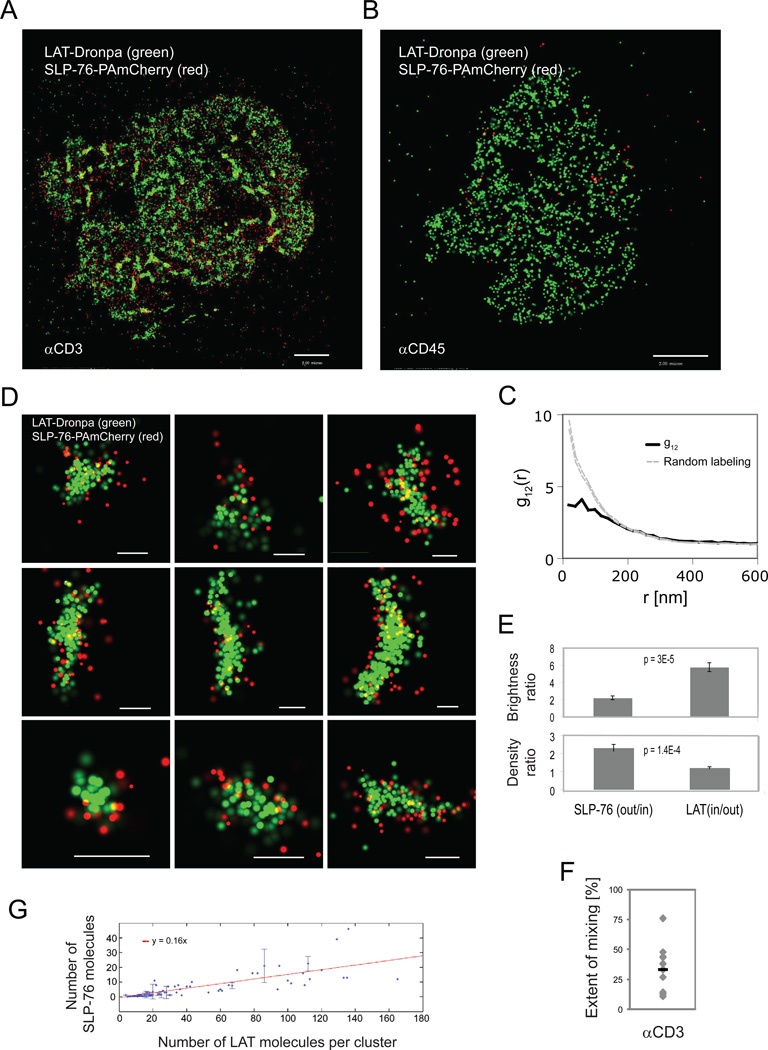
Nanoscale organization of SLP-76 is revealed at the rim of LAT clusters upon TCR stimulation
SLP-76 is a cytosolic adapter protein that is recruited to LAT phosphotyrosines via binding of the adapter, Gads (Bunnell et al., 2006). By imaging Jurkat T cells expressing LAT-Dronpa and SLP-76-PAmCherry with two-color PALM, we found that SLP-76 was efficiently recruited to the PM and to LAT nanoclusters only upon cell activation (Figure 6A,B), but in a pattern markedly different from either Grb2 or PLC-γ1. The bivariate correlation function of LAT and SLP-76 strongly departed from the homogeneous mixing model at short distances below 160nm, and agreed with this model only at longer distances (Figure 6C). Taking a closer look at LAT clusters at this short length-scale, we found that SLP-76 was enriched at the rims of LAT clusters (Figure 6D-F) with an intensity contrast (SLPout/SLPin)(LATin/LATout) of about 16-fold (Figure 6E). Thus, LAT-nucleated clusters exhibit nanoscale organization as defined by the distinctive localization of SLP-76. Importantly, a similar contrast was found by two-color PALM imaging of LAT and SLP-76 with reversed colors, i.e. SLP-76-Dronpa and LAT-PAmCherry (data not shown). The extent of mixing between the two molecules averaged at 33 ±7 % (Figure 6F), further showing partial molecular segregation and indicating that most of the SLP-76 molecules lie outside of LAT microclusters. Finally, we used the clustering algorithm to demonstrate that LAT clusters recruit SLP-76 with a linear efficiency as a function of LAT cluster size (Figure 6G). However, we could not determine whether SLP-76 molecules arrange at the periphery of the predominant and smallest LAT clusters as the number of SLP-76 molecules that were associated with these clusters was too small. Two-color PALM imaging of SLP-76 and TCRζ showed no mixing of these two molecules, thus demonstrating that the nanoscale structure observed between LAT and SLP-76 is specific to them (Figure S4A). Additionally, by imaging either Grb2-Dronpa or SLP-76-Dronpa with LAT-2CA-PAmCherry, we found that LAT-2CA did not mix with these proteins (Figure S4B). Since LAT-2CA did not incorporate into intact LAT clusters (Figure 4F,G), this observation shows that phosphorylation-dependent interactions of LAT require localization within intact clusters at the PM.
Figure 6. Nano-scale organization of SLP-76 is revealed at the rim of LAT clusters upon TCR stimulation.
Two-color PALM images of a LAT-Dronpa and SLP-76-PAmCherry expressing Jurkat T cell (A) on αCD3- (B) αCD45-coated coverslips and (C) Bivariate correlation curve (dashed grey lines indicate 95% confidence levels of a random labeling model). (D) Zoomed images of individual LAT clusters showing preferential organization of SLP-76 at the rims of LAT clusters. Bars – 200nm. (E) Top - ratio of LAT and SLP-76 brightness in the rim vs. inner part of the cluster, as identified by image processing (n=10 clusters). Bottom - density ratios of LAT and SLP-76 as calculated by dividing the brightness measurements with the matching areas of the rim and inner part of the cluster (n=10 clusters). (F) The extent of mixing between LAT and SLP-76 (n=10 cells). (G) SLP-76 molecules were associated with LAT clusters using the clustering algorithm (see Analyses section in SI and Figure S1M,N for further details). The number of SLP-76 molecules in LAT clusters is plotted as a function of LAT cluster size (in copy number). A linear fit indicates linear efficiency of recruiting SLP-76 molecules to LAT clusters as a function of LAT cluster size. Maximal probability density values (as detailed in Figure 1B) for LAT-Dronpa and SLP-76-PAmCherry – (A) 310 and 310 molecules/µm2, respectively; (D) 320 and 310 molecules/µm2. Bars – 2 µm.
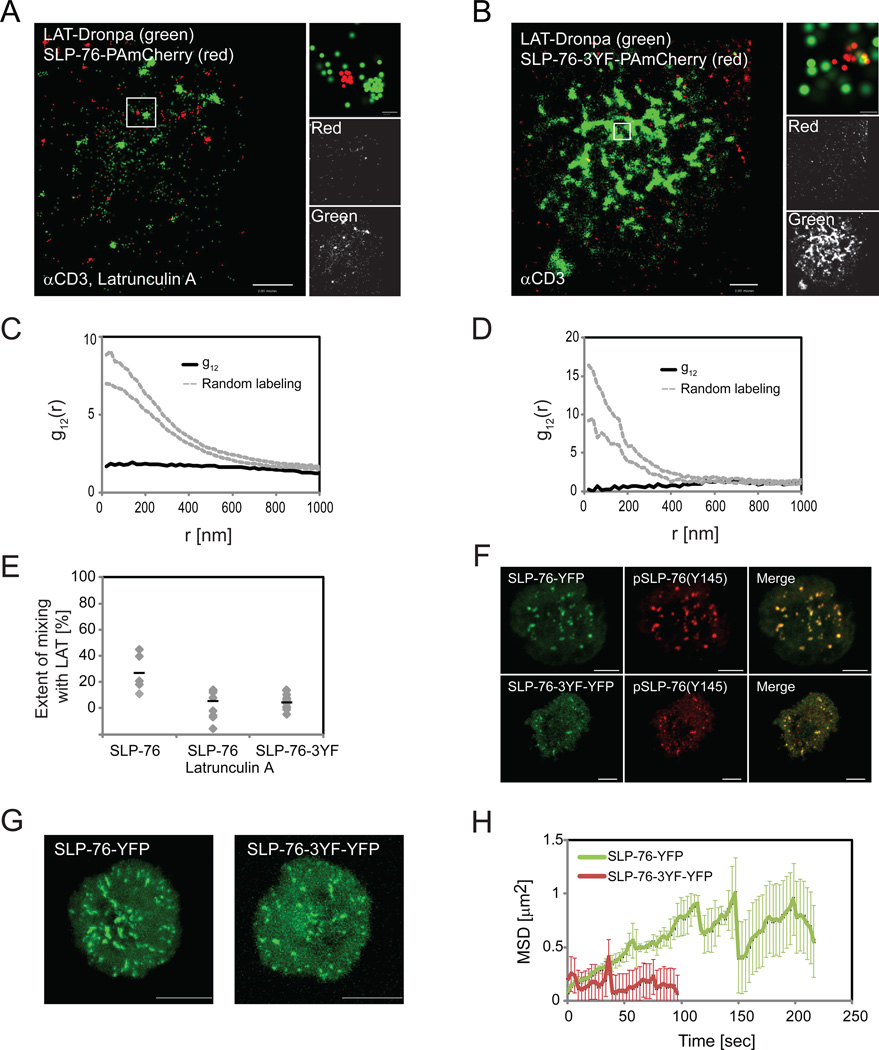
The nanoscale organization of SLP-76 depends on actin polymerization and facilitates SLP-76 cluster dynamics
SLP-76 links LAT clusters to the process of actin polymerization via interactions with the effector proteins Nck, Vav-1 and Wasp (Barda-Saad et al., 2010; Bubeck Wardenburg et al., 1998). To study the possible relationship between the nanoscale organization of SLP-76 and LAT with actin, we disrupted SLP-76 and actin interactions in two different ways. First, we treated Jurkat E6.1 cells with Latrunculin A, a drug that inhibits actin polymerization (Morton et al., 2000). Second, we mutated three tyrosines on SLP76, namely Y113, 128 and 145, to phenylalanines, thus blocking both phosphorylation of these sites and binding of Nck, Vav-1 and the tyrosine kinase Itk (Bunnell et al., 2006). We observed that in both experiments, SLP-76 failed to organize at the periphery of LAT clusters (Figure 7A,B) and the two molecules showed a very low extent of mixing (Figure 7C,D,E) although univariate analysis showed that LAT and SLP-76 could still cluster individually at the PM (Figure S5A,B). Interestingly, treatment with Latrunculin significantly affected LAT clustering (Figure S5B) but not clustering of SLP-76 (Figure S5A). Thus, actin polymerization is required for establishment of the LAT-SLP-76 nanostructure.
Figure 7. Nano-scale organization of SLP-76 depends on actin and facilitates regulation of SLP-76 clusters.
Two-color PALM images of (A) a LAT-Dronpa and SLP-76-PAmCherry-expressing Jurkat T cells treated with 300nM Latrunculin A and (B) a LAT-Dronpa and SLP-76-3YF-PAmCherry-expressing Jurkat T cells. (A,B) cells were imaged on αCD3-coated coverslips. (C,D) The bivariate correlation curves related to cells in panels A and B in cells treated with (C) Latrunculin A or (D) between LAT and the SLP76-3YF mutant (Dashed grey lines indicate 95% confidence level of a random labeling model). (E) The extent of mixing derived from imaging multiple cells as in panel A (n=8) and panel B (n=10) and cells expressing LAT-Dronpa and SLP-76-PAmCherry (n=5). (F,G,H) Jurkat T cells, transiently expressing SLP-76-YFP or SLP-76-3YF-YFP, were imaged by confocal microscopy as they spread on αCD3-coated coverslips. (F) Confocal images of fixed cells expressing SLP-76-YFP or SLP-76-3YF-YFP and stained with αphosphoSLP-76(Y145). (G) Images are shown summing all frames from Maximal intensity projection (MIP) movies (see related Movies S1 and S2). These images describe SLP-76 cluster movement across the plasma membrane for SLP-76 or SLP-76-3YF. (H) Individual clusters containing SLP-76 or SLP-76-3YF were identified and tracked in the MIP movies using SlideBook (3i) as described in the methods section. An average of the mean squared displacement (MSD) over time was calculated for multiple trajectories from multiple cells (SLP-76, n=2496 from 19 cells; SLP-76-3YF, n=233 from 10 cells as a significantly lower number of clusters could be detected for the SLP-76-3YF expressing cells). Maximal probability density values (as detailed in Figure 1B) for LAT-Dronpa and SLP-76-PAmCherry – (A) 400 and 240 molecules/µm2, respectively; (B) 640 and 520 molecules/µm2. Bars – 2 µm.
Abrogating SLP-76 cluster dynamics, and in particular, blocking SLP-76 internalization or decreasing its persistence in clusters has been correlated with impaired TCR-dependent cell function, including defects in TCR induced Ca++ flux, CD69 upregulation, and nuclear factor of activated T cells (NF-AT) activation (Barr et al., 2006; Bunnell et al., 2006). To study the functional role of the nanoscale organization of SLP-76 and LAT we tracked SLP-76-3YF clusters at the PM of cells and compared their dynamics to the dynamics of WTSLP-76 clusters by confocal microscopy. By imaging SLP-76-3YF-YFP together with immunofluorescence staining for pSLP-76 on Y145, we also found that WTSLP-76 and the SLP-76-3YF mutant co-localized to the same clusters (Figure 7F). Strikingly, clusters containing the SLP-76-3YF mutant were less mobile and less persistent than clusters with only WTSLP-76 clusters (Figure 7G,H and supplement movies M1 and M2). By univariate analysis we found a small increase in clustering of SLP-76-3YF compared to WTSLP-76 (Figure S5C), and an insignificant decrease in LAT clustering (Figure S5D). These data indicate that the SLP-76-3YF mutant has a dominant negative effect and could abrogate both the intact nanostructure of LAT and SLP-76 as well as the dynamics of the clusters in spite of the presence of WTSLP-76 in these clusters. Thus, in combination with previous studies (Barr et al., 2006; Bunnell et al., 2006), our results strongly suggest that LAT-SLP-76 nanostructure is required for intact TCR-dependent cellular function.
Discussion
Using PALM imaging of Jurkat, and PBT, and live and fixed cells together with extensive spatial analyses, we observed a continuous size distribution of LAT in nanoclusters. Most nanoclusters were comprised of a few molecules each and accounted for the majority of LAT molecules at the PM, either within or outside microclusters. Thus, we set out to study the formation mechanism, role in signaling, and organization of upstream and downstream molecules of the newly identified LAT nanoclusters.
Two-color PALM imaging and bivariate correlation analyses enabled us to resolve complex nanoscale relationships between several different proteins. We found that LAT is clustered within lamellae or membrane folds that first engage the activating surface of the coverslip. Indeed, previous scanning electron micrograph (SEM) images of the interface between T cells and antigen presenting cells (APCs) have captured similar protrusions that mediate early contact between the cells (Gomez et al., 2007). This complexity of contact morphology gives rise to apparent first order clustering, which has not been previously appreciated (Lillemeier et al., 2010; Lillemeier et al., 2006; Wilson et al., 2001). The clustering of LAT and other proteins of interest discussed throughout this study is thus considered a second-order clustering mechanism observed at the sites of first cellular contact with the activating surface.
We further defined the role of critical LAT amino acids that control protein-protein interactions and protein-lipid interactions in the formation of LAT nanoclusters. Recent studies have emphasized the role of protein-protein interactions in LAT clustering leaving the transport of LAT to the cell surface as the sole function of protein-lipid interactions (Hundt et al., 2009). We show here the complementary role of both protein-protein and protein-lipid interactions in the sorting of LAT into intact LAT clusters at the PM. As expected, mutations that block cluster formation also block interactions of downstream signaling molecules. We propose that the full size distribution of LAT is dependent on intact nanocluster assembly since the mutations we studied also fail to form microclusters (Bunnell et al., 2006; Hundt et al., 2009). Finally, the self-association of LAT4YF mutants or of TCRζ suggests the ability of different molecules to sort into separate nanoclusters in the PM.
Previous studies of TCR and LAT distribution using PALM and electron microscopy (EM) have suggested that TCR and LAT form domains ('protein islands') at the PM with larger sizes than the smallest nanoclusters reported here (Lillemeier et al., 2010; Lillemeier et al., 2006; Wilson et al., 2001). This interpretation is not supported by our results. We note differences in how signal intensity was used to threshold and thus exclude molecules in these other studies. The choice of statistical techniques also resulted in differences in scoring signaling clusters. Additionally, we believe that consideration of the heterogeneity of membrane contact with the coverslip has consequences for the analysis. These important differences are further elaborated in the Analyses part of the Methods section of the SI. We conclude that, in several ways, the previous analyses made it difficult to recognize the dominance of very small clusters (namely dimers, trimers, etc.) over the cluster size distributions, while overestimating the importance of considerably larger clusters. Other studies (Purbhoo et al., 2010; Williamson et al., 2011) focus on the presence of LAT in large vesicular structures but we note that the timing of activation in these studies (10 min) is far longer than in ours. We focussed on the basal state of molecules and their initial changes within seconds to a few minutes of activation, when phosphorylation of LAT and its binding partners peak.
Analysis of the PM distribution of the TCR using biochemical and EM techniques has shown a range of size distribution down to the nanoscale much as we now see with LAT and as we confirm for the TCRζ chain under non-stimulating conditions (Schamel et al., 2005). The relationship of immunuoreceptor (TCR and FcRε) clusters to LAT clusters has been studied extensively using immuno-EM (Lillemeier et al., 2010; Lillemeier et al., 2006; Wilson et al., 2001). In contrast to our observation about the TCR and LAT, these studies showed that before cell activation LAT and the two immunoreceptors exist in segregated domains that come together upon cell activation without mixing. However, multiple concerns have been raised regarding the interpretation of immuno-EM results in molecular clustering (D'Amico and Skarmoutsou, 2008). These concerns include the possibility of false clustering of nano-gold particles due to the attachment of multiple primary or secondary antibodies to single target molecules, the steric hindrance by one type of nano-gold label on another in experiments that use multiple immuno-gold labels, and alterations of molecular distributions and PM structure due to procedures of sample preparation such as fixation with glutaraldehyde, sample drying and membrane ripping. PALM microscopy of intact cells has been suggested as a way to overcome these caveats.
Using two-color PALM, we also observed separate pools of LAT and TCR. However, in contrast to previous studies, we identified mixed pools of LAT and TCR molecules prior to and then following cell activation. Importantly, these areas of overlap and mixture have functional significance, as they are likely to be the most efficient locations for the phosphorylation of LAT by activated ZAP-70. We further addressed this finding by imaging ZAP-70 with either TCRζ or LAT. Indeed, TCRζ and ZAP-70 showed very high co-localization consistent with the binding of ZAP-70 to phosphorylated TCRζ. LAT and ZAP-70, in contrast, were in spatially separated pools with partial mixing similar to the spatial separation of TCRζ and LAT. Thus we suggest that these areas in which LAT overlaps with TCRζ and activated ZAP-70 act as critical “hot spots” for LAT phosphorylation by ZAP-70 bound to TCRζ or in near proximity to it. These sites may very well change dynamically with time and might also serve as the location where protein binding to LAT occurs. They thus may be the sites of signal relay and amplification. It should be mentioned that in our imaging assay LAT can freely diffuse at the PM of T cells and ZAP-70 can freely diffuse in the cytosol and bind or detach from TCRζ (Douglass and Vale, 2005; Sloan-Lancaster et al., 1998). TCRζ could be potentially mobile with partial independence from the rest of the TCR chains (Ono et al., 1995), although the CD3 chains of the TCR molecules at the PM are immobilized on the αCD3-coated coverslip. Thus, our findings regarding the nanoscale organization of TCRζ, ZAP-70 and LAT relative to each other directly relate to physiologic function.
We further showed that LAT nanoclusters were viable for signaling by demonstrating that the downstream adapter Grb2 mixed with LAT in a homogeneous manner. Since the distribution of LAT is dominated by clusters as small as dimers and trimers, Grb2 must associate with the very smallest of nanoclusters upon T cell activation. This conclusion contrasts with previous studies that emphasized the role of large microcluster formation for efficient LAT signaling (Lillemeier et al., 2010). If LAT becomes activated at ‘hot spots’ proximal to TCR clusters, one might wonder how activated LAT (marked here by Grb2 recruitment) homogeneously distributes between LAT clusters. An obvious explanation is that the high mobility of LAT, which has been shown at the PM of both non-activated and activated T cells (Douglass and Vale, 2005) leads to the rapid exchange of LAT molecules between clusters of all sizes. The enzyme PLC-γ1, whose activity downstream of LAT is crucial to T cell activation, is also efficiently incorporated into LAT clusters. However, PLC-γ1 seemed to get recruited into isolated clusters without mixing with all of the LAT-rich domains at the PM. The reason for this preferential clustering might relate to the cooperative binding of PLC-γ1 to LAT with molecules such as SLP-76 and Gads (Braiman et al., 2006; Houtman et al., 2006); however, a detailed account of PLC-γ1 organization awaits further study.
Surprisingly, using two-color PALM imaging we found a nanoscale organization of SLP-76 at the rim of LAT microclusters. To our knowledge, this is the first observation of multi-molecular nanoscale organization in signaling complexes. Previous studies have suggested a role of the cytoskeleton in the organization of signaling molecules at the PM (Harwood and Batista, 2010a). Indeed, we further showed that the nanoscale organization of LAT and SLP-76 depended on polymerized actin, since the organization was disrupted by perturbations that affected the link between SLP-76 and actin. Finally, we showed that abrogating nanoscale organization of LAT and SLP-76 correlated with impaired mobility of SLP-76 microclusters at the cell surface. Based on these findings and previous work (Barr et al., 2006; Bunnell et al., 2006), we conclude that the nanoscale organization LAT and SLP-76 is required for intact TCR-dependent cell functions. We predict that other signaling systems at the PM are organized in nanoclusters and expect that further super-resolution studies will lead to important insights into the structure and function of these critical signaling aggregates.
To conclude, using state-of-the art imaging techniques, we were able to show that LAT resides in nanoclusters at the PM under non-stimulating conditions and observe a modest growth in cluster size upon cell stimulation. We also showed unexpected results regarding the formation mechanism, functional role and structure of the newly identified LAT nanoclusters. Some of these observations include: (i) the distribution of the TCR and LAT at the PM demonstrating mixing in the basal state with a decrease in mixing following T cell activation, (ii) the distribution of ZAP-70 compared to that of TCRζ and LAT suggests that the sites of partial mixing of the latter two molecules are where ZAP-70 phosphorylates LAT, (iii) the complementary role of protein-protein and protein-lipid interactions in LAT nanocluster formation, (iv) the ability of LAT nanoclusters, as small as dimers, to signal without large-scale aggregation, (v) the colocalization of PLC-γ1 with LAT in isolated clusters, (vi) the localization of SLP-76 to the periphery of clusters indicating that these nanoclusters have structure, and (vii) the dependence of this structure on actin and the relationship of LAT-SLP-76 nanostructure to cluster dynamics. Our observations show the complexity of organization of signaling complexes and the mechanisms that shape their arrangement at the PM at the level of single molecules. These findings greatly extend our understanding of signal transduction at the PM. In addition, these observations can serve as a model for many signaling processes that occur at the PM and other cellular compartments.
Methods
Sample preparation
Fluorescent conjugates of proteins of interest and the photoactivatable proteins Dronpa or PAmCherry were created by the replacement of previously tagged constructs by digestion and ligation of new inserts. DNA was introduced into E6.1 Jurkat T cells or PBTs, isolated from healthy donors (Barda-Saad et al., 2005), using the LONZA nucleofector shuttle system. Transiently transfected cells were sorted for positive expression of PAmCherry or Dronpa chimeras and imaged within 24 hours (PBTs) or 48-72 hours (Jurkats) from transfection or used to establish stable cell lines (Jurkats). Sorting was performed by the NCI flow cytometry core facility.
The preparation of coverslips for imaging spread cells followed a previously described technique (Bunnell et al., 2003) and as further described in the SI. Briefly, sorted cells were dropped on glass coverslips coated with 0.01% poly-L-lysine (Sigma) and either stimulatory or non-stimulatory antibodies at a concentration of 10 µg/ml (unless specified otherwise). Cells were resuspended in imaging buffer and dropped onto the coated coverslips, incubated at 37°C for the specific spreading time (typically 3 min) and fixed with 2.4% PFA for 30 min at 37°C.
Imaging
Confocal images were taken with a 510 LSCM confocal microscope using a 63X, 1.4 NA objective (Zeiss). PALM imaging was performed on a previously described homemade TIRF microscope (Betzig et al., 2006) and on a second system based on a TIRF microscope (Nikon). For live cell PALM imaging, the set-up was modified for the delivery of high excitation power (x16 times the power flux used for fixed cell imaging) and fast readout (at 800 frame/sec), as described in the SI. PALM images were analyzed using a previously described algorithm to identify peaks and group them into functions that reflect the positions of single molecules (Betzig et al., 2006). The acquisition sequence typically consisted of 5-10 min of PALM image acquisition in a single channel or 15-20 min in two channels, or 5min at 800 frame/sec for live cell PALM imaging. Custom algorithms were then applied to further study the position map of the identified molecules, as detailed in the SI.
To image molecules with two-color PALM, we created chimeric constructs of molecules with Dronpa or PAmCherry. We made use of the ~50-fold difference in activation intensities between Dronpa and PAmCherry to image them sequentially. We first activated Dronpa with ~0.3mW of 405nm illumination. After the Dronpa population was completely bleached, we switched detection channels and activated PAmCherry with up to 10mW of 405nm light. Fiduciary markers (0.1 μm gold particles or TetraSpec microspheres; Invitrogen) and affine transformations were used to register images taken in the two channels.
Further information regarding materials, methods and algorithms can be found in the SI.
Supplementary Material
Acknowledgments
The authors would like to thank B. J. Taylor at the NCI Flow Cytometry Core Facility, Zeiss, H. Hess (HHMI, Janelia Farm) for providing the PALM software, W. Losert (University of Maryland) for multiple discussions on data analyses and T. Wiegand (Helmholtz Centre for Environmental Research - UFZ) for providing us his point-pattern analyses software. We thank P. Sengupta, M. Renz and T. Jovanovic-Talisman (NIH/NICHD) for reading the manuscript. This research was supported by the Intramural Research Programs of the National Cancer Institute (The Center for Cancer Research) and of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- Balagopalan L, Barr VA, Sommers CL, Barda-Saad M, Goyal A, Isakowitz MS, Samelson LE. c-Cbl-mediated regulation of LAT-nucleated signaling complexes. Molecular and Cellular Biology. 2007;27:8622–8636. doi: 10.1128/MCB.00467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopalan L, Coussens NP, Sherman E, Samelson LE, Sommers CL. The LAT story: a tale of cooperativity, coordination, and choreography. Cold Spring Harb Perspect Biol. 2010;2:a005512. doi: 10.1101/cshperspect.a005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barda-Saad M, Braiman A, Titerence R, Bunnell SC, Barr VA, Samelson LE. Dynamic molecular interactions linking the T cell antigen receptor to the actin cytoskeleton. Nature Immunology. 2005;6:80–89. doi: 10.1038/ni1143. [DOI] [PubMed] [Google Scholar]
- Barda-Saad M, Shirasu N, Pauker MH, Hassan N, Perl O, Balbo A, Yamaguchi H, Houtman JC, Appella E, Schuck P, et al. Cooperativ interactions at the SLP-76 complex are critical for actin polymerization. EMBO J. 2010;29:2315–2328. doi: 10.1038/emboj.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr VA, Balagopalan L, Barda-Saad M, Polishchuk R, Boukari H, Bunnell SC, Bernot KM, Toda Y, Nossal R, Samelson LE. T-cell antigen receptor-induced signaling complexes: Internalization via a cholesterol-dependent endocytic pathway. Traffic. 2006;7:1143–1162. doi: 10.1111/j.1600-0854.2006.00464.x. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Braiman A, Barda-Saad M, Sommers CL, Samelson LE. Recruitment and activation of PLC gamma 1 in T cells: a new insight into old domains. Embo Journal. 2006;25:774–784. doi: 10.1038/sj.emboj.7600978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck Wardenburg J, Pappu R, Bu JY, Mayer B, Chernoff J, Straus D, Chan AC. Regulation of PAK activation and the T cell cytoskeleton by the linker protein SLP-76. Immunity. 1998;9:607–616. doi: 10.1016/s1074-7613(00)80658-5. [DOI] [PubMed] [Google Scholar]
- Bunnell SC, Barr VA, Fuller CL, Samelson LE. High-Resolution Multicolor Imaging of Dynamic Signaling Complexes in T Cells Stimulated by Planar Substrates. Sci STKE. 2003:P18. doi: 10.1126/stke.2003.177.pl8. [DOI] [PubMed] [Google Scholar]
- Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, Samelson LE. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. Journal of Cell Biology. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnell SC, Singer AL, Hong DI, Jacque BH, Jordan MS, Seminario MC, Barr VA, Koretzky GA, Samelson LE. Persistence of cooperatively stabilized signaling clusters drives T-cell activation. Molecular and Cellular Biology. 2006;26:7155–7166. doi: 10.1128/MCB.00507-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campi G, Varma R, Dustin ML. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J Exp Med. 2005;202:1031–1036. doi: 10.1084/jem.20051182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebecauer M, Spitaler M, Serge A, Magee AI. Signalling complexes and clusters: functional advantages and methodological hurdles. Journal of Cell Science. 2010;123:309–320. doi: 10.1242/jcs.061739. [DOI] [PubMed] [Google Scholar]
- D'Amico F, Skarmoutsou E. Quantifying immunogold labelling in transmission electron microscopy. J Microsc. 2008;230:9–15. doi: 10.1111/j.1365-2818.2008.01949.x. [DOI] [PubMed] [Google Scholar]
- Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Kumar K, Mecleiros RB, Shimizu Y, Leibson PJ, Billadeau DD. Formins regulate the actin-related protein 2/3 complex-independent polarization of the centrosome to the immunological synapse. Immunity. 2007;26:177–190. doi: 10.1016/j.immuni.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder T. Lipid raft domains and protein networks in T-cell receptor signal transduction. Current Opinion in Immunology. 2004;16:353–359. doi: 10.1016/j.coi.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Harding A, Hancock JF. Ras nanoclusters: Combining digital and analog signaling. Cell Cycle. 2008;7:127–134. doi: 10.4161/cc.7.2.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood NE, Batista FD. The Cytoskeleton Coordinates the Early Events of B-cell Activation. Cold Spring Harb Perspect Biol. 2010a doi: 10.1101/cshperspect.a002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood NE, Batista FD. Early events in B cell activation. Annu Rev Immunol. 2010b;28:185–210. doi: 10.1146/annurev-immunol-030409-101216. [DOI] [PubMed] [Google Scholar]
- Houtman JC, Yamaguchi H, Barda-Saad M, Braiman A, Bowden B, Appella E, Schuck P, Samelson LE. Oligomerization of signaling complexes by the multipoint binding of GRB2 to both LAT and SOS1. Nat Struct Mol Biol. 2006;13:798–805. doi: 10.1038/nsmb1133. [DOI] [PubMed] [Google Scholar]
- Hundt M, Harada Y, De Giorgio L, Tanimura N, Zhang W, Altman A. Palmitoylation-dependent plasma membrane transport but lipid raft-independent signaling by linker for activation of T cells. J Immunol. 2009;183:1685–1694. doi: 10.4049/jimmunol.0803921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci U S A. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton WM, Ayscough KR, McLaughlin PJ. Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat Cell Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- Ono S, Ohno H, Saito T. Rapid Turnover of the Cd3-Zeta Chain Independent of the Tcr-Cd3 Complex in Normal T-Cells. Immunity. 1995;2:639–644. doi: 10.1016/1074-7613(95)90008-x. [DOI] [PubMed] [Google Scholar]
- Purbhoo MA, Liu HB, Oddos S, Owen DM, Neil MAA, Pageon SV, French PMW, Rudd CE, Davis DM. Dynamics of Subsynaptic Vesicles and Surface Microclusters at the Immunological Synapse. Science Signaling. 2010;3 doi: 10.1126/scisignal.2000645. [DOI] [PubMed] [Google Scholar]
- Schamel WWA, Arechaga I, Risueno RM, van Santen HM, Cabezas P, Risco C, Valpuesta JM, Alarcon B. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J Exp Med. 2005;202:493–503. doi: 10.1084/jem.20042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Sloan-Lancaster J, Presley J, Ellenberg J, Yamazaki T, Lippincott-Schwartz J, Samelson LE. ZAP-70 association with T cell receptor zeta (TCR zeta): Fluorescence imaging of dynamic changes upon cellular stimulation. Journal of Cell Biology. 1998;143:613–624. doi: 10.1083/jcb.143.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommers CL, Park CS, Lee J, Feng CG, Fuller CL, Grinberg A, Hildebrand JA, Lacana E, Menon RK, Shores EW, et al. A LAT mutation that inhibits T cell development yet induces lymphoproliferation. Science. 2002;296:2040–2043. doi: 10.1126/science.1069066. [DOI] [PubMed] [Google Scholar]
- Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy (vol 6, pg 153, 2009) Nature Methods. 2009;6:311–311. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kadlecek TA, Au-Yeung BB, Goodfellow HE, Hsu LY, Freedman TS, Weiss A. ZAP-70: an essential kinase in T-cell signaling. Cold Spring Harb Perspect Biol. 2010;2:a002279. doi: 10.1101/cshperspect.a002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson DJ, Owen DM, Rossy J, Magenau A, Wehrmann M, Gooding JJ, Gaus K. Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nature Immunology. 2011;12:655–662. doi: 10.1038/ni.2049. [DOI] [PubMed] [Google Scholar]
- Wilson BS, Pfeiffer JR, Surviladze Z, Gaudet EA, Oliver JM. High resolution mapping of mast cell membranes reveals primary and secondary domains of Fc epsilon RI and LAT. Journal of Cell Biology. 2001;154:645–658. doi: 10.1083/jcb.200104049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BS, Steinberg SL, Liederman K, Pfeiffer JR, Surviladze Z, Zhang J, Samelson LE, Yang LH, Kotula PG, Oliver JM. Markers for detergent-resistant lipid rafts occupy distinct and dynamic domains in native membranes. Molecular Biology of the Cell. 2004;15:2580–2592. doi: 10.1091/mbc.E03-08-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T, Sakata-Sogawa K, Kobayashi W, Hiroshima M, Hashimoto-Tane A, Tokunaga M, Dustin ML, Saito T. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat Immunol. 2005;6:1253–1262. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- Zhang WG, Trible RP, Samelson LE. LAT palmitoylation: Its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity. 1998;9:239–246. doi: 10.1016/s1074-7613(00)80606-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.