Summary
Nickel is toxic to all forms of life, but the mechanisms of cell damage are unknown. Indeed, environmentally relevant nickel levels (8 µM) inhibit wild-type Escherichia coli growth on glucose minimal medium. The same concentration of nickel also inhibits growth on fructose, but not succinate, lactate, or glycerol; these results suggest that fructose-1,6-bisphosphate aldolase (FbaA) is a target of nickel toxicity. Cells stressed by 8 µM Ni(II) for 20 min lost 75% of their FbaA activity, demonstrating that FbaA is inactivated during nickel stress. Furthermore, over-expression of fbaA restored growth in glucose minimal medium supplemented with 8 µM Ni(II), thus confirming that FbaA is a primary target of nickel toxicity. This class II aldolase has an active site zinc and a non-catalytic zinc nearby. Purified FbaA lost 80 % of its activity within 2 min when challenged with 8 µM Ni(II). Nickel-challenged FbaA lost 1.3 zinc and gained 0.8 nickel per inactivated monomer. FbaA mutants (D144A and E174A) affecting the non-catalytic zinc were resistant to nickel inhibition. These results define the primary site of nickel toxicity in E. coli as the class II aldolase FbaA through binding to the non-catalytic zinc site.
Keywords: metal substitution, metalloprotein, glycolysis, allosteric regulation, glutathione
Introduction
Nickel is toxic to all forms of life. In humans, inhalation of nickel particulates has been linked to carcinomas while exposure to elemental nickel is responsible for dermatological allergy (Barceloux, 1999; Das et al., 2008; Kasprzak et al., 2003). Elevated concentrations of nickel in the environment occur either naturally in metalliferous soils or through industrial pollution. Nickel-resistant plants sequester the metal as a protective mechanism, leading to nickel hyperaccumulation (Yusuf et al., 2011). By contrast, nickel-resistant bacteria, such as Cupriavidus metallidurans and Alcaligenes xylosoxidans, survive nickel-contaminated environments because they contain plasmid-encoded efflux pumps (Liesegang et al., 1993; Schmidt and Schlegel, 1994). How nickel poisons cells is poorly understood, with several mechanisms of nickel toxicity hypothesized [reviewed in (Macomber and Hausinger, 2011)]: 1) nickel substitutes for a required metal in a metalloprotein (Chen et al., 1984; Kalliri et al., 2005; Niemirowicz et al., 2007); 2) nickel binds to catalytic residues of non-metalloenzymes (Ghosh et al., 1999; Louwrier and Knowles, 1996; Meredith and Woodard, 2003); 3) nickel binds to amino acid side chains separate from the active site and acts as an allosteric inhibitor (Sato et al., 1998; Seffernick et al., 2002); or 4) nickel promotes the production of reactive oxygen species (Geslin et al., 2001; Shi et al., 1993).
Although toxic at high concentrations, nickel also is required by many microorganisms where it serves as a cofactor in urease, [NiFe] hydrogenase, glyoxylase, and several other enzymes (Kaluarachchi et al., 2010; Li and Zamble, 2009; Mulrooney and Hausinger, 2003). To assure adequate provision of this metal, cells often contain nickel specific uptake systems that may include ATP-binding cassette systems (e.g., NikABCDE) (Li and Zamble, 2009; Navarro et al., 1993) or members of the nickel/cobalt transporter (NiCoT) family (Eberz et al., 1989; Eitinger et al., 2005; Li and Zamble, 2009). In organisms requiring nickel, the binding affinities for the regulators of nickel uptake and efflux are poised to maintain a nickel concentration that allows enzyme metallation but prevents toxicity (Iwig et al., 2006). Nickel also can enter cells via non-specific pathways such as using the CorA magnesium importer and these nonspecific transporters are likely the route of nickel uptake in metal replete conditions (Navarro et al., 1993; Niegowski and Eshaghi, 2007).
This study elucidates the primary mechanism of nickel toxicity in Escherichia coli. Like many other bacteria, this microorganism contains a chromosomally-encoded nickel-efflux pump (RcnA) that transports Ni(II) and Co(II) from the cytoplasm into the periplasm, presumably driven by the proton gradient (Rodrigue et al., 2005). Elimination of rcnA increases the sensitivity of E. coli to nickel-dependent growth inhibition and leads to the accumulation of cellular nickel (Rodrigue et al., 2005). RcnA is regulated by the nickel-binding transcriptional repressor RcnR (Iwig et al., 2006; Iwig et al., 2008), thus allowing for nickel homeostasis. The presence of a chromosomally-encoded RcnA/RcnR nickel defense system in E. coli suggests that this microorganism routinely encounters toxic levels of this metal. Here, the major site of nickel damage in E. coli is shown to be an allosteric binding site of the class II fructose-1,6-bisphosphate aldolase, FbaA.
Results
Nickel is toxic to E. coli at environmentally relevant concentrations
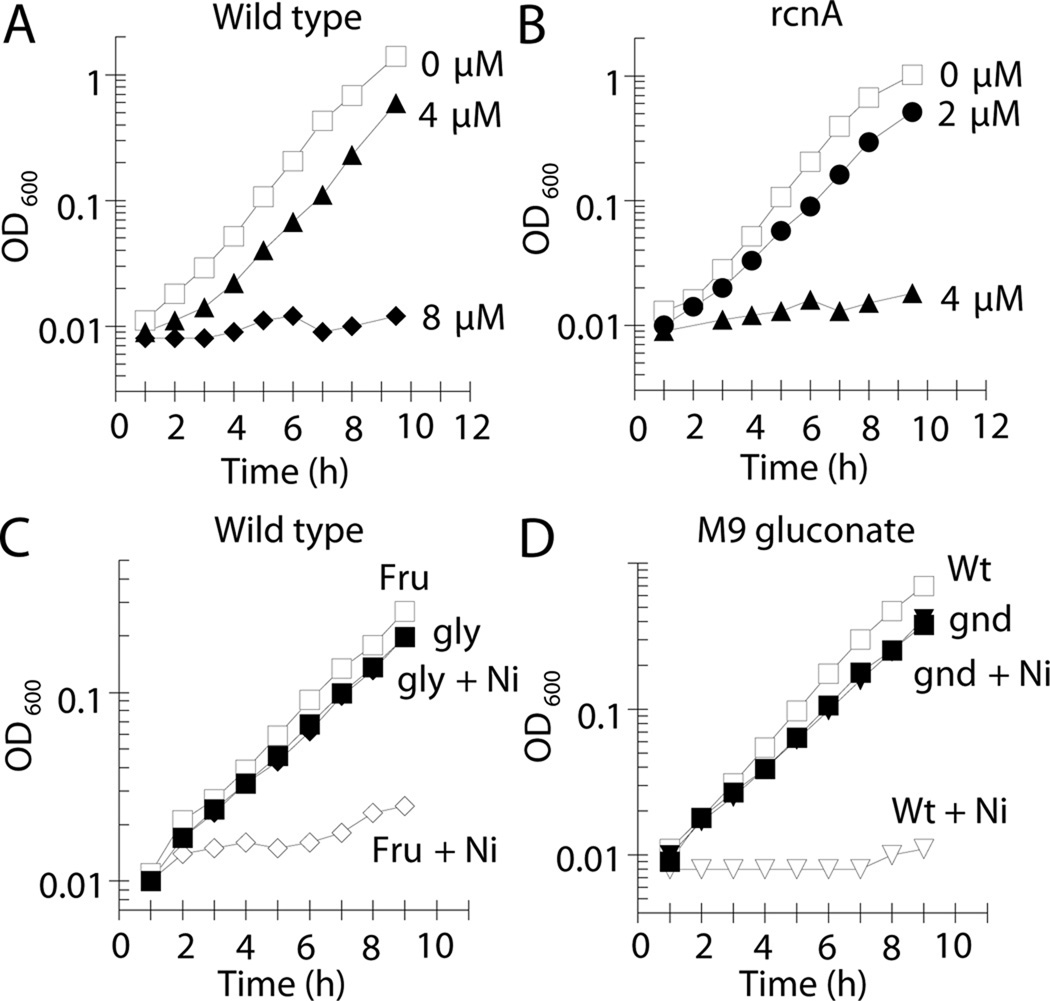
Low micromolar levels of nickel inhibited the growth of wild-type E. coli (MG1655) in M9 glucose medium (Fig. 1A). When wild-type cells were grown for 1 h with 8 µM nickel they contained 140 µM total cellular nickel, as determined by inductively-coupled plasma optical emission spectroscopy (ICP-OES). Surprisingly, cells grown without nickel supplementation (~1 µM nickel in M9 glucose medium) contained 30 µM of this metal. These results demonstrate that cells can accumulate an order of magnitude more nickel than the concentration in the extracellular environment, a situation noted previously with several metals (Outten and O'Halloran, 2001). In order to combat this cellular accumulation of nickel and thereby protect against toxicity, E. coli evolved the inner-membrane RcnA nickel-efflux system (Rodrigue et al., 2005). As expected from prior studies that grew cells on a variety of media types (Koch et al., 2007; Rodrigue et al., 2005; Wang et al., 2011), elimination of rcnA sensitized E. coli (LEM201) to this metal ion (Fig 1B).
Fig. 1.
Nickel is toxic at micromolar levels and sensitivity is dependent on the carbon source. Cells were grown aerobically at 37 °C. (A) Wild-type cells (MG1655) and (B) rcnA mutant cells (LEM201) were grown on M9 glucose medium and challenged with 0 µM (□), 2 µM (●), 4 µM (▲), or 8 µM Ni(II) (♦). (C) Wild-type cells (MG1655) were grown in M9 medium containing fructose (open symbols) or glycerol (closed symbols) and challenged with 0 µM (squares) or 8 µM (diamonds) Ni(II). (D) Wild-type cells (MG1655, open symbols) and gnd mutant cells (LEM202, closed symbols) were grown in M9 gluconate medium containing 0 µM (squares) or 16 µM (crucibles) Ni(II). The data are representative of three independent experiments.
Nickel blocks glycolysis
Nickel ion-dependent inhibition of E. coli cell growth did not lead to a decrease in cell viability as would be expected for a bacteriocidal agent. At a nickel concentration that prevented the growth of wild-type and rcnA cultures (16 µM) the number of viable cells for both strains was unchanged after 4 h of exposure (Fig. S1), consistent with a bacteriostatic agent; therefore, the site of primary toxicity of nickel is likely to be a block in metabolism.
To determine how nickel inhibits growth, wild-type cells were grown with various carbon sources in order to bypass a potential nickel-dependent block in central metabolism (Fig. S2). Wild-type cells were equally sensitive to nickel when grown with fructose as for growth on glucose (Fig. 1C). In contrast, cells grown on succinate, lactate, or glycerol were resistant to nickel (Fig. 1C & Fig. S3). These results did not arise from secondary effects on nickel transport when grown on different substrates, for the nickel content of the glucose- and glycerol-grown cultures exposed to 8 µM for an hour were the same (140 µM) as determined by ICP-OES. These results suggest that nickel blocks glycolysis between the entrance of fructose carbon and glycerol carbon, consistent with nickel inhibiting fructose-1,6-bisphosphate aldolase.
Nickel inhibits the activity of FbaA in vivo
E. coli contains two fructose-1,6-bisphosphate aldolases: FbaA and FbaB. FbaA, a class II aldolase that employs zinc to stabilize the catalytic intermediate, is absolutely required for glycolysis (Böck and Neidhardt, 1966; Marsh and Lebherz, 1992; Stribling and Perham, 1973). FbaB, a class I aldolase that utilizes a catalytic lysine residue (Marsh and Lebherz, 1992), participates in gluconeogenesis and is not expressed when cells are grown on glucose (Scamuffa and Caprioli, 1980; Stribling and Perham, 1973). Thus, nickel-dependent growth inhibition on glucose is likely due to loss of FbaA activity.
Evidence that nickel inhibits FbaA was corroborated by the use of mutants. Previous studies demonstrated that temperature-sensitive fbaA mutants fail to grow on gluconate at non-permissive temperatures (Böck and Neidhardt, 1966; Schreyer and Böck, 1973). While a majority of the gluconate carbon proceeds through the Entner-Doudoroff pathway (Fig. S4), some enters the pentose phosphate pathway via 6-phosphogluconate dehydrogenase (6PGD) (Schreyer and Böck, 1973). A portion of the resulting ribulose-5-phosphate is converted to fructose-6-phosphate, phosphorylated, and the accumulating fructose-1,6-bisphosphate causes the production of ppGpp with the subsequent cessation of growth (Schneider and Gourse, 2003). When carbon flow from the Entner-Doudoroff pathway to the pentose phosphate pathway is eliminated through a gnd mutation (the gene encoding 6PGD), the temperature-sensitive fbaA mutant is able to grow with gluconate at the non-permissive temperature (Schreyer and Böck, 1973). Nickel inhibition of FbaA should confer nickel sensitivity to cells growing on gluconate. Indeed, 16 µM nickel inhibited the growth of wild-type cells on this carbon source (Fig. 1D). Furthermore, elimination of gnd (LEM202) restored growth on gluconate in the presence of 16 µM nickel (Fig. 1D). These results are consistent with nickel inhibition of FbaA.
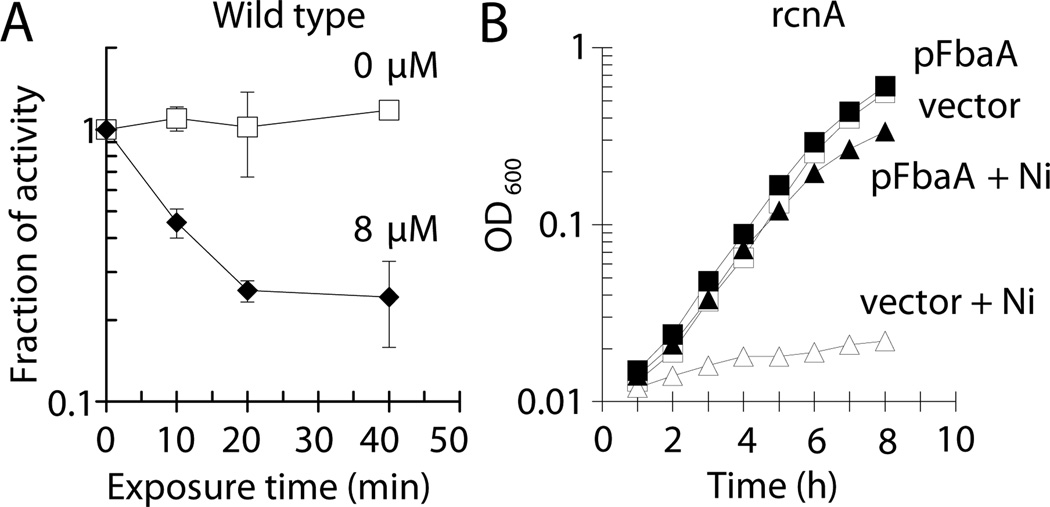
Wild-type cells exposed to nickel in vivo exhibited reduced FbaA activity, as measured in their cell-free extracts, compared to untreated cells (Fig. 2A) For example, incubation with 8 µM nickel for 20 min led to >70% reduction of activity. The rapid loss of activity suggests that extant enzyme was damaged by nickel. To confirm that loss of this aldolase activity was responsible for nickel-dependent growth inhibition, FbaA was over-produced on a medium copy plasmid (pWKS30, 6 – 8 copies per cell (Wang and Kushner, 1991)) in an rcnA mutant, chosen to eliminate any complications arising from the induction of this efflux pump. Nickel (4 µM) inhibited the growth of the rcnA mutant containing the vector control (LEM233), whereas rcnA cells over-expressing fbaA (LEM234) were able to grow (Fig. 2B). Together, these data demonstrate that nickel-dependent growth inhibition is due to the reduction of FbaA activity.
Fig. 2.
Nickel toxicity occurs by inhibition of fructose-1,6-bisphosphate aldolase. Cells were grown aerobically in M9 glucose medium at 37 °C. (A) At OD600 ~0.1, wild-type cells (MG1655) were challenged with 0 µM (□) or 8 µM (♦) Ni(II) and FbaA activity was measured in cell-free extracts after selected time points. (B) rcnA mutant cells containing the vector (pWKS30; LEM233; open symbols) or pFbaA (pLEM1; LEM234; closed symbols) were challenged with 0 µM (squares) or 4 µM (diamonds) Ni(II) and further growth was monitored. The data are representative of three independent experiments.
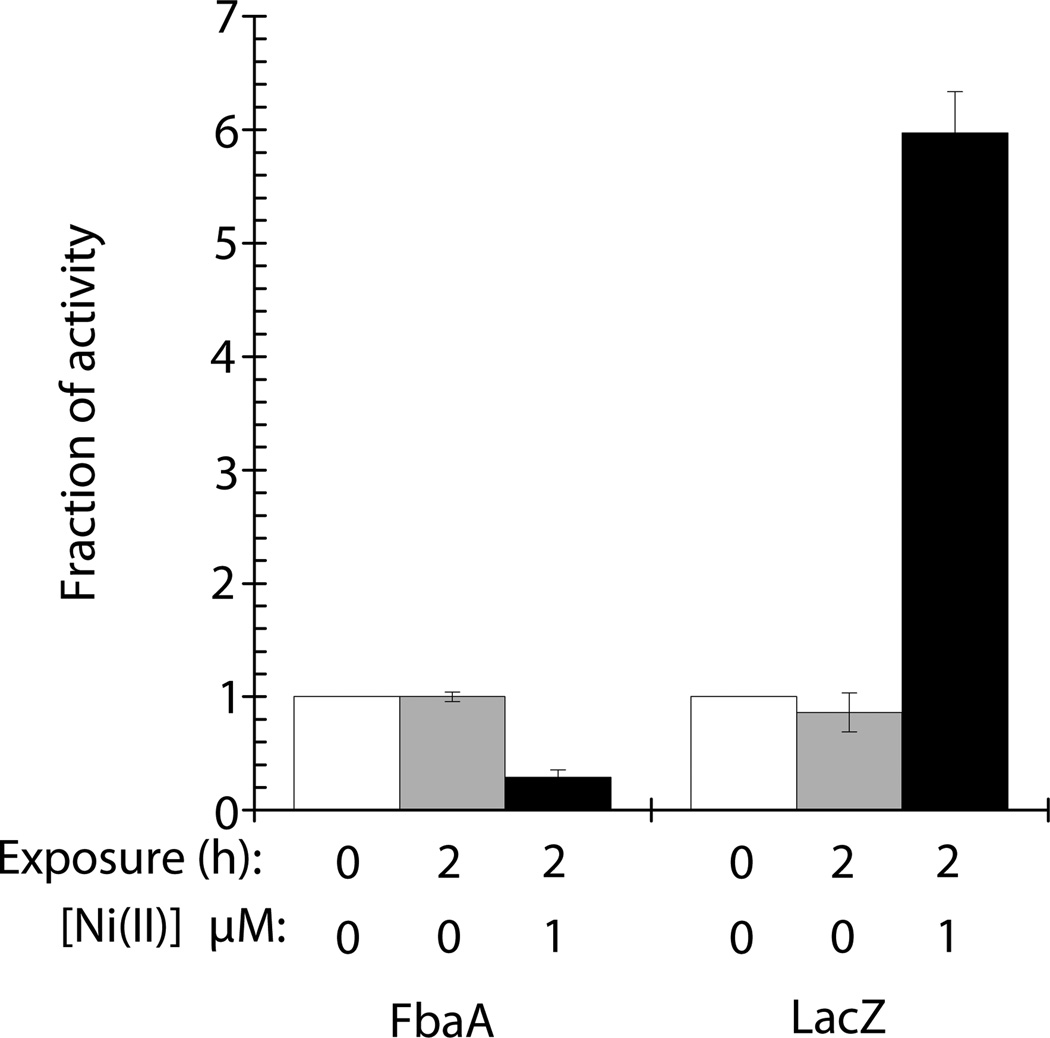
With FbaA as a primary target of nickel toxicity, one would anticipate that induction of the nickel-defense system would occur prior to or concomitant with the loss of aldolase activity. As expected, when rcnA mutant cells containing a PrcnA-lacZ fusion (LEM273) were exposed to 1 µM nickel for 2 h, the level of LacZ activity increased by 6-fold whereas the fructose-1,6-bisphosphate activity decreased by 70% (Fig. 3). This result indicates that the nickel defense system is induced concomitantly with FbaA inhibition.
Fig. 3.
RcnA is induced concomitantly with nickel-dependent FbaA damage. LEM273 (rcnA lacZ PrcnA-lacZ) were grown in M9 glucose medium to OD600 of 0.1. Cultures were split and 0 µM NiCl2 (gray bars) or 1 µM NiCl2 (black bars) was added. Cultures were incubated aerobically at 37 °C for 2 hours. White bars represent enzyme activity prior to the culture being split. Error bars represent the standard deviation of three independent experiments.
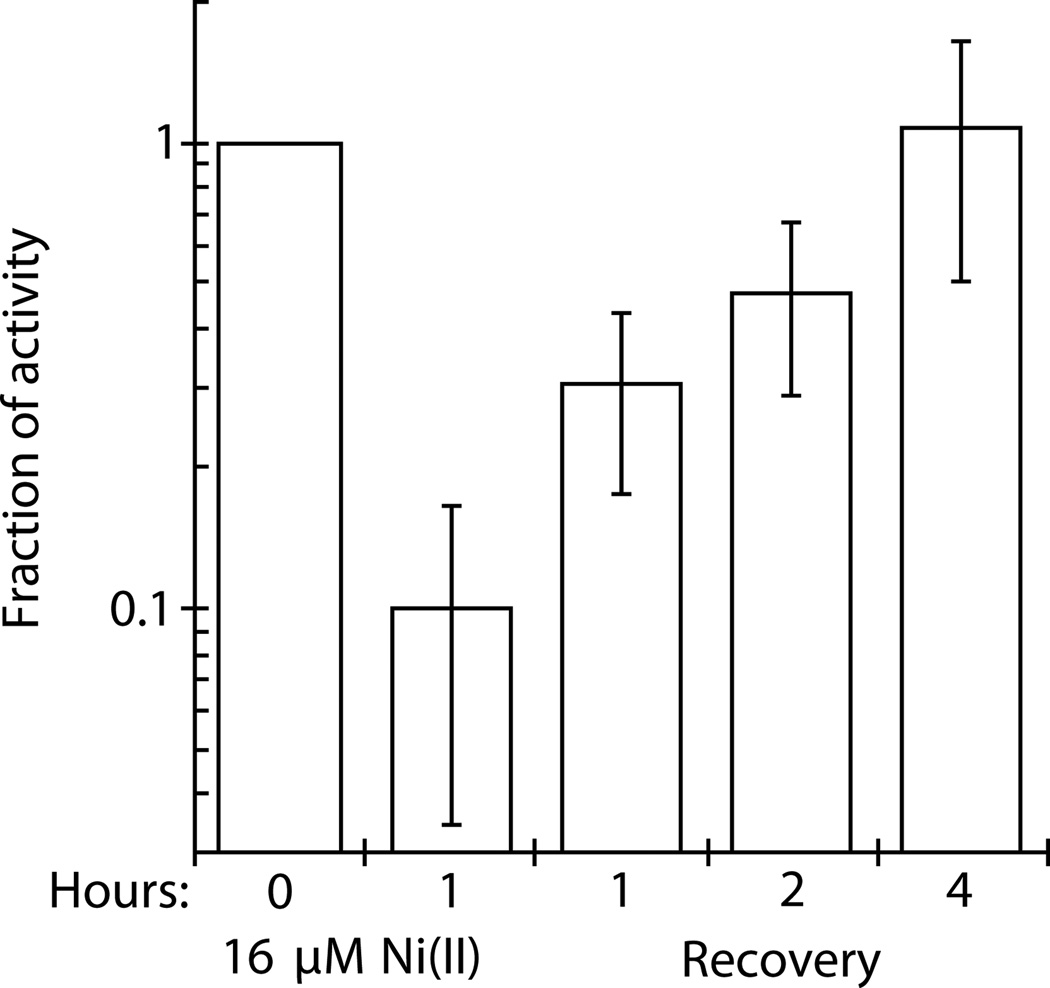
To test whether nickel irreversibly inhibits FbaA in vivo, cells were incubated with 16 µM nickel, chloramphenicol was added to block protein synthesis, and cells were resuspended in fresh medium containing the antibiotic. After 4 h incubation without nickel, during which time the OD600 was essentially unchanged providing assurance that protein synthesis was inhibited, the initial FbaA activity was completely restored (Fig. 4). This result suggests that nickel does not irreversibly damage FbaA; however, whether reactivation of nickel-damaged FbaA is an active or passive process has yet to be determined.
Fig. 4.
Nickel-damaged FbaA is reactivated in vitro. Wild-type MG1655 cells were grown aerobically in M9 glucose medium at 37 °C. At OD600 ~0.1, the cells were challenged with 16 µM Ni(II). After an hour exposure, chloramphenicol (150 µg ml−1) was added to inhibit protein synthesis. Cells were washed with M9/glucose/chloramphenicol medium to remove nickel, resuspended in the same medium, incubated aerobically at 37 °C, and assayed for FbaA activity at the times indicated. Error bars represent the standard deviation of three independent experiments.
Nickel inhibits FbaA by binding to a secondary zinc binding site
The activity of purified FbaA was inhibited in a time- and concentration-dependent manner by incubation with low micromolar concentrations of nickel (Fig. 5A & B), confirming the in vivo results. The addition of copper also led to significant levels of inhibition, whereas zinc slightly activated at low concentrations while modestly inhibiting at greater concentrations (Fig. S5). By contrast, calcium, magnesium, and manganese had no effects on FbaA activity and cobalt slightly activated the enzyme (Fig. S5).
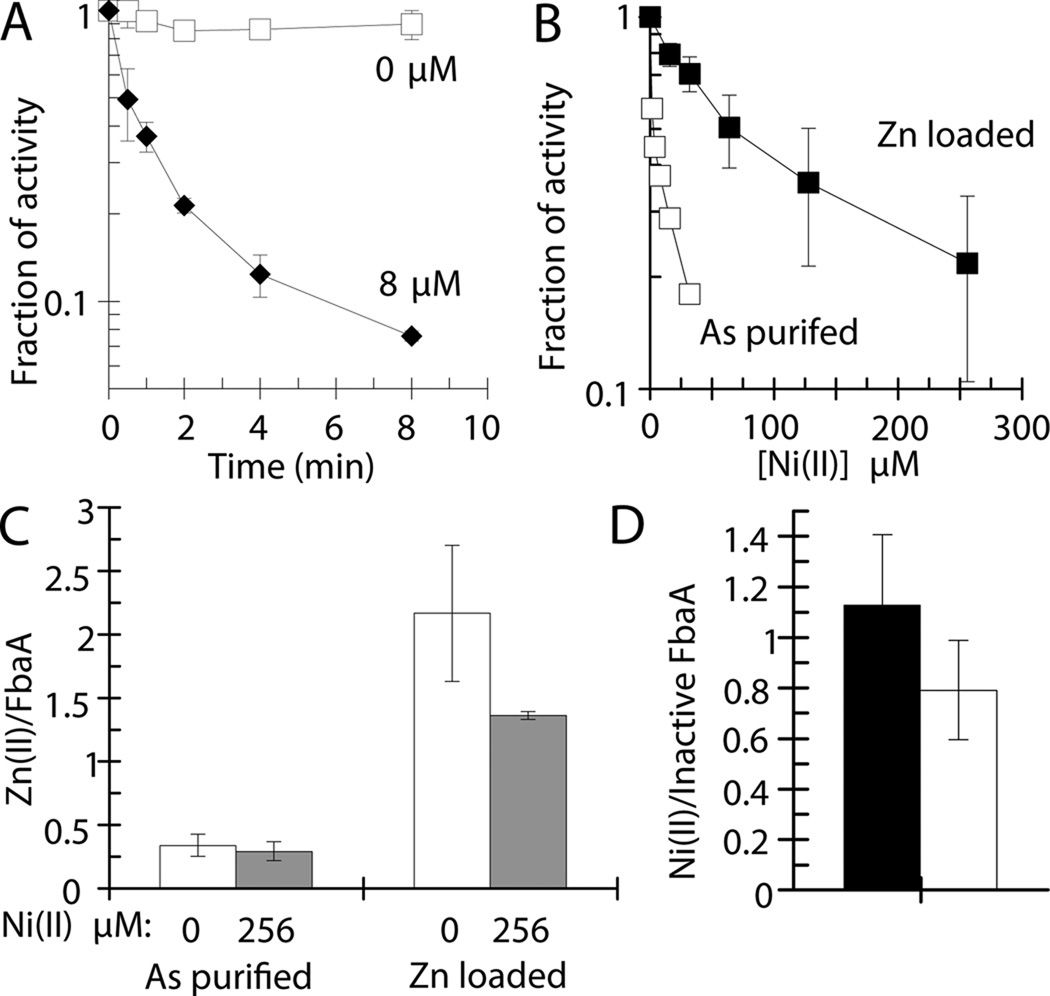
Fig. 5.
FbaA is inhibited by nickel, whereas zinc-loaded enzyme is resistant to nickel damage. Purified FbaA was challenged with nickel aerobically at room temperature (25 °C). (A) FbaA (50 nM) was incubated with 0 µM (□) or 8 µM (♦) Ni(II) and enzyme activity was assessed at the indicated times. (B) FbaA as purified (90 nM) (□) or Zn-loaded (30 nM) (■) was incubated with the indicated concentrations of Ni(II) for 5 min and assayed. (C) The zinc contents of FbaA were determined by ICP-OES for FbaA as purified and after Zn-loading, including samples challenged with 0 µM (white) or 256 µM Ni(II) (grey) for 10 min. (D) The nickel content of FbaA was determined by ICP-OES for FbaA as purified (black) and after Zn-loading (white), both challenged with 256 µM Ni(II) for 10 min. (C & D) In each case, unbound metal was removed by gel filtration. Error bars represent the standard deviation of three independent experiments.
FbaA binds two zinc atoms; one is coordinated by His110, His226, and His264 (this numbering corresponds to that of protein as purified, following removal of the initial methionine by aminopeptidase) and is used to catalyze the carbon-carbon bond cleavage of fructose-1,6-bisphosphate; whereas the second metal ion is coordinated via Asp144, Glu174, and Glu181 and has no known function (Fig. S6)(Hall et al., 1999). It was possible that nickel inhibits aldolase by displacing the catalytic zinc and sequestering the coordinating histidine residues or by displacing the non-essential zinc, through binding to the carboxylate side chains, and inhibiting allosterically. In either case, there should be a net loss of zinc with a concomitant gain of nickel. Purified FbaA [1.8 U (mg protein)−1] was shown by ICP-OES to contain up to 0.3 equivalents of zinc and 0.2 equivalents of nickel per FbaA monomer. This protein was incubated with 0 or 256 µM nickel (i.e., to more than 5000-fold the enzyme concentration) for 5 min, with the latter sample resulting in >90% loss of FbaA activity. The samples were desalted to remove unbound metals and the metal contents re-examined. The enzyme incubated with nickel lost only minor amounts of zinc and gained 1.1 nickel per inactivated FbaA (Fig. 5C & D). To fully metallate FbaA with zinc, the purified enzyme was incubated with this metal under slightly denaturing conditions. After removing unbound zinc by dialysis with EDTA, FbaA possessed 3.8-fold increased activity [6.9 U (mg protein)−1; i.e., the same order of magnitude as reported earlier (Berry and Marshall, 1993)] and contained 2.3 zinc per monomer (Fig. 5C). Of note, the 3.8-fold increase in specific activity of the zinc-metallated sample correlates well to a 3.3-fold increase in occupancy of the catalytic site (from 0.3 equivalents to fully loaded). The zinc-treated FbaA exhibited enhanced resistance to low-micromolar levels of nickel (Fig. 5B), suggesting that occupancy of both zinc sites in FbaA partially protected against nickel-dependent inhibition. Incubation of fully-metallated FbaA with 256 µM nickel for 10 min (leading to 60% inactivation) reduced the zinc content by 0.8 per FbaA while increasing the nickel content to 0.8 per inactive FbaA (Fig. 5C & D). These data demonstrate that nickel displaces some, but not all, zinc when inhibiting the fully metallated enzyme, consistent with nickel substitution at one of the two zinc sites.
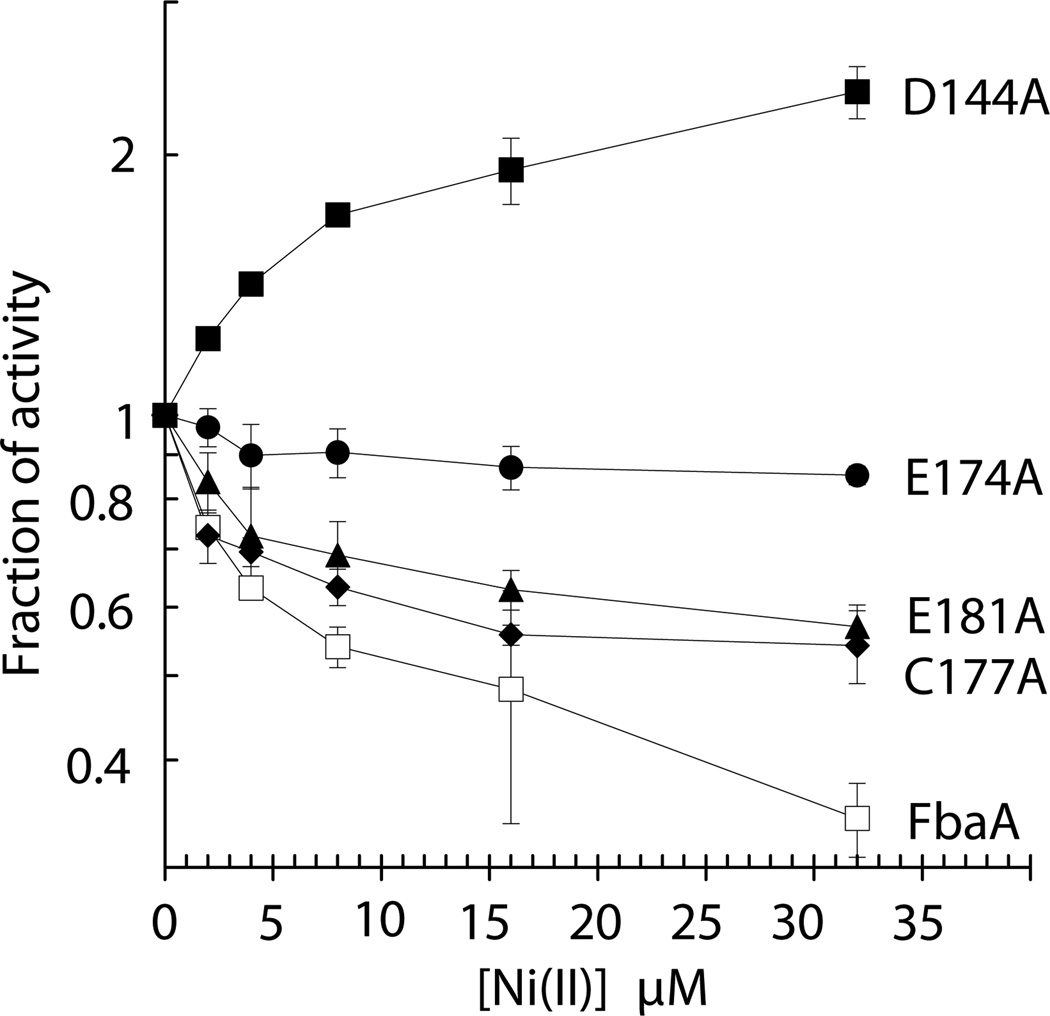
To further define the inhibitory site of nickel binding to FbaA, variants of the protein were created. The catalytic site was left unchanged. The Asp144, Glu174, and Glu181 amino acids ligating the secondary zinc atom along with a nearby Cys177 residue were changed to alanines by mutagenesis of the corresponding codons. The activities of the His-tagged FbaA and its variant proteins [4.6, 0.6, 0.2, 0.8, and 3.0 U (mg protein)−1, respectively, after in vitro metallation with zinc] were examined as a function of nickel concentration. When compared to the wild-type enzyme, the C177A and E181A variants had small increases in nickel resistance, the E174A enzyme was insensitive to nickel, and the D144A sample exhibited enhancement of enzymatic activity by added nickel (Fig. 6). Control samples comparing wild-type and His-tagged versions of FbaA showed no changes in nickel sensitivity up to 32 µM nickel (data not shown). This result is likely due to nickel being super stoichiometric to the enzyme, thus indicating that the inhibitory patterns of the mutant proteins are unlikely to be affected by the affinity tag under our experimental conditions. These results demonstrate that nickel inhibits FbaA by binding to the noncatalytic zinc site.
Fig. 6.
Nickel inactivates FbaA at the secondary zinc binding site. As purified His-tagged FbaA (400 nM; 0.4 U/mg; □) and its corresponding D144A (200 nM; 0.6 U/mg; ■), E174A (300 nM; 0.1 U/mg; ●), C177A (25 nM; 0.4 U/mg; ♦), and E181A (1000 nM; 0.08 U/mg; ▲) variants were challenged with the indicated nickel concentrations for 5 min aerobically at room temperature (25 °C) and tested for remaining activity. Error bars represent the standard deviation of three independent experiments.
Cellular metabolites modulate nickel-dependent FbaA inactivation in vitro and in vivo
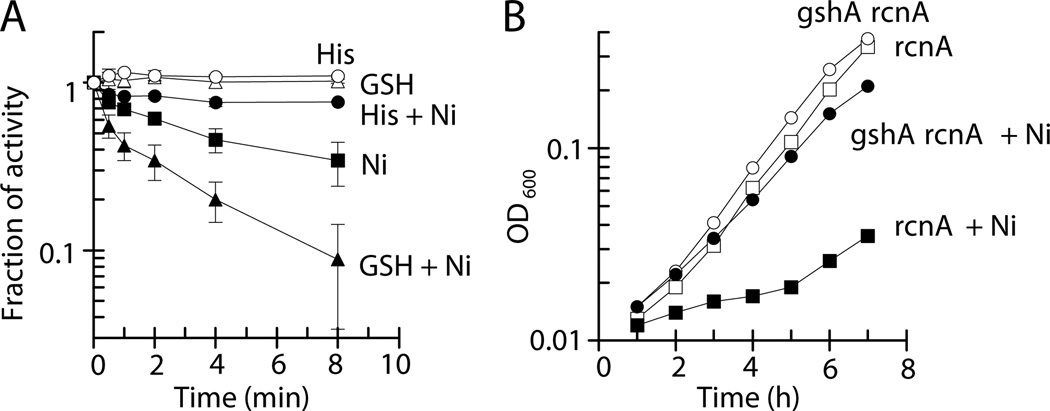
The in vivo pool of nickel available to inhibit aldolase is expected to be governed by the concentrations of other cellular metabolites. The effects of two such compounds on inhibition of FbaA were examined in vitro. Histidine (His), a well-studied ligand of nickel (Kowalik-Janowska et al., 2007), when present at 100 µM protected FbaA against the metal (16 µM), leading to only ~20% loss of FbaA activity compared to the 70% loss of enzyme activity for solutions lacking the metal ligand (Fig. 7A). Surprisingly, the addition of reduced glutathione (GSH) enhanced the nickel-dependent inhibition of FbaA (Fig. 7A). This latter effect was verified in vivo; i.e., E. coli rcnA mutants incapable of synthesizing glutathione (gshA) were less sensitive to nickel-dependent growth inhibition than GSH+ cells (Fig. 7B). This enhancement of FbaA nickel inhibition by glutathione contrasts with its protective effect noted for several other toxic metals such as copper, zinc, and cadmium (Helbig et al., 2008; Macomber and Imlay, 2009).
Fig. 7.
Cellular metabolites affect nickel-dependent damage to FbaA. (A) As purified FbaA (20 nM) was challenged with 0 µM (open symbols) or 16 µM (closed symbols) Ni(II) in the absence (squares) or presence of 100 µM His (circles) or 1 mM reduced glutathione (GSH; triangles) aerobically at 25 °C, and the enzyme was assayed at the indicated times. The error bars represent the standard deviation of three independent experiments. (B) Mutant E. coli cells LEM201 (rcnA, squares) and LEM267 (gshA rcnA, circles) were grown aerobically at 37 °C in M9 glucose medium and challenged with 0 µM (open symbols) or 1 µM (closed symbols) Ni(II). The data are representative of three independent experiments.
Discussion
Fructose-1,6-bisphosphate aldolase is the target of nickel inhibition in E. coli
FbaA is a member of the class II aldolases found throughout eubacteria, in some archaea, and in fungi (Marsh and Lebherz, 1992). These enzymes play important roles in metabolism, including FbaA’s central function in glycolysis. Several lines of evidence establish that FbaA is the site of damage by nickel in E. coli: cell growth is hindered by nickel when using substrates dependent on the glycolytic aldolase, inhibition is reduced by overexpression of fbaA, direct assays of cellular FbaA activity reveal suppression by nickel, and purified FbaA is inhibited by this metal.
The inhibition of FbaA by nickel is quite specific, with no effect observed for magnesium, manganese, or calcium, and a slight increase in activity by added cobalt (cobalt-substituted enzymes often exhibit hyperactivity compared to their zinc forms) (Mulrooney and Hausinger, 2002). The cupric form of copper also resulted in substantial inhibition of the enzyme; however, this metal is unlikely to be a physiological inhibitor because cytoplasmic copper is primarily cuprous. Added zinc had only modest effects on the activity of the enzyme. At low concentrations zinc slightly activated FbaA, presumably associated with the partial activation of apoprotein present with the enzyme as purified; however, full loading of zinc into the enzyme required incubation with low concentrations of a denaturant. This enhanced metallation approach produced enzyme with several fold enhanced activity, whereas simply providing high concentrations of zinc led to very slight inhibition of FbaA.
For nickel to inhibit FbaA in vivo, the metal must associate with the enzyme in the presence of many other cellular components. Cytoplasmic nickel-binding compounds could hinder this association by sequestering the metal or they could enhance nickel toxicity by facilitating the inhibitory interaction. The toxicity of nickel for FbaA is reduced by His, an effective ligand of the metal, likely by simple competition. By contrast, inhibition of FbaA by nickel is potentiated by GSH, consistent with this metabolite acting to specifically donate the metal to aldolase. This in vitro result was confirmed by in vivo studies with mutant cells incapable of synthesizing GSH which were more resistant to inhibition by nickel, suggesting that GSH facilitates the intracellular movement of nickel. An alternative explanation is that, in a gshA mutant, zinc can readily compete with nickel for enzyme binding due to the absence of zinc-chelation by GSH (Díaz-Cruz et al., 1998; Gelinsky et al., 2003), though this is unlikely to occur in the in vitro experiment.
The fundamental importance of FbaA coupled with its sensitivity to nickel likely led to the requirement of a nickel defense system in E. coli. RcnR was estimated to have an affinity for nickel of < 25 nM (Iwig et al., 2008), therefore rcnA should be induced prior to the accumulation of toxic intracellular nickel levels. Consistent with this hypothesis, rcnA transcription is induced by nickel at concentrations that are lower than those which cause the debilitating loss of FbaA activity (Fig. 3).
Mechanism of FbaA inactivation
E. coli FbaA binds both a catalytic zinc and a secondary zinc near the active site (Hall et al., 1999) (Fig. S6). After fully loading the enzyme with zinc, inhibitory nickel was shown to displace one zinc atom. Significantly, the fully zinc-loaded sample was more resistant to nickel inhibition than the as-purified enzyme with its substoichiometric zinc. These results demonstrate that inhibition by nickel is associated with its binding to one of the zinc sites. The identity of the nickel binding site was established as the secondary zinc site by the protective effect of the D144A and E174A mutations that are likely to reduce nickel binding to the second site. Nickel bound at the secondary zinc site is likely to affect the residues that coordinate the catalytic zinc atom; e.g., binding of inhibitory nickel possibly leads His110 to swing away from its normal position to bind this metal, thus changing the coordination of the catalytic zinc atom (Hall et al., 1999). Indeed, the absence of nickel inhibition for the D144A and E174A proteins could be explained by increased flexibility of the secondary site so that nickel binds without perturbing the position of His110 or the activity of the catalytic zinc atom.
Conclusion
Although nickel is well established as a toxic and carcinogenic metal, prior to this study no direct mechanism for its toxicity had been elucidated. The limited ability of microbes to manipulate their environment makes them uniquely sensitive to fluctuations in metal concentrations, so they are ideal organisms to study the mechanisms of metal toxicity. We combined physiological, genetic, biochemical, and mutagenic studies of E. coli to demonstrate that nickel toxifies the cells by displacing zinc from a secondary binding site of the metalloprotein FbaA.
Experimental procedures
Bacterial Growth
Lysogeny broth (LB), terrific broth (TB), and base M9 minimal salts were of standard composition (Miller, 1972). Where indicated, media were supplemented with 0.2% glucose, 0.2 % fructose, 0.4% gluconate, 40 mM succinate, 40 mM glycerol, or 40 mM D,L-lactate. Media were supplemented with 100 µg ml−1 ampicillin, 20 µg ml−1 chloramphenicol, or 30 µg ml−1 kanamycin when antibiotic selection was needed. Overnight cultures were diluted to OD600 of 0.01 and cells were grown at 37 °C to an OD600 of ~0.1 before analyses.
Strain Constructions
Strains and plasmids are listed in Table 1. Mutations were introduced into strains by P1 transduction and selected with the appropriate antibiotic. To construct the pLEM1 plasmid, the fbaA open reading frame was PCR-amplified from E. coli MG1655 by using the primers listed in Table S1. The PCR product was digested with EcoRI and SacI, then cloned into pWKS30 (Wang and Kushner, 1991) behind the lac promoter. To construct a plasmid encoding an N-terminal His6-tagged FbaA, the fbaA open reading frame was PCR-amplified from E. coli MG1655, digested with BamHI and XhoI and cloned into the pET28b vector behind the T7 promoter (pLEM5). The cloned fbaA was confirmed by restriction analysis and sequenced (Davis Sequencing). Site-directed mutants of FbaA were constructed by Quickchange site-directed mutagenesis (Stratagene) using pLEM5 as the template. The primers used to construct the D144A, E174A, C177A, and E181A variants of FbaA are listed in Table S1. The plasmids and mutant fbaAs were confirmed by restriction analysis and sequencing.
TABLE 1.
E. coli strains and plasmids used
| Strains | Genotype | Source |
|---|---|---|
| BL21 (DE3) | dcm ompT hsdS(rB− mB−) gal λ(DE3) | Stratagene |
| Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) rph-1 | ||
| BW25113 | Δ(rhaD-rhaB)568 hsdR514 | (Datsenko and Wanner, 2000) |
| JW2093-1 | BW25113 plus ΔrcnA731::kan | (Baba et al., 2006) |
| JW2011-2 | BW25113 plus Δgnd-727::kan | (Baba et al., 2006) |
| LEM201 | MG1655 plus ΔrcnA731::kan | This study |
| LEM202 | MG1655 plus Δgnd-727::kan | This study |
| LEM232 | MG1655 plus pLEM1 | This study |
| LEM233 | MG1655 plus ΔrcnA731::kan pWKS30 | This study |
| LEM234 | MG1655 plus ΔrcnA731::kan pLEM1 | This study |
| LEM267 | MG1655 plus ΔrcnA731::kan ΔgshA::cat | This study |
| LEM273 | MG1655 plus Δ(lacZ1)1 ΔrcnA731 pJI114 | This study |
| LEM276 | BL21 (DE3) plus pLEM5 | This study |
| LEM277 | BL21 (DE3) plus pLEM6 | This study |
| LEM278 | BL21 (DE3) plus pLEM7 | This study |
| LEM279 | BL21 (DE3) plus pLEM8 | This study |
| LEM280 | BL21 (DE3) plus pLEM9 | This study |
| MG1655 | F− λ− wild type | (Neidhardt, 1996) |
| SJ99 | As MG1655 plus Δ(lacZ1::cat)1 | (Anjem et al., 2009) |
| SSK112 | As BW25113 plus ΔgshA::cat | Jim Imlay |
| Plasmids | ||
| pET28b | PT7 His6-tag kmr | Novagen |
| pJI114 | pACYC163 plus rcnR-PrcnA-lacZ | (Iwig et al., 2006) |
| pLEM1 | pWKS30 plus fbaA inserted into SacI-EcoRI. | This study |
| pLEM5 | pET28b plus fbaA inserted into XhoI-BamHI. His6-tagged. | This study |
| pLEM6 | D144A mutant of pLEM5. | This study |
| pLEM7 | E174A mutant of pLEM5. | This study |
| pLEM8 | C177A mutant of pLEM5. | This study |
| pLEM9 | E181A mutant of pLEM5. | This study |
| pWKS30 | Plac polylinker Amr | (Wang and Kushner, 1991) |
In vivo experiments
To determine cellular metal content, cultures were grown aerobically at 37 °C in M9 glucose or glycerol medium. At an OD600 of 0.1, cultures were split and treated with 0 µM or 8 µM Ni(II). After 1 h exposure, cells were washed twice with ice-cold 50 mM Tris/1 mM EDTA, pH 7.5, and once with ice-cold 50 mM Tris, pH 7.5. Cells were sonicated and the cell debris was removed by centrifugation at 6000 g for 10 min, 4 °C. The metal contents were measured by ICP-OES (Center for Applied Isotope Studies, University of Georgia).
Cell viability after nickel exposure was determined by plate counts. Cells were grown aerobically in M9 glucose medium at 37 °C. At each time point, aliquots were diluted into LB medium, plated onto LB, and incubated aerobically overnight at 37 °C.
The ability of fbaA overexpression to suppress nickel toxicity was determined by using strains LEM233 and LEM234. Cells were grown overnight in M9 glucose containing 100 µg ml−1 ampicillin and 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cultures were diluted into M9 glucose/ampicillin/IPTG medium to an OD600 of 0.01 and grown aerobically at 37 °C.
The correlation between nickel-dependent induction of the RcnA efflux pump and loss of FbaA activity was determined by using LEM273. Cells were grown in M9 glucose medium to OD600 of 0.1. Cultures were split and 0 µM NiCl2 or 1 µM NiCl2 was added. Cultures were incubated aerobically at 37 °C for 2 h. Cells were washed twice in ice-cold 50 mM Tris, pH 7.5, extracts were prepared, and enzymes were assayed as described below.
The in vivo repair of nickel-damaged FbaA was measured in wild-type cells in order to allow efflux of intracellular nickel by RcnA. Wild-type MG1655 cells were grown aerobically in M9 glucose medium to an OD600 of ~0.1, exposed to 16 µM nickel for 1 h, treated with chloramphenicol (150 µg ml−1), centrifuged, washed twice, and resuspended in M9/glucose/chloramphenicol medium that was pre-warmed to 37 °C. The resulting culture was incubated at 37 °C aerobically and aliquots were removed at intervals to assay FbaA activity.
Enzyme Assays
To prepare cell-free extracts, cells were washed twice, resuspended in buffer, lysed by sonication, and cell membranes were removed by centrifugation at 100,000 × g for 1 h at 4 °C. All enzyme reactions were monitored aerobically at 25 °C. An FbaA coupled assay (without triose phosphate isomerase) (Baldwin et al., 1978) and β-galactosidase (Miller, 1972) activities were measured according to the cited methods. Protein concentrations were determined by using the Coomassie protein assay reagent (Bio-Rad) with bovine serum albumin as the standard.
In vitro studies
FbaA was purified from MG1655 containing pLEM1 by using a modification of a known protocol (Baldwin et al., 1978). Lem232 was grown overnight in TB/glucose/0.5 mM IPTG at 37 °C, disrupted, and membrane-free cell extracts were loaded onto a DEAE-Sepharose column and eluted by a 0–250 mM gradient of NaCl in 50 mM Tris, pH 7.5. The fractions containing the highest activity were pooled and further separated on a Q-Sepharose column that was eluted by 0–250 mM gradient of NaCl in 50 mM Tris, pH 7.5. Pooled fractions were loaded onto a hydroxyapatite column and eluted with a 5–250 mM gradient of sodium phosphate in 150 mM NaCl, pH 6.8. Finally, pooled fractions were loaded onto a phenyl-Sepharose column and eluted by a decreasing 750-0 mM KCl gradient in 50 mM Tris, pH 7.5. The fractions containing the highest activity were concentrated (Amicon). Purification of the FbaA was confirmed by its sensitivity to 100 µM EDTA. FbaA preparations were ≥70% pure on the basis of denaturing polyacrylamide gel electrophoresis.
Site-directed mutants and a control sample of FbaA were purified by immobilized metal ion affinity chromatography. Cells were grown overnight in LB/0.5 mM IPTG at 37 °C. Cell extracts were loaded onto a Ni-Sepharose column and weakly bound proteins were removed with 30 mM imidazole/500 mM NaCl/20 mM sodium phosphate, pH 7.4. His6-tagged FbaA and its variants were eluted by using 500 mM imidazole/500 mM NaCl/20 mM sodium phosphate, pH 7.4. Purified FbaA samples were ≥95% pure on the basis of denaturing gel electrophorectic analysis.
In vitro inactivation of purified FbaA was conducted aerobically at 25 °C using enzyme dissolved in buffer (50 mM Tris-HCl, pH 7.5). Where indicated, 100 µM EDTA, 1 mM GSH, or 100 µM His was present.
FbaA, His-FbaA, D144A, E174A, C177A, and E181A were metallated with zinc after partial denaturation (Kirby et al., 1980). Purified protein was dialyzed twice against 20 mM 8-hydroxy-5-quinolinesulfonic acid/2.5 M guanidinium chloride/5 mM Tris/ 0.1 mM EDTA, pH 3.8 at 4 °C for 12 h. Apoprotein was metallated by dialyzing twice against 5 mM Tris/0.1 mM ZnCl2/10 mM dithiothreitol, pH 7.8, at 4 °C for 12 h. Unbound zinc was removed by dialyzing twice against 5 mM Tris/0.1 mM EDTA, pH 7.8, at 4 °C for 12 h. EDTA was removed by dialyzing twice against 50 mM Tris, pH 7.5 at 4°C for 12 h. All dialysis buffer volumes were ≥500-fold over enzyme volumes.
Where indicated, gel filtration chromatography was used to remove unbound metal ions from protein preparations. PD10 columns (GE Healthcare) were equilibrated with 50 mM Tris-HCl, pH 7.5, prior to and after protein loading.
Supplementary Material
Acknowledgements
We thank Peter Chivers, Jim Imlay, Richard Lenski, and Carrie Vanderpool for strains, plasmids, and phage, and Robert Britton for use of equipment. This study was supported by the National Institutes of Health Grant DK045686.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
References
- Anjem A, Varghese S, Imlay JA. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherchia coli. Molec Microbiol. 2009;72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gen knockout mutants: The Keio collection. Molec Syst Biol. 2006 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin SA, Perham RN, Stribling D. Purification and characterization of the class II D-fructose 1,6-bisphosphate aldolase from Escherichia coli (Crooke's strain) Biochem J. 1978;169:633–641. doi: 10.1042/bj1690633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barceloux DG. Nickel. J Toxicol Clin Toxicol. 1999;37:239–258. doi: 10.1081/clt-100102423. [DOI] [PubMed] [Google Scholar]
- Berry A, Marshall KE. Identification of zinc-binding ligands in the class II fructose-1,6-bisphosphate aldolase of Escherichia coli. FEBS Lett. 1993;318:11–16. doi: 10.1016/0014-5793(93)81317-s. [DOI] [PubMed] [Google Scholar]
- Böck A, Neidhardt FC. Properties of a mutant of Escherichia coli with a temperature-sensitive fructose-1,6-diphosphate aldolase. J Bacteriol. 1966;92:470–476. doi: 10.1128/jb.92.2.470-476.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YP, Dilworth MJ, Glenn AR. Aromatic metabolism in Rhizobium trifolii -- protocatechuate 3,4-dioxygenase. Arch Microbiol. 1984;138:187–190. [Google Scholar]
- Das KK, Das SN, Dhundasi SA. Nickel, its adverse health effects & oxidative stress. Indian J Med Res. 2008;128:412–425. [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Cruz MS, Mendieta J, Monjonell A, Tauler R, Esteban M. Study of zinc-binding properties of glutathione by differential pulse polarography and multivariate curve resolution. Journal of Inorganic Biochemistry. 1998;70:91–98. [Google Scholar]
- Eberz G, Eitinger T, Friedrich B. Genetic determinants of a nickel-specific transport system are part of the plasmid-encoded hydrogenase gene cluster in Alcaligenes eutrophus. J Bacteriol. 1989;171:1340–1345. doi: 10.1128/jb.171.3.1340-1345.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitinger T, Suhr J, Moore L, Smith JAC. Secondary transporters for nickel and cobalt ions: Theme and variations. Biometals. 2005;18:399–405. doi: 10.1007/s10534-005-3714-x. [DOI] [PubMed] [Google Scholar]
- Gelinsky M, Vogler R, Vahrenkamp H. Zinc complexation of glutathione and glutathione-derived peptides. Inorganica Chimica Acta. 2003;334:230–238. [Google Scholar]
- Geslin C, Llanos J, Prieur D, Jeanthon C. The manganese and iron superoxide dismutases protect Escherichia coli from heavy metal toxicity. Res Microbiol. 2001;152:901–905. doi: 10.1016/s0923-2508(01)01273-6. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Sadhukhan PC, Chaudhuri J, Ghosh DK, Mandal A. Purification and properties of mercuric reductase from Azotobacter chroococcum. J Appl Microbiol. 1999;86:7–12. [Google Scholar]
- Hall DR, Leonard GA, Reed CD, Watt CI, Berry A, Hunter WN. The crystal structure of Escherichia coli class II fructose-1, 6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity. J Mol Biol. 1999;287:383–394. doi: 10.1006/jmbi.1999.2609. [DOI] [PubMed] [Google Scholar]
- Helbig K, Bleuel C, Krauss GJ, Nies DH. Glutathione and transition-metal homeostasis in Escherichia coli. J Bacteriol. 2008;190:5431–5438. doi: 10.1128/JB.00271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwig JS, Rowe JL, Chivers PT. Nickel homeostasis in Escherichia coli - the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol Microbiol. 2006;62:252–262. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- Iwig JS, Leitch S, Herbst RW, Maroney MJ, Chivers PT. Ni(II) and Co(II) sensing by Escherichia coli RcnR. J Am Chem Soc. 2008;130:7592–7606. doi: 10.1021/ja710067d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliri E, Grzyska PK, Hausinger RP. Kinetic and spectroscopic investigation of CoII, NiII, and N-oxalylglycine inhibition of the FeII/alpha-ketoglutarate dioxygenase, TauD. Biochem Biophys Res Commun. 2005;338:191–197. doi: 10.1016/j.bbrc.2005.08.223. [DOI] [PubMed] [Google Scholar]
- Kaluarachchi H, Chung KCC, Zambel DB. Microbial nickel proteins. Natural Product Reports. 2010;27:681–694. doi: 10.1039/b906688h. [DOI] [PubMed] [Google Scholar]
- Kasprzak KS, Sunderman FW, Jr, Salnikow K. Nickel carcinogenesis. Mutat Res. 2003;533:67–97. doi: 10.1016/j.mrfmmm.2003.08.021. [DOI] [PubMed] [Google Scholar]
- Kirby T, Blum J, Kahane I, Fridovich I. Distinguishing between Mn-containing and Fe-containing superoxide dismutases in crude extracts of cells. Arch Biochem Biophys. 1980;201:551–555. doi: 10.1016/0003-9861(80)90544-5. [DOI] [PubMed] [Google Scholar]
- Koch D, Nies DH, Grass G. The RcnRA (YohLM) system of Escherichia coli: A connection between nickel, cobalt, and iron homeostasis. Biometals. 2007;20:759–771. doi: 10.1007/s10534-006-9039-6. [DOI] [PubMed] [Google Scholar]
- Kowalik-Janowska T, Kozlowski H, Farkas E, Sóvágó I. Nickel ion complexes of amino acids and peptides. In: Sigel A, Sigel H, Sigel RKO, editors. Metal Ions in Life Sciences. Vol. 2. New York, NY: John Wiley & Sons, Ltd.; 2007. pp. 63–107. [Google Scholar]
- Li Y, Zamble DB. Nickel homeostasis and nickel regulation: An overview. Chem Rev. 2009;109:4617–4643. doi: 10.1021/cr900010n. [DOI] [PubMed] [Google Scholar]
- Liesegang H, Lemke K, Siddiqui RA, Schlegel HG. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J Bacteriol. 1993;175:767–778. doi: 10.1128/jb.175.3.767-778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwrier A, Knowles CJ. The purification and characterization of a novel D(−)-specific carbamoylase enzyme from an Agrobacterium sp. Enzyme Microbial Technol. 1996;19:562–571. [Google Scholar]
- Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L, Hausinger RP. Mechanisms of nickel toxicity in microorganisms. Metallomics. 2011 doi: 10.1039/c1mt00063b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JJ, Lebherz HG. Fructose-bisphosphate aldolases: An evolutionary history. Trends Biochem Sci. 1992;17:110–113. doi: 10.1016/0968-0004(92)90247-7. [DOI] [PubMed] [Google Scholar]
- Meredith TC, Woodard RW. Escherichia coli YrbH is a D-arabinose 5-phosphate isomerase. J Biol Chem. 2003;278:32771–32777. doi: 10.1074/jbc.M303661200. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Mulrooney SB, Hausinger RP. Metal ion dependence of recombinant Escherichia coli allantoinase. J Bacteriol. 2002;185:126–134. doi: 10.1128/JB.185.1.126-134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrooney SB, Hausinger RP. Nickel uptake and utilization by microorganisms. FEMS Microbiol Rev. 2003;27:239–261. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
- Navarro C, Wu LF, Mandrand-Berthelot MA. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol. 1993;9:1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. [Google Scholar]
- Niegowski D, Eshaghi S. The CorA family: structure and function revisited. Cell Mol Life Sci. 2007;64:2564–2574. doi: 10.1007/s00018-007-7174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemirowicz G, Parussini F, Agüero F, Cazzulo JJ. Two metallocarboxypeptidases from the protozoan Trypanosoma cruzi belong to the M32 family, found so far only in prokaryotes. Biochem J. 2007;401:399–410. doi: 10.1042/BJ20060973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten CE, O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- Rodrigue A, Effantin G, Mandrand-Berthelot MA. Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J Bacteriol. 2005;187:2912–2916. doi: 10.1128/JB.187.8.2912-2916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Okubo A, Yamazaki S. Characterization of a multi-copper enzyme, nitrous oxide reductase, from Rhodobacter sphaeroides f. sp. denitrificans. J Biochem. 1998;124:51–54. doi: 10.1093/oxfordjournals.jbchem.a022096. [DOI] [PubMed] [Google Scholar]
- Scamuffa MD, Caprioli RM. Comparison of the mechanisms of two distinct aldolases from Escherichia coli grown on gluconeogenic substrates. Biochim Biophys Acta. 1980;614:583–590. doi: 10.1016/0005-2744(80)90247-8. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Schlegel HG. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J Bacteriol. 1994;176:7045–7054. doi: 10.1128/jb.176.22.7045-7054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, Gourse RL. Changes in the concentrations of guanosine 5'-diphosphate 3'-diphosphate and the initiating nucleoside triphosphate account for inhibition of rRNA transcription in fructose-1,6-diphosphate aldolase (fda) mutants. J Bacteriol. 2003;185:6192–6194. doi: 10.1128/JB.185.20.6192-6194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreyer R, Böck A. Phenotypic suppression of a fructose-1,6-diphosphate aldolase mutation in Escherichia coli. J Bacteriol. 1973;115:268–276. doi: 10.1128/jb.115.1.268-276.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seffernick JL, McTavish H, Osborne JP, de Souza ML, Sadowsky MJ, Wackett LP. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP is a metalloenzyme. Biochemistry. 2002;41:14430–14437. doi: 10.1021/bi020415s. [DOI] [PubMed] [Google Scholar]
- Shi X, Dalal NS, Kasprzak KS. Generation of free radicals in reactions of Ni(II)-thiol complexes with molecular oxygen and model lipid hydroperoxides. J Inorg Biochem. 1993;15:211–225. doi: 10.1016/0162-0134(93)80026-6. [DOI] [PubMed] [Google Scholar]
- Stribling D, Perham RN. Purification and characterization of two fructose diphosphate aldolases from Escherichia coli (Crookes' strain) Biochem J. 1973;131:833–841. doi: 10.1042/bj1310833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- Wang S, Wu Y, Outten FW. Fur and the novel regulator YqjI control transcription of the ferric reductase gene yqjH in Escherichia coli. J Bacteriol. 2011;193:563–574. doi: 10.1128/JB.01062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf M, Fariduddin Q, Hayat S, Ahmad A. Nickel: An overview of uptake, essentiality and toxicity in plants. Bull Environ Contam Toxicol. 2011;86:1–17. doi: 10.1007/s00128-010-0171-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.