Abstract
Loss of FMR1 gene function results in fragile X syndrome (FXS), the most common heritable form of intellectual disability. The protein encoded from this locus (FMRP) is an RNA binding protein thought to primarily act as a translational regulator, however recent studies implicate FMRP in other mechanisms of gene regulation. Here, we demonstrate that the Drosophila fragile X homolog (dFMR1) biochemically interacts with the A-to-I RNA editing enzyme, dADAR. We found that dAdar and dfmr1 mutant larvae exhibit distinct morphological neuromuscular junction (NMJ) defects. Epistasis experiments based on these phenotypic differences suggest that dAdar acts downstream of dfmr1 and that dFMR1 modulates dADAR activity. Furthermore, sequence analyses revealed that loss or overexpression of dFMR1 affects editing efficiency on certain dADAR targets with defined roles in synaptic transmission. These results link dFMR1 with the RNA editing pathway and suggest that proper NMJ synaptic architecture requires modulation of dADAR activity by dFMR1.
Keywords: Fragile X syndrome, dFMR1, dADAR, RNA editing, Neuromuscular junction
Fragile X syndrome (FXS) is the most common heritable form of intellectual impairment and a known genetic cause of autism. Individuals with this disease exhibit learning disabilities and an array of behavioral and cognitive deficits, in addition to abnormal synaptic development and function1. The most prevalent genetic aberration associated with FXS arises from a CGG repeat in the 5′ untranslated region (UTR) of the fragile X mental retardation 1 gene (FMR1). Expansion of this region results in hypermethylation and subsequent transcriptional silencing of the FMR1 gene, preventing expression of the FMRP protein1.
Previous studies suggest that translational regulation by FMRP is essential for aspects of neuronal function and synaptic development. FMRP is an RNA binding protein that co-sediments with ribonucleoprotein (RNP) particles and translating polyribosomes2–5. In vitro and in vivo analyses suggest that FMRP functions as a translational repressor6,7, however recent studies demonstrate that FMRP can also positively regulate expression of certain target mRNAs8,9. Moreover, FMRP is present at synapses, where localized protein translation occurs, and is rapidly translated upon metabotropic glutamate receptor (mGluR) stimulation10. Therefore, it is proposed that FMRP regulates synaptic-localized protein translation and subsequently affects synaptic plasticity. How misregulation of this process leads to the synaptic and cognitive defects observed in FXS, however, still remains unclear.
Although the role for FMRP in translational regulation has been well studied, recent findings suggest that FMRP also affects other aspects of post-transcriptional gene regulation. FMRP regulates stability of certain transcripts, such as PSD-9511, and also regulates activity-dependent mRNA transport and localization in neurons12–14. In addition, FMRP interacts with multiple pathways that regulate gene expression, including the RNA interference (RNAi) pathway15–20. Collectively, these studies suggest multiple roles for FMRP in post-transcriptional gene regulation.
In this report, we present findings demonstrating that the Drosophila FMRP homolog (dFMR1) acts in the dADAR (Drosophila adenosine deaminase acting on RNA)-mediated RNA editing system. Through a tandem affinity purification screen, we identified dADAR as a protein that associates with dFMR1. ADARs act to catalytically deaminate adenosine (A) to inosine (I) residues in double-strand RNA templates21. Because inosine is interpreted as guanosine by the translational machinery, A-to-I editing in coding mRNAs may lead to the incorporation of amino acids not directly encoded by the genomic template21. Proper ADAR function is critical for neuronal integrity, as loss-of-function (lof) mutations in several model organisms result in severe neurological defects or lethality22–24, and mRNAs edited by ADARs are enriched for those encoding ion channels and genes important for synaptic transmission and architecture25.
To determine if dfmr1 and dAdar genetically interact, we took advantage of the neuromuscular junction (NMJ) system, where dFMR1 has previously been shown to function26. We found that dADAR expression and function is essential for normal NMJ morphology and that dAdar acts downstream of dfmr1 for proper NMJ synaptic architecture. Sequence analyses of dADAR substrates revealed that dFMR1 affects editing efficiency of particular transcripts involved in synaptic transmission. These findings demonstrate a novel mechanism by which FMRP affects gene regulation and function through its association with dADAR-dependent RNA editing.
Results
dFMR1 biochemically interacts with dADAR
To explore the biochemical function of dFMR1, we used a combined strategy of tandem affinity purification (TAP) followed by mass spectrometry analysis to identify novel interacting proteins27. We generated S2 cell lines expressing a recombinant form of the dFMR1 protein with a carboxy-terminal (C-terminal) TAP tag (dFMR1-cTAP) (Supplemental Fig. 1). Extracts from S2 cells expressing dFMR1-cTAP or cTAP alone were used for the TAP pulldown and eluates were subjected to SDS-PAGE electrophoresis (Supplemental Fig. 1).
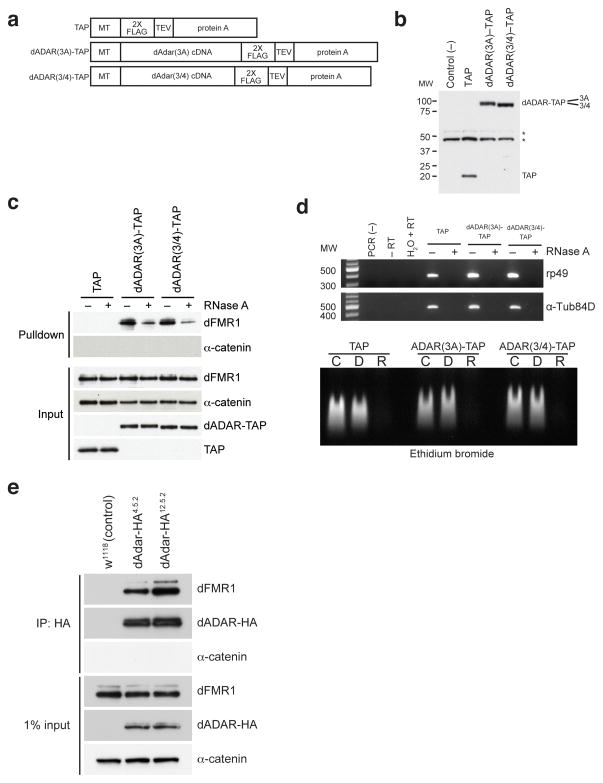
One protein that was identified in the dFMR1-cTAP-expressing eluates was dADAR, the A-to-I RNA editing enzyme. To verify the interaction between dFMR1 and dADAR, we transfected S2 cells with constructs that express two differentially spliced dADAR isoforms with a modified TAP tag28,29 (Fig. 1a–b). Utilizing a modified TAP method with dADAR-TAP-expressing lysates, Western analyses revealed that dFMR1 co-purifies with both isoforms of dADAR-TAP, but not with control samples (Fig. 1c), indicating that dFMR1 and dADAR associate in S2 cells.
Figure 1. dFMR1 biochemically interacts with dADAR in Drosophila S2 cell culture and in vivo.
(a) Structure of TAP (consisting of 2X FLAG and protein A sequences separated by a TEV cleavage site), dADAR(3A)-TAP and dADAR(3/4)-TAP constructs used to generate stable S2 cell lines. Constructs are under control of an inducible metallothionein (MT) promoter. (b) Western analysis showing expression of constructs in transfected S2 cells. Untransfected S2 cells were used as a negative control for the FLAG antibody. Astericks denote non-specific bands present in all samples that were detected by the anti-FLAG antibody. Molecular weight (MW) on left is measured in kilodaltons (kDa). (c) Eluates from TAP pulldown followed by TEV cleavage show that dFMR1 associates with dADAR-TAP in the presence of RNase A. Samples treated or untreated with RNase A are designated as (+) or (−), respectively. α-catenin was used as a loading control and does not associate with dADAR-TAP. A FLAG antibody was used to detect TAP constructs in input lanes. (d) RT-PCR analysis (upper panel) and ethidium bromide staining of total RNA (lower panel) on RNase A-treated and control lysates, showing efficient RNA degradation in RNase-treated samples. For RT-PCR analysis (upper panel), samples treated or untreated with RNase A are designated as (+) or (−), respectively. Primers against ribosomal protein 49 (rp49) and α-Tubulin84D (α-Tub84D) were used for PCR amplification. Molecular weight marker (MW) denotes size migration in basepairs (bp). For ethidium bromide staining of total RNA (lower panel), total RNA from TAP, dADAR(3A)-TAP, and dADAR(3/4)-TAP lysates were treated with DNase I (D), RNase A (R) or were untreated (C). (e) Co-IP experiments performed on head lysates prepared from w1118 (control) and two independent endogenously tagged dADAR-HA fly lines, dAdar-HA4.5.2 and dAdar-HA12.5.2. An HA antibody was used to detect dADAR-HA and α-catenin was used as a loading control and negative control for the co-IP.
To determine if this interaction is dependent on RNA, the TAP pulldown was performed in the presence of ribonuclease A (RNase A). dFMR1 reproducibly co-purified with dADAR-TAP in samples treated with RNase A, but to a lesser degree when compared to untreated samples (Fig. 1c). RT-PCR amplification and ethidium bromide staining of total RNA isolated from extracts used for the pulldown confirmed efficient removal of RNA from the RNase-treated samples (Fig. 1d). Because the interaction is not ablated by RNase A treatment, these results suggest that the association between dADAR and dFMR1 is possibly enhanced by the presence of RNA. It is also possible that a subset of dFMR1 pulled down in our TAP purification represents a pool that is not interacting with dADAR, but is simply tethered to RNAs that are also bound by dADAR.
To confirm that dFMR1 and dADAR interact in vivo, we performed co-immunoprecipitation (co-IP) experiments using two independent fly lines that contain an HA tag inserted in the endogenous dAdar open reading frame (dAdar-HA4.5.2 and dAdar-HA12.5.2)30. Co-IP experiments using adult head lysates revealed that dFMR1 associates with dADAR-HA in the two independent dAdar-HA lines, but not in control samples (Fig. 1e), demonstrating an interaction between dADAR and dFMR1 in vivo.
We next sought to determine where dFMR1 and dADAR associate within the cell, as both proteins exhibit different localization patterns at the sub-cellular level. Whereas dADAR primarily resides in the nucleus where A-to-I deamination normally occurs, FMRP is mostly observed in the cytoplasm despite containing nuclear localization and export signals31,32. Because treatment of COS cells with leptomycin B (LMB), an inhibitor of exportin 1-dependent nuclear export, is able to trap FMRP in the nucleus33, we assessed dFMR1 and dADAR sub-cellular localization in S2 cells treated with LMB to increase the amount of dFMR1 in the nucleus (Supplemental Fig. 2). As previously shown30,33, we observed nuclear-specific localization of dADAR in S2 cells expressing dADAR-TAP recombinant protein as well as an increase in the amount of dFMR1 within the nucleus by approximately 2.4 fold relative to vehicle control-treated cells, verifying that dFMR1 and dADAR are both expressed within the nucleus (Supplemental Fig. 2). Furthermore TAP purification using dADAR-TAP-expressing S2 cells treated with LMB increased the amount of dFMR1 pulled down with dADAR-TAP by at least 2 fold compared to control samples (Supplemental Fig. 2). Based on these results, we predict that dFMR1 and dADAR associate within the nucleus.
dAdar mutant larvae exhibit morphological NMJ defects
To explore the physiological significance of the dFMR1:dADAR biochemical interaction, we utilized the neuromuscular junction (NMJ) system in third instar larvae. Previous studies indicate that loss and overexpression of dFMR1 give rise to opposing defects in NMJ synaptic architecture15,26. Although NMJ defects in dAdar mutants have never been identified, loss of ADAR expression leads to severe behavioral and neuronal defects in Drosophila and other model systems22–24. Furthermore, many characterized dADAR substrates encode for proteins that function in synaptic transmission at the NMJ34.
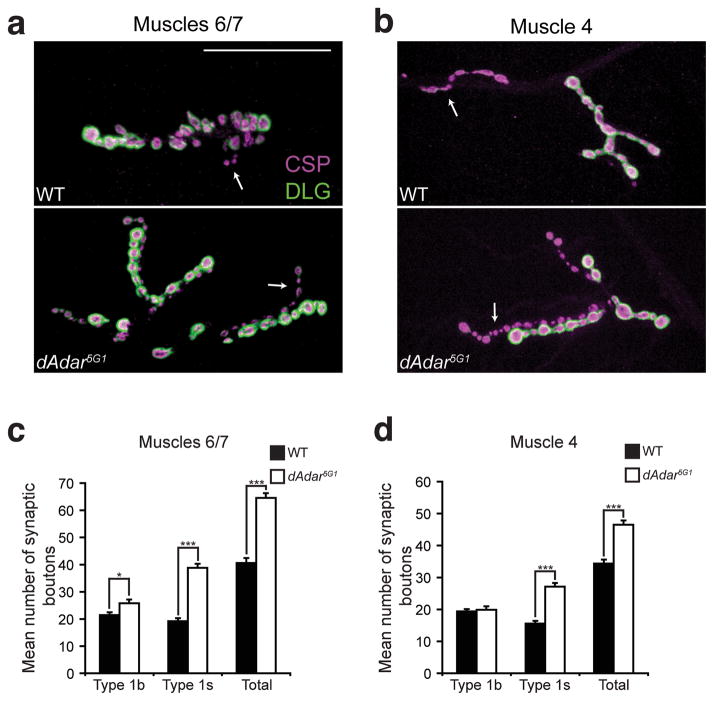
Analyses of dAdar5G1 null larvae22 revealed morphological NMJ defects in two different muscles analyzed (Fig. 2a–b). dAdar5G1 mutant larvae exhibited a 59% and 35% increase in the total number of type 1 synaptic boutons compared to wild-type (WT) samples in muscles 6/7 (Fig. 2c) and muscle 4 (Fig. 2d), respectively. Quantification of branching in muscles 6/7 also revealed that dAdar5G1 mutants exhibit an increase in branching compared to control samples (6.2 ± 0.2 in dAdar5G1 compared to 3.5 ± 0.2 in WT; n≥16, p<0.001). To further examine the dAdar5G1 mutant phenotype, we next quantified the two subclasses of type 1 synaptic boutons (type 1s and type 1b) which exhibit different electrophysiological properties35. We found that the increase in type 1 synaptic boutons in dAdar5G1 mutants arises largely from an increase in type 1s synaptic boutons, whereas type 1b synaptic boutons are subtly affected, if at all, when compared to control genotypes (Fig. 2c–d). From these data, we conclude that loss of dADAR expression leads to defects in NMJ synaptic architecture.
Figure 2. dAdar5G1 mutants exhibit NMJ defects in third instar larvae.
(a,b) Confocal images of muscles 6/7 (a) or muscle 4 (b) from WT (w1118) and dAdar5G1 L3 larvae. Presynaptic vesicles were stained for Cysteine-string protein (CSP, magenta) and Discs-large (DLG, green) was used as a postsynaptic marker. Type 1b and 1s boutons were distinguished by 1) the intensity of DLG staining, as DLG fluorescence is more intense on type 1b boutons relative to type 1s boutons, and 2) size, as type 1b boutons are larger than type 1s boutons. DLG staining was observed on type 1s synaptic boutons in all genotypes, however images were taken to keep visualization of DLG on type 1s boutons low to distinguish between the bouton subclasses. No noticeable differences in CSP and DLG intensity were observed across genotypes. Arrows indicate type 1s synaptic boutons. Scale bar represents 50 μm. (c,d) Quantification of average number of type 1b, type 1s, and total type 1 synaptic boutons in muscles 6/7 (c) or muscle 4 (d) of WT (black bars) or dAdar5G1 null (white bars) larvae. All images and quantification were performed using abdominal hemisegment A3. n≥16 for each genotype. Error bars denote s.e.m. *p<0.05, ***p<0.0001, WT vs. dAdar5G1 by Student’s t-test.
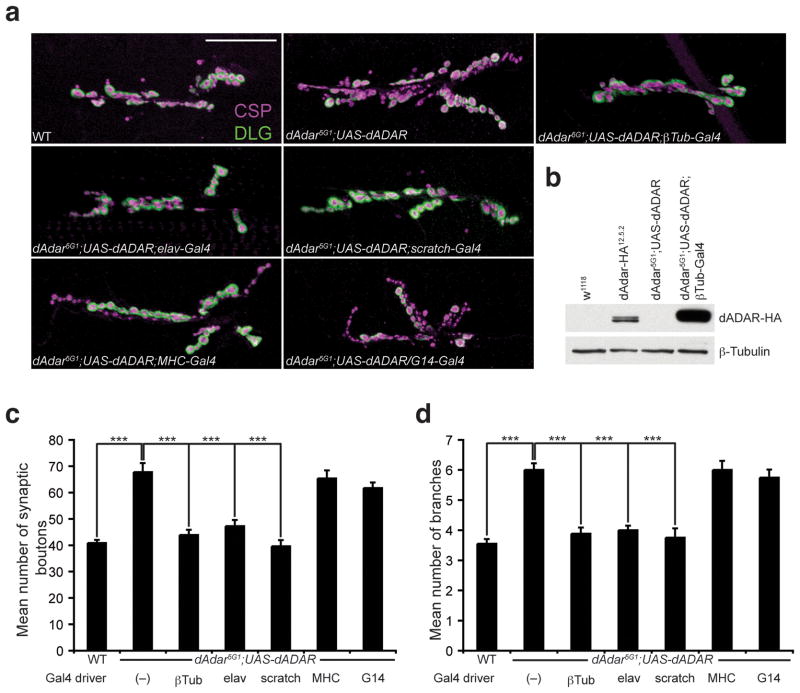
To validate the dAdar5G1 mutant phenotype and to determine where dADAR is required for normal NMJ synaptic architecture, we used the Gal4/UAS system to express dADAR in tissue-specific regions (Fig. 3a). To verify that the NMJ defects observed in dAdar5G1 mutant larvae are due to an absence of dADAR expression and not due to genetic background effects, we expressed dADAR ubiquitously from a wild-type UAS-dADAR transgene using a β-Tubulin-Gal4 (βTub-Gal4) driver, which expressed dADAR to higher levels compared to endogenous dADAR (Fig. 3b). Ubiquitous expression of dADAR completely rescued the NMJ phenotype observed in dAdar5G1 larvae with respect to synaptic bouton number and branching (Fig. 3c–d and Supplemental Table 1), supporting our findings that dADAR expression is required for aspects of NMJ morphology.
Figure 3. Neuronal expression of dADAR is sufficient for normal NMJ synaptic architecture.
(a) Confocal images of the NMJ from the following genotypes: WT, dAdar5G1 larvae carrying a wild-type UAS-dADAR transgene (dAdar5G1;UAS-dADAR), dAdar5G1;UAS-dADAR;βTub-Gal4 (ubiquitous expression), dAdar5G1;UAS-dADAR;elav-Gal4 (neuronal expression), dAdar5G1;UAS-dADAR;scratch-Gal4 (neuronal expression), dAdar5G1;UAS-dADAR;MHC-Gal4 (muscle expression), and dAdar5G1;UAS-dADAR/G14-Gal4 (muscle expression). Larvae were stained for CSP (presynaptic, magenta) and DLG (postsynaptic, green). No noticeable differences in CSP and DLG intensity were observed across genotypes. Scale bar represents 50 μm. (b) Western analysis on L3 brain extracts from the following genotypes: w1118 (negative control for the HA antibody), dAdar-HA12.5.2 (for endogenous levels), dAdar5G1;UAS-dADAR and dAdar5G1;UAS-dADAR;βTub-Gal4. An antibody against HA was used to detect dADAR-HA expression (upper panel). β-Tubulin (lower panel) was used as a loading control. (c,d) Quantification of the average number of type 1 synaptic boutons (c) and synaptic branching (d) in muscles 6/7 for all genotypes. All images and quantification were performed using abdominal hemisegment A3, muscles 6/7. n≥16 for each genotype. Error bars denote s.e.m. ***p<0.001 analyzed by one-way ANOVA, p<0.0001 overall, Tukey-Kramer post-test. See Supplemental Table 1 for control genotypes.
We next determined if pre- or postsynaptic expression of dADAR was sufficient to rescue the morphological phenotypes observed in dAdar5G1 mutant larvae. We found that neuronal expression of dADAR using the elav-Gal4 and scratch-Gal4 drivers rescued the NMJ defects observed in dAdar5G1 mutants (Fig. 3c–d and Supplemental Table 1). In contrast, postsynaptic expression of dADAR in muscle using either the myosin heavy chain-Gal4 (MHC-Gal4) or G14-Gal4 drivers failed to rescue the dAdar5G1 NMJ phenotype (Fig. 3c–d and Supplemental Table 1). We therefore conclude that neuronal expression of dADAR is sufficient for proper NMJ synaptic architecture. These findings parallel published reports with respect to the spatial requirements of dFMR1 for proper NMJ morphology36.
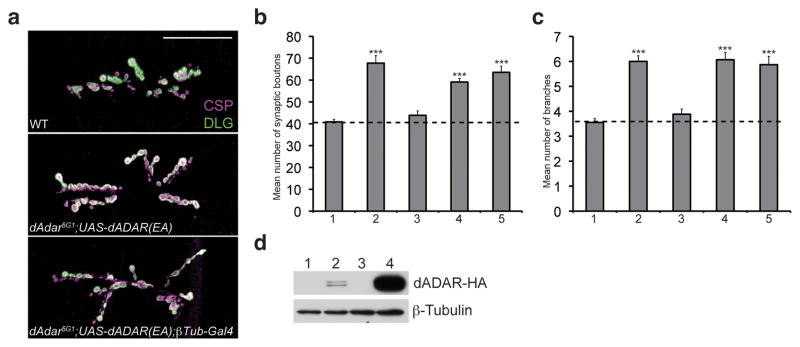
To determine if normal NMJ synaptic architecture is dependent on the A-to-I RNA editing-function of dADAR, we took advantage of an active site glutamate (E) to alanine (A) substitution in motif I of the catalytic deaminase domain that severely reduces catalytic activity37. Ubiquitous expression of the UAS-dADAR(EA) transgene in an dAdar mutant background results in minimal rescue in locomotor activity and significantly reduced levels of editing37, but can still compete with wild-type dADAR isoforms for RNA substrates38. We crossed the UAS-dADAR(EA) transgene into the dAdar5G1 null background to determine if the catalytically inactive dADAR(EA) protein would rescue the dAdar5G1 NMJ defects. We found that the dADAR(EA) transgene driven with a βTub-Gal4 driver was unable to rescue the NMJ defects observed in dAdar5G1 mutant larvae (Fig. 4a–c) despite being expressed at higher levels compared to endogenous dADAR (Fig. 4d). These results demonstrate that functional deaminase activity of dADAR is required for normal NMJ synaptic architecture.
Figure 4. Deaminase activity by dADAR is essential for normal NMJ synaptic architecture.
(a) Confocal images of the NMJ from the following genotypes: WT, dAdar5G1 larvae carrying a UAS-dADAR(EA) transgene to express a catalytic mutant form of dADAR (dAdar5G1;UAS-dADAR(EA)), and dAdar5G1;UAS-dADAR(EA);βTub-Gal4. Larvae were stained for CSP (presynaptic, magenta) and DLG (postsynaptic, green). No noticeable differences in CSP and DLG intensity were observed across genotypes. Scale bar represents 50 μm. (b,c) Quantification of the average number of type 1 synaptic boutons (b) and synaptic branches (c) in muscles 6/7 for WT (1), dAdar5G1;UAS-dADAR(WT) (2), dAdar5G1;UAS-dADAR(WT);βTub-Gal4 (3), dAdar5G1;UAS-dADAR(EA) (4), and dAdar5G1;UAS-dADAR(EA);βTub-Gal4 (5). All images and quantification were performed using abdominal hemisegment A3, muscles 6/7. n≥16 for each genotype. Error bars denote s.e.m. ***p<0.001 analyzed by one-way ANOVA, p<0.0001 overall, Tukey-Kramer post-test. (d) Western analysis on lysates purified from w1118 (1), dAdar-HA12.5.2 (2), dAdar5G1;UAS-dADAR(EA) (3), and dAdar5G1;UAS-dADAR(EA);βTub-Gal4 (4) L3 larval brains. As observed with the wild-type dADAR transgene (see Fig. 3b), dADAR(EA) expression was higher compared to endogenous dADAR levels. Upper panel shows dADAR-HA expression and β-Tubulin (lower panel) served as a loading control.
dAdar is epistatic to dfmr1 with respect to NMJ morphology
As our results demonstrated that dADAR expression and catalytic activity are required for normal NMJ synaptic architecture, and previous studies reported NMJ defects in dfmr1 mutants15,26, we directly compared the dfmr1 and dAdar5G1 mutant phenotypes. Quantitative analyses using dAdar5G1 and dfmr13 mutant animals revealed that loss of either dADAR or dFMR1 expression results in a similar increase in the number of type I synaptic boutons (66.6 ± 1.9 in dAdar5G1 and 63.3 ± 2.3 in dfmr13 compared to 40.8 ± 2.3 in WT; n≥16, p<0.001) and branching (6.2 ± 0.2 in dAdar5G1 and 6.0 ± 0.2 in dfmr13 compared to 3.5 ± 0.2 in WT; n≥16, p<0.001) compared to wild-type controls. However, morphological analyses indicated that the phenotypes observed in the single mutants are distinct from each other (Fig. 5a). To examine the phenotypic difference in more detail, we quantified the morphological synaptic defects based two categories: subclasses of type 1 synaptic boutons and primary branch length. Although dAdar mutants exhibit a greater increase in type 1s synaptic boutons than type 1b boutons in L3 larvae compared to controls, loss of dFMR1 expression leads to similar increases in both type 1b and type 1s synaptic boutons compared to wild-type samples (Fig. 5b). In addition, previous reports26 and our analyses found that dfmr13 mutant larvae exhibit an increase in the length of the primary nerve branch when compared to control samples, whereas the primary branch length in dAdar5G1 mutants is largely unaffected in comparison to control larvae (Fig. 5c). We therefore conclude that although both dADAR and dFMR1 affect NMJ synaptic architecture, the single mutant phenotypes are distinct from one another.
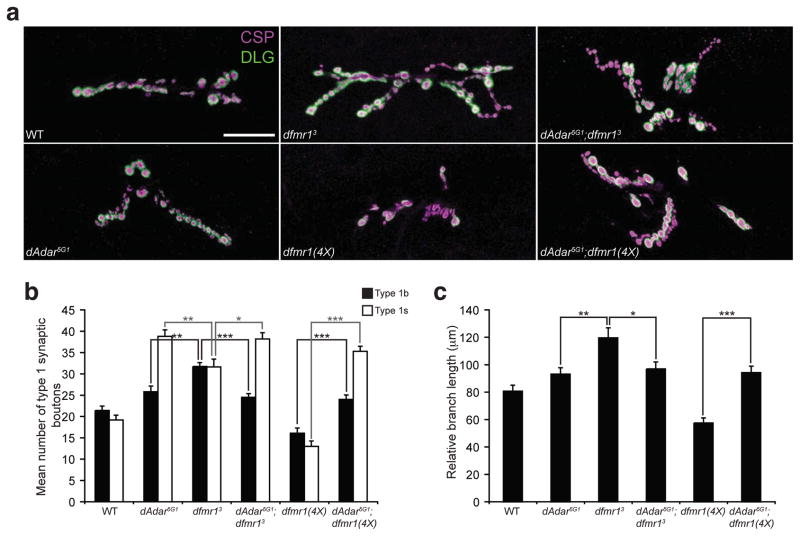
Figure 5. dAdar and dfmr1 genetically interact.
(a) Confocal images of L3 larval NMJs from the following genotypes: WT, dAdar5G1, dfmr13, dfmr1(4X), dAdar5G1;dfmr13 and dAdar5G1;dfmr1(4X). L3 larvae were stained for CSP (presynaptic, magenta) and DLG (postsynaptic, green). Type 1b and type 1s synaptic boutons were distinguished as described in Fig. 2. No noticeable differences in CSP and DLG intensity were observed across genotypes. Scale bar represents 50 μm. (b) Genetic studies demonstrating that dAdar is epistatic to dfmr1 with respect to synaptic bouton formation. Quantification of average number of type 1b (black bars) and type 1s (white bars) synaptic boutons for genotypes shown in (a). (c) Genetic studies demonstrating that dAdar is epistatic to dfmr1 with respect to primary branch length. Relative primary branch length was quantified for all genotypes shown in (a) using anti-HRP staining. The length of the primary branch length was normalized to the length of the abdominal hemisegment, and mean relative primary branch length was measured and plotted for each genotype. All images and quantification were performed using muscles 6/7, hemisegment A3. n≥16 for each genotype. Error bars denote s.e.m. *p<0.05, **p<0.01, ***p<0.001, analyzed by one-way ANOVA, p<0.0001 overall, Tukey-Kramer post-test.
We utilized the distinct NMJ phenotypes in dfmr1 and dAdar mutants to perform genetic epistasis experiments. Comparison of the dAdar5G1;dfmr13 double mutant larvae to the dAdar5G1 and dfmr13 single mutants revealed that the dAdar5G1;dfmr13 double null larvae exhibit an dAdar5G1 single mutant-like phenotype with respect to the sub-classes of type 1 synaptic boutons (Fig. 5b) and primary branch length (Fig. 5c). Overexpression of dFMR1 leads to NMJ morphological defects characterized by a reduction in the number of synaptic boutons, branching, and primary branch length compared to control samples15,26. Quantitative analyses of dAdar5G1;dfmr1(4X) larvae revealed that loss of dADAR activity in dfmr1(4X) overexpression mutants also results in an dAdar5G1 null phenotype with respect to type 1s and type 1b synaptic boutons (Fig. 5b), primary branch length (Fig. 5c), and the number of synaptic branches (data not shown). Thus, neither loss nor overexpression of dFMR1 has any impact on NMJ morphology in the absence of dADAR expression. From these results we conclude that dfmr1 and dAdar genetically interact and that dAdar is epistatic to dfmr1 with respect to NMJ synaptic architecture. Because we observe a physical interaction between dFMR1 and dADAR, we propose that dfmr1 and dAdar are in a similar pathway with respect to NMJ synaptic development, and that dAdar functions downstream of dfmr1.
To better characterize the genetic interaction between dAdar and dfmr1 at the NMJ, we examined NMJ architecture in trans-heterozygous genotypes. We reduced dAdar dosage in a dfmr1 lof or gain-of-function (gof) background and quantified the number of type 1s and type 1b synaptic boutons in various dAdar;dfmr1 combinatorial genotypes. Strikingly, reduction of dAdar dosage in a dfmr13 mutant background genetically suppressed the dfmr13 null NMJ phenotype to wild-type with respect to the number of type 1 synaptic boutons (Fig. 6). We also reduced dAdar dosage in dfmr1(4X) overexpressing flies but saw neither rescue nor enhancement of the dfmr1(4X) phenotype (Supplemental Fig. 3). These results further support the model that dAdar is downstream to dfmr1 with respect to NMJ synaptic architecture. Because the dfmr13 NMJ phenotype was rescued by reducing dAdar dosage, we predict that loss of dFMR1 expression leads to aberrant dADAR activity, which can be prevented by reducing dAdar dosage.
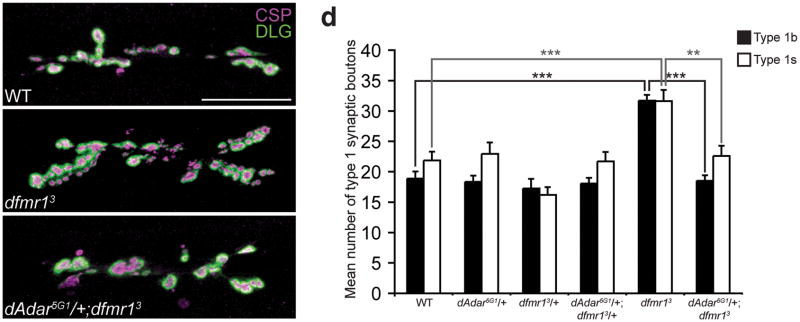
Figure 6. Reduction of dAdar dosage rescues the dfmr13 null NMJ defects in L3 larvae.
(a–c) Confocal images of WT (a), dfmr13 (b), and dAdar5G1/+;dfmr13 (c) L3 larval NMJs. Larvae were stained for CSP (presynaptic, magenta) and DLG (postsynaptic, green). Type 1b and type 1s synaptic boutons were distinguished as described in Fig. 2. No noticeable differences in CSP and DLG intensity were observed across genotypes. Scale bar represents 50 μm. (d) The reduction of dAdar dosage rescues the dfmr1 synaptic bouton phenotype, as revealed by quantification of average number of type 1b (black bars) and type 1s (white bars) synaptic boutons for trans-heterozygous genotypes using the dAdar5G1 and dfmr13 mutant alleles. All images and quantification were performed using muscles 6/7, hemisegment A3. n≥16 for each genotype. Error bars denote s.e.m. **p<0.01, ***p<0.001, analyzed by one-way ANOVA, p<0.0001 overall, Tukey-Kramer post-test.
dFMR1 modulates dADAR-dependent RNA editing activity
Our genetic analyses suggest that dFMR1 modulates dADAR function with respect to NMJ development. Therefore, we next explored how dFMR1 is acting on dADAR activity to regulate NMJ architecture. Because dFMR1 can act as a translational regulator, we first examined dADAR expression and localization in dfmr1 mutant backgrounds. Western and immunohistochemistry analyses failed to reveal a detectable change in both dADAR-HA expression and localization in dfmr1 lof and gof backgrounds compared to wild-type (Supplemental Fig. 4). Similarly, Western analyses on all genotypes used for the epistasis experiments revealed that dFMR1 was expressed at expected levels for all genotypes (Supplemental Fig. 5), indicating to us that the genetic interaction between dFMR1 and dADAR is not due to alterations in expression levels of the two proteins.
To determine if dADAR’s editing function is influenced by dFMR1, we examined editing efficiency of known dADAR substrates in dfmr1 mutant backgrounds. We found that altering dFMR1 expression leads to differential effects on editing efficiency in five of the six transcripts analyzed: lap, Caα1D, shab, stn-B, and syt-1 (Fig. 7a). Interestingly, editing levels at two transcripts analyzed (lap and Caα1D) exhibited a clear bi-directional change in response to reduced or increased dFMR1 expression (Fig. 7a–b). Editing patterns at other sites showed non-linear changes in response to changes in dFMR1 expression levels. For example, editing levels of sites 3 and 5 of the shab potassium channel were significantly higher in controls compared to dfmr13 null and dfmr1(4X) overexpressing larvae (Fig. 7a).
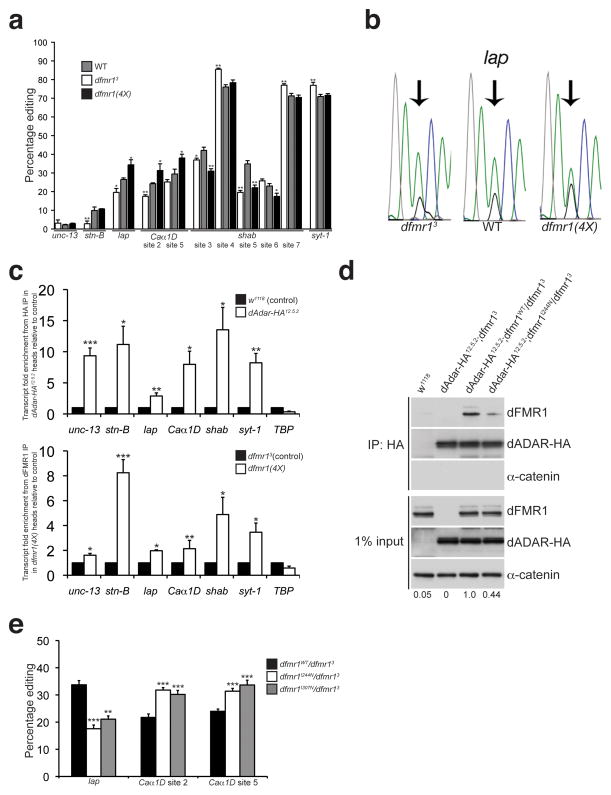
Figure 7. dFMR1 modifies dADAR function and affects A-to-I editing efficiency.
(a) Percentage of editing observed in samples from WT (gray bars), dfmr13 (white bars), and dfmr1(4X) (black bars) larvae was quantified and graphed for the following transcripts: unc-13, stoned-B (stn-B), lap, Caα1D, shab, and synaptotagmin-1 (syt-1). n = 3–7 individual RT-PCR reactions for each site. *p<0.05, **p<0.01, WT vs. dfmr13 and WT vs. dfmr1(4X) were analyzed with Mann Whitney-U test. Error bars denote s.e.m. (b) Representative electropherograms for the lap edited adenosine sequenced from WT, dfmr13, and dfmr1(4X) whole larval cDNA. Arrows point to edited sites analyzed. Green peak represents unedited (A) site, and black peak represents edited (G) site. (c) dADAR and dFMR1 associate with edited transcripts in vivo. RNA immunoprecipitation of transcripts associating with dADAR-HA or dFMR1 in adult head lysates. Fold enrichment of transcripts in dAdar-HA12.5.2 samples relative to w1118 was performed for the dADAR-HA RNA immunoprecipitation in the upper graph, and fold enrichment of transcripts in dfmr1(4X) overexpressing flies relative to dfmr13 null flies is shown for the dFMR1 RNA immunoprecipitation (lower graph). TBP was used as an unedited, non-specific transcript. Quantification of transcript enrichment was performed by using quantitative RT-PCR and fold enrichment was normalized to fold change of actin mRNA. Results represent four independent immunoprecipitation experiments for each HA and dFMR1 RNA IP and quantitative RT-PCR experiments was performed using three technical replicates. *p<0.05, **p<0.01, ***p<0.001, analyzed with Student’s t-test. Error bars denote s.e.m. (d) A mutation in the KH1 RNA binding domain of dFMR1 reduces the robustness of the dFMR1:dADAR biochemical interaction. co-IP experiments were performed with flies containing a wild-type dFMR1 construct (dfmr1WT) or a construct containing a point mutation in the dFMR1 KH1 domain (dfmr1I244N) that were crossed to the dAdar-HA12.5.2;dfmr13 fly line. w1118 flies served as a negative control for the HA antibody. An HA antibody was used to detect dADAR-HA and α-catenin was used as a loading control and negative control for the co-IP. Expression levels of dFMR1 in the IP samples were normalized to dFMR1 input levels and average fold change relative to dAdar12.5.2;dfmr1WT/dfmr13 is denoted below each lane. Experiment was performed three times. (e) Mutations in the dFMR1 KH domains affect editing of lap and Caα1D. Percentage of editing observed in dfmr1WT/dfmr13 (black bars), dfmr1I244N/dfmr13 (white bars), and dfmr1I307N/dfmr13 (gray bars) whole larvae. n = 3 8 individual RT-PCR reactions for each site. **p<0.01, ***p<0.001, dfmr1WT/dfmr13 vs. dfmr1I244N/dfmr13 and dfmr1WT/dfmr13 vs. dfmr11307N/dfmr13 were analyzed with Mann Whitney-U test. Error bars denote s.e.m.
We next ascertained whether dFMR1 associates with transcripts that are edited by dADAR using RNA immunoprecipitation followed by quantitative RT-PCR. As a positive control, we first demonstrated that dADAR-HA associates with the six edited transcripts used for the sequencing analysis (Fig. 7c). We observed an approximate 2.8–13.5 fold enrichment of edited transcripts in the dAdar-HA12.5.2 immunoprecipitated complexes compared to control samples, but no significant enrichment of a control unedited transcript TBP (Fig. 7c). We further found that Caα1D, lap, stn-B, shab, unc-13, and syt-1 were all enriched in the dFMR1-immunoprecipitated complexes in dfmr1(4X) head extracts compared to dFMR1-deficient extracts by between 1.6–8.2 fold (Fig. 7c), demonstrating that dFMR1 and dADAR-HA associate with similar transcripts in vivo.
Because we found that the presence of RNA enhances the biochemical association between dFMR1 and dADAR, we next sought to determine if dFMR1’s ability to bind RNA impacts its ability to associate with dADAR as well as modulate dADAR’s editing activity. We crossed transgenic flies containing point mutations in the RNA-binding KH domains of dFMR1 (dfmr1I244N and dfmr1I307N)39 to dAdar-HA12.5.2;dfmr13 flies and performed co-IP experiments to first determine if a reduction in RNA binding affects dFMR1’s ability to associate with dADAR. We found that a point mutation in the KH1 domain (I244N) reduces the abundance of dFMR1 pulled down with dADAR-HA by approximately 60% compared to control samples expressing a wild-type dFMR1 genomic construct (Fig. 7d). Similar assays were performed with the I307N mutation, but our results were inconclusive due to instability of the mutant dFMR1 protein during the pulldown procedure (data not shown). This finding parallels our RNase A experiments presented earlier and indicates that RNA binding by dFMR1 enhances the dFMR1:dADAR interaction.
We next assessed editing efficiency in the dfmr1 mutant strains to determine if a reduction in RNA binding by dFMR1 affects dADAR’s function. We sequenced lap and Caα1D, as both transcripts exhibited bidirectional effects on editing in the dfmr1 lof and gof backgrounds (Fig. 7a). We predicted that a decrease in RNA binding by dFMR1 would mimic the dfmr13 null phenotype, where we observed a decrease in editing in lap and Caα1D compared to control samples (Fig. 7a). Indeed, we found that both the dfmr1I244N and dfmr1I307N mutations in the dFMR1 KH domains decreased the percentage of editing in the lap transcript (Fig. 7e). In contrast, each point mutation in the dFMR1 KH domains increased editing at sites 2 and 5 of Caα1D (Fig. 7e), suggesting that mutations in the dFMR1 KH motifs lead to differential effects on editing at particular transcripts, which we discuss below. Collectively, these data suggest that dFMR1 is able to associate with dADAR substrates and modify dADAR activity by affecting the editing efficiency of a subset of edited transcripts.
Discussion
The genetic and molecular analyses in this study lead us to propose that modulation of dADAR activity by dFMR1 is important for NMJ synaptic architecture. The epistatic relationship of these two genes, the requirement of RNA editing by dADAR for normal NMJ morphology, and genetic suppression of the dfmr1 lof NMJ defects all support a model in which dFMR1 affects the editing activity of dADAR. Molecular analyses of dADAR substrates support this prediction, as we found that both loss and overexpression of dFMR1 result in changes in editing efficiency in several dADAR-dependent editing sites. While the changes in editing observed in dfmr1 mutant whole larvae are not large (eg: a ~15% change in editing was observed for lap), they are statistically significant, and we propose that these changes would likely be larger if analyses could be performed using mRNA prepared from isolated neurons or synapses rather than whole larvae. In addition, despite a few transcripts that are highly edited throughout development, dADAR function during developmental stages is relatively low compared to its high editing activity in pupal and adult stages29. Therefore, we cannot rule out a larger effect of dFMR1 levels on dADAR substrates with lower efficiency editing sites.
In considering how dFMR1 affects editing, an important clue might come from the fact that both dFMR1 and dADAR are RNA binding proteins that associate with secondary and higher order RNA structures. FMRP can bind to two separate complex RNA structures that are believed to allow for specificity of FMRP-associated transcripts: the RGG domain in the C-terminus of FMRP protein interacts with an intramolecular G quartet stem loop RNA structure whereas the KH2 domain associates with a complex tertiary kissing complex RNA structure40,41. Similarly, the dADAR family of proteins contains several double-stranded RNA binding domains (dsRBD) and requires duplex RNA structures to identify, bind to, and function on its target RNAs21. The RNA structure required for dADAR activity, however, can vary from a simple hairpin to complex pseudoknot structures.
Additionally, our immunoprecipitation experiments indicate that dFMR1 and dADAR associate on common RNA targets. The dADAR:dFMR1 biochemical interaction was reduced through both a decrease in the amount of RNA in our lysates using RNase A as well as by mutating the KH domains of dFMR1, suggesting to us that the ability for dFMR1 to bind to RNA plays an important role in its association with dADAR. Molecular analyses of lap and Caα1D in the dFMR1 RNA binding-mutants further support this theory, albeit, differential effects were observed with respect to the two transcripts analyzed. It is possible that dFMR1 associates with these two particular transcripts via different RNA binding motifs. Although the analogous I307N mutation in mammals reduces FMRP’s ability to associate with both poly(U)-rich sequences and large RNP complexes42,43, FMRP can still bind to RNA, including transcripts containing G-quartet structures, through an intact RGG RNA binding motif42. Therefore, we propose that the I244N and I307N mutations in dFMR1 reduce particular dFMR1:dADAR complexes associating with certain edited transcripts while concurrently enriching for dFMR1:dADAR complexes associating with the dFMR1 RGG box. Further studies delving into the importance of each RNA binding domain in both dFMR1 and dADAR will give more insight into the biochemical and functional interaction between these two proteins.
Based on our results, we predict that dFMR1 and dADAR can associate in a common complex and converge on similar RNA substrates. Because the effect that dFMR1 has on the editing efficiency is context dependent, we propose that the association of dFMR1 with dADAR has no net positive or negative effect on the editing activity of dADAR, but instead maintains a balance of dADAR activity. At sites that demonstrate enhanced editing in the presence of dFMR1, dFMR1 could promote editing by either recruiting dADAR to the site via its own RNA binding activity, or it could help form and/or stabilize RNA structures that create a site for editing by dADAR. At sites that are negatively affected by the presence of dFMR1, we propose that the RNA binding activity of dFMR1 interferes with the formation of a substrate for dADAR (Supplemental Fig. 6).
Our analyses revealed several transcripts whose level of editing is regulated by the interaction between dFMR1 and dADAR, however at this time we do not know how many such transcripts are key to the proper formation of the NMJ. Although many identified dADAR targets encode for proteins that function in synaptic transmission at the NMJ34 and mutations in several dADAR substrates (eg: syt-1, lap, and unc-13) affect NMJ synaptic architecture and/or function44–46, how editing is affecting the function of most of these gene products remains unknown. It is also important to note that a role for dFMR1 in translational regulation is already proposed to be important for proper NMJ development through its interaction with the microtuble-associated protein (MAP1B) homolog futsch26. Collectively, these studies suggest that both dADAR and dFMR1 play multifaceted roles at the NMJ.
In summary, we demonstrate that dFMR1 physically and genetically interacts with dADAR-dependent RNA editing. This is the first study to report a disease-associated protein that associates with and modulates A-to-I RNA editing. In addition, our findings introduce a novel function for FMRP with respect to neuronal architecture and expands FMRP’s predicted role as a translational regulator. Understanding all the mechanisms by which FMRP functions to regulate synaptic development and function is essential to better understand the pathogenesis of the FXS symptoms, and consequently can lead to effective therapeutic treatments for people afflicted with this disease.
Methods
Genetics and fly stocks
Fly stocks were raised on standard cornmeal/yeast/molasses/agar medium at 25°C. w1118 (Bloomington) was used as the wild-type control stock. The dfmr13 allele was generated by P-element excision and was characterized as a molecular null allele47 and the dfmr1(4X)15,47 overexpression fly line contains a dfmr1 genomic rescue construct on the second chromosome that increases dFMR1 expression 2–4 fold compared to wild-type15. The dAdar5G1 null fly line was generated by P-element mutagenesis and excision which deleted the entire coding region of the genomic dAdar locus22. Flies containing the wild-type UAS-dADAR or UAS-dADAR(EA) transgene37 were crossed into the dAdar5G1 null background for the Gal4/UAS experiments. The ubiquitous βTub-Gal4 driver was obtained from Bloomington. The neuronal drivers used in this study were elav-Gal4 (Bloomington) and scratch-Gal4 (kindly provided by Cynthia Hughes). The MHC-Gal4 and G14-Gal4 muscle drivers used for the rescue experiments were kindly provided by Greg Bashaw. The dAdar4.5.2 and dAdar12.5.2 fly lines contain the HA sequence in the endogenous dAdar locus (inserted by homologous recombination) to express an HA-tag in the C-terminus of the endogenous dADAR protein30. Transgenic fly lines expressing either the control or RNA binding mutant forms of dFMR1 in a genomic rescue fragment on the third chromosome (dfmr1WT, dfmr1I244N, and dfmr1I307N)39 were a generous gift from Tom Dockendorff.
Constructs
The pUAST-dfmr1 construct was generated by cloning the 14.5 kb dfmr1 genomic rescue construct47 into the pUAST transformation vector. The pUAST-dfmr1-cTAP vector was then generated by inserting the TAP tag48 into a unique site into exon 12 of the dfmr1 genomic construct in the pUAST-dfmr1 vector. The pUAST-dfmr1-cTAP and pUAST-cTAP (a kind gift from the Artavanis-Tsakonas lab) constructs were transfected in with the inducible pMT-Gal4 construct (a kind gift from Greg Bashaw) and pCoBlast (Invitrogen) for stable cell line selection.
To generate the dADAR-TAP constructs, dAdar(3A) and dAdar(3/4) cDNA were PCR amplified with the addition of EcoRI and BamHI restriction sites on the forward and reverse primers, respectively. The TAP tag was cut out of pINX-C-FF-ZZ-B (kindly provided by Russ Carstens) using BamHI and AclI. Both were gel purified (Qiagen) and ligated into pCoBlast-MT (a kind gift from Greg Bashaw) cut with EcoRI and AccI to produce pCoBlast-dAdar-TAP. To generate the TAP control construct, TAP sequence was PCR amplified using BamHI and an ATG start site on the forward primer and an AclI restriction site on the reverse primer and ligated into pCoBlast-Mtn cut with BamHI and AccI to produce pCoBlast-TAP. See below for oligonucleotide sequences.
Antibodies
For immunohistochemistry, the following primary antibodies were used at the given concentrations: 1:50 mouse-anti-CSP (6D6, Developmental Studies Hybridoma Bank [DSHB]), 1:1000 mouse-anti-DLG (4F3, DSHB), 1:200 Texas Red-conjugated-anti-HRP (Jackson ImmunoResearch), 1:100 mouse-anti-dFMR1 (6A15)43, 1:500 chicken-anti-protein A (CPA 65A, Immunology Consultants Laboratory), 1:50 mouse-anti-elav (DSHB), and 1:50 rabbit-anti-HA (Y-11, Santa Cruz Biotechnology). For Western analysis, the following primary antibodies were used at the following concentrations: 1:5000 mouse-anti-dFMR1, 1:1000 rat-anti-α-catenin (DCAT 1, DSHB), 1:10000 mouse-anti-β-Tubulin (E7, DSHB), 1:2000 mouse-anti-HA (HA.11, Covance), 1:5000 anti-FLAG M2-HRP (A8592, Sigma), and 1:5000 chicken-anti-protein A. Secondary antibodies were obtained from either Molecular Probes or Jackson ImmunoResearch Laboratories, rehydrated according to the manufacturer’s instructions and used at a concentration of 1:200 for immunofluorescence staining and 1:2000 for Western analyses.
Immunohistochemistry
For larval immunostaining, fly stocks were transferred daily and grown at 25°C in a 12:12 hour light dark (LD) cycle. Wandering third instar larvae were dissected and filleted in saline buffer (128 mM NaCl, 2 mM KCl, 4 mM MgCl2, 35.5 mM sucrose, 5 mM Hepes pH 7.4, 0.1 mM CaCl2) and fixed for 30 min in 4% paraformaldehyde, washed in 1X PBT (1X PBS, 0.3% TritonX 100), and blocked for 1 hr at room temperature (RT) in 5% normal goat serum diluted in 1X PBT. Larvae were incubated in primary antibody overnight at 4°C. The next day, larvae were washed in 1X PBT and incubated in secondary antibody for 3 hrs at RT. Following final washes in 1X PBT, larvae were mounted in Vectashield (Vector Labs). Third instar larval brains were stained and imaged as previously described30.
Tandem affinity purification using dFMR1-cTAP-expressing cells
For dFMR1-TAP pulldown experiments, approximately 500 mL of S2 cells stably expressing pUAST-dfmr1-cTAP or pUAST-cTAP were induced with CuSO4 (final concentration 0.5 mM) 24 hrs prior to harvest. All manipulations were performed at 4°C or on ice unless otherwise specified. Cells were harvested by centrifugation for 5 min at 200g, room temperature (RT) and washed 2X in 5 mL ice-cold 1X PBS. Pellet was resuspended in 5X volume of Buffer A (10 mM Hepes pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1X PIC) and incubated on ice for 15 min followed by dounce homogenization 40X. Extracts were spun at 700g, 20 min to collect cytoplasmic lysates. Pellet was resuspended in 3X volume of Buffer B (20 mM Hepes, pH 7.9, 20% glycerol, 0.5% NP40, 200 mM KCl, 0.5 mM DTT, 1X PIC) and dounce homogenized 40X. Extracts were spun 10 min at 10,000g to isolate nuclear extract. Cytoplasmic and nuclear extracts were combined for TAP procedure. The TAP protocol was performed as previously described49 with the following modifications: Protease inhibitors without EDTA (P8340, Sigma) were used in all steps. IgG sepharose fast flow (Amersham) bead incubation with S2 cell extracts was carried out overnight, as was the calmodulin resin binding to the TEV eluate. Following SDS-PAGE electrophoresis on a 4–12% Bis-Tris gel (Invitrogen), gels were stained with silver stain (Silverquest kit) and bands were excised and destained according to the manufacturer’s protocol. Isolated proteins were identified using mass spectrometry carried out by the University of Pennsylvania School of Medicine Proteomics Facility50.
Tandem affinity purification (TAP) assay for dADAR-TAP-expressing cells
For dADAR-TAP pulldown assays, Drosophila S2 cells expressing pCoBlast-dADAR-3A-TAP, pCoBlast-dADAR-3/4-TAP, or pCoBlast-TAP constructs were used for the TAP assay. All manipulations were performed at 4°C or on ice. Cells were collected by centrifugation for 5 min at 100g. TAP purification was performed as previously described28 with the following modifications: After overnight incubation with IgG sepharose, beads were washed in TEV cleavage buffer five times, with the fourth wash being supplemented with RNase A (Roche, final concentration 0.05 μg/μl) for 1 hr. TEV cleavage was performed for 4 hrs and complexes were collected and run on SDS-PAGE followed by Western analysis. TAP assay was repeated at least three times.
Co-Immunoprecipitation (co-IP) assay
Adult fly heads were collected using liquid nitrogen. All manipulations were performed at 4°C or on ice. Heads were homogenized in 1.2 mL IP buffer (150 mM NaCl, 0.1% NP40, 20 mM Hepes pH 7.4, 2 mM MgCl2, 1 mM DTT, 1X PIC). Lysates were spun down twice at 13000 rpm to remove debris. To pre-clear extract, 30 μl rProtein G agarose beads (Invitrogen) were added to lysates and incubated for 30 min. 2 μg of anti-HA antibody (Y-11, Santa Cruz Biotechnology) was added to approximately 5 mg of lysate and incubated for 2 hrs, followed by incubation with 30 μl rProtein G agarose beads overnight. Following five washes using 20x IP buffer, the complexes were eluted using LDS buffer (Invitrogen) and samples were run on SDS-PAGE followed by Western analysis. Co-IP experiments were repeated at least three times.
RNA immunoprecipitation
Adult heads were homogenized in 1.2 mL RNase-free IP buffer (150 mM NaCl, 0.1% NP40, 20 mM Hepes pH 7.4, 2 mM MgCl2, 1 mM DTT, 2X PIC). Extracts were spun down twice at 13000 rpm, 4°C to remove debris and pre-cleared using 50 μl Protein A and/or G Dynabeads (Invitrogen) at 4°C for 30 min. Approximately 3 mg of lysate was incubated for 3 hours to overnight with 50 μl Dynabeads conjugated to rabbit-anti-HA antibody (Y-11, for dADAR-HA IP) or mouse-anti-dFMR1 (6A15 for dFMR1 IP) for 1.5 hrs, 1 mg/mL yeast tRNA (Invitrogen), 1 mg/mL BSA, and 300 U RNasIn (Promega). Following eight washes using 20x IP buffer, the immunoprecipitated RNA was purified using TRI-reagent (Ambion) followed by RNeasy-spin columns (Qiagen) as previously described15, and cDNA was generated using Superscript III and random hexamers (Invitrogen). Quantitative RT-PCR was performed with the Mx 3000 system (Agilent Technologies) using Brilliant III SYBR Green QPCR master mix (Agilent Technologies). For quantitative analysis, the Ct values obtained from the Mx 3000 software were normalized to the input cDNA and control genotypes. Data is represented as the fold difference above control genotypes relative to input cDNA using the following equation: 2^[(CtIgG–CtInput)control genotype–(CtIgG–CtInput)experimental genotype]. Fold change of experimental samples relative to control genotypes was then normalized to fold change observed for actin RNA. Statistical analysis was performed using Student’s t-test using InStat (GraphPad Software). Immunoprecipitation experiments were performed four times and three technical replicates were used for QPCR assays. See below for oligonucleotide sequences.
RNA editing analysis and statistical analysis
RNA extractions from WT, dfmr13, and dfmr1(4X) L3 larvae were performed using TRIzol reagent (Invitrogen) using manufacturer’s protocol. Analysis of editing efficiency was previously described30. Statistical analysis measuring percentage of editing was performed using a Mann-Whitney U-test (InStat).
RNA purification and RT-PCR
Total RNA was extracted from ~1.5 μg of S2 cell extract (for RNase A experiments) or from RNA IP samples using TRI Reagent (Ambion) using manufacturer’s protocol with the following modifications: After addition of isopropanol and incubation step, sample was further purified using columns from RNeasy Mini Kit (Qiagen) with on-column treatment with DNase I (Qiagen) for 20 min at RT. cDNA was generated using random hexamers and Superscript III (Qiagen).
Western analysis
For larval brain and head Western analysis, 2–4 L3 larval brains or 5–10 adult fly heads were dissected in saline buffer or 1X Robbs buffer (55 mM sodium acetate, 40 mM potassium acetate, 100 mM sucrose, 10 mM glucose, 1.2 mM MgCl2, 1 mM CaCl2, 100 mM HEPES pH 7.5), respectively. Lysate preparation, SDS-PAGE, Western analysis, and quantification were performed as previously described15.
Microscopy and statistical analysis
For quantification of synaptic boutons, analysis of muscles 6/7 or muscle 4 in abdominal segment A3 was performed. Confocal images were collected on a Leica TCS SP confocal microscope and represent maximal projections from serial sections spanning the entire NMJ. At least 16 hemisegments were quantified for each genotype per experiment. For Fig. 2, mean number of synaptic boutons and branching for WT and dAdar5G1 mutant samples were analyzed with Student’s t-test using InStat (GraphPad Software). The mean number of synaptic boutons and branching were analyzed with one-way ANOVA followed by a Tukey-Kramer multiple comparisons post-test using InStat for Fig. 3–6. For Fig. 5, primary branch length was normalized to the length of muscles 6/7 for each hemisegment and analyzed by one-way ANOVA followed by a Tukey-Kramer multiple comparisons post-test using InStat.
Leptomycin B experiments
S2 cells stably expressing dADAR-TAP constructs were induced with CuSO4 (final concentration of 0.5 mM) overnight (~16–20 hrs). The next day, cells were treated with either leptomycin B (final concentration 30nM) or methanol (vehicle control) for 4 hours at room temperature (RT). For immunofluorescence analysis, cells were fixed for 10 min in 2% paraformaldehyde, washed three times in 1X PBS, permeabilized in 1X PBT-X (0.1% Triton X-100) for 2 min, and incubated with blocker solution (3% BSA, 1% PVP-10, 1% PVP-40, 0.1% PVP-360) for 10 min, RT. Cells were incubated with primary antibody (1:100 mouse-anti-dFMR1 and 1:500 chicken-anti-protein A diluted in 1X PBS) overnight at 4°C. The following day, cells were washed 2 times in blocker solution, once in 1X PBS and incubated with secondary antibody (at a final concentration of 1:200) for 30 min, RT. Cells were treated with 1μM TO-PRO-3 for 10 min, RT, followed by 3 washes in blocker solution. Following final washes, cells were mounted in Vectashield. Confocal images were collected on a Leica TCS SP confocal microscope using sequential scans. Laser intensity, gain and offset were kept constant for dFMR1 imaging for all samples. Pixel intensity of nuclear dFMR1 staining was quantified using ImageJ and statistical analysis was performed using InStat (Mann-Whitney test). For tandem affinity purification experiments, cells were harvested after 4 hr leptomycin B or vehicle control treatment and TAP was performed as described above.
Oligonucleotide sequences
Cloning primer sequences:
EcoRI-dAdar-F1 5′ AAGAATTCATGTTAAACAGCGCTAATAACAATTCT 3′;
BamHI-dAdar-R1 5′ AAGGATCCTTCGGCAAGACCGAACTC 3′;
BamHI-TAP-F3 5′ AAGGATCCATGGCGGCCGCCGATTACAAG 3′;
AclI-TAP-R1 5′ AAAACGTTTTAATTCGCGTCTACTTTCGG 3′;
RT-PCR primer sequences:
rp49.RT.F1 5′ ATGCTAAGCTGTCGCACAAA 3′;
rp49.RT.R1 5′ ATGGTGCTGCTATCCCAATC 3′;
αtub84D.RT.F1 5′ GAATCGTTTAATCGGCCAAA 3′;
αtub84D.RT.R1 5′ GTGGGTGGCTGGTAGTTGAT 3′;
QPCR primer sequences:
lap.QT.F2 5′ GCCCAGTTCGTTGTTAGATGC 3′;
lap.QT.R2 5′ GCTGGTAGAAGACAGGGCAGA 3′;
Caa1D.QT.F2 5′ GAAAACCGCAACCAGACAGAC 3′;
Caa1D.QT.R2 5′ ATGGTTTCCACTCCACAATGC 3′;
unc-13.QT.F1 5′ TCGGCCAGTGATAGATTTGCT 3′;
unc-13.QT.R1 5′ TGGACTCGATGCAGGAAAGTT 3′;
syt-1.QT.F2 5′ TCGTCATCCTAGAGGCCAAGA 3′;
syt-1.QT.R2 5′ AAGGGTTGAGGGTGCATTTTT 3′;
shab.QT.F2 5′ GGGCGGGTATTACAATGACAA 3′;
shab.QT.R2 5′ AACACCGCATATGCAACACAC 3′;
stn-B.QT.F2 5′ AAGCAGTTCTCGGTCTGCATC 3′;
stn-B.QT.R2 5′ GATCTCATCAACGGCGGTTAC 3′;
TBP.QT.F1 5′ TTATTTTCAGCTCCGGCAAGA 3′;
TBP.QT.R1 5′ GAGCCGACCATGTTTTGAATC 3′;
actin.sybr.F 5′ CCACGCCATCCTTCGTCTAG 3′;
actin.sybr.R 5′ GCCATCTCCTGCTCGAAGTC 3′.
Supplementary Material
Acknowledgments
We thank current and former members of the Jongens lab for helpful discussions and technical support, especially Rebecca Beerman for preliminary studies with the leptomycin B experiments, Stephanie Cruz, and Christine Dubowy. We also thank Meera Sundaram and Dustin Hancks for critical reading of the manuscript. We are grateful to Russ Carstens, Greg Bashaw, Tom Dockendorff, Cynthia Hughes, Klaus Kaestner, Spyros Artavanis-Tsakonas, the Bloomington Stock Center and the Developmental Studies Hybridoma Bank for fly strains and reagents. We also thank Chao-Xing Yuan and the University of Pennsylvania School of Medicine Proteomics Facility for help with the mass spectrometry results. This work was supported by a Predoctoral Training Grant in Genetics 5T32GM008216 to B.B., NIMH MH086705 and NIGMS GM086902 grants to T.A.J, and the Ellison Medical Foundation Senior Scholar award to R.A.R.
Footnotes
Competing Interests Statement
The authors declare no financial conflict of interest.
Author Contributions
B.B. performed the experiments shown in Figs. 1–7 as well as Supplemental Figs. 2–5, generated the figures, and wrote the manuscript. J.E.J. and Y.A.S. generated the dAdar-HA4.5.2 and dAdar-HA12.5.2 fly lines used in this study, performed experiments shown in Fig. 7, helped generate the figure, and contributed to the writing of this manuscript. J.E.J also performed experiments for, and generated part of, Supplemental Fig. 4. A.S-R.P performed the experiments shown in Supplemental Fig. 1, generated the figure, identified the initial findings for this manuscript, and contributed to the writing of this manuscript. R.A.R. and T.A.J. contributed to the conclusions drawn from all figures, provided intellectual input, contributed portions of the funding, and contributed to the writing of this manuscript.
References
- 1.O’Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- 2.Feng Y, et al. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 3.Corbin F, et al. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum Mol Genet. 1997;6:1465–1472. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- 4.Khandjian EW, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- 5.Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum Mol Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- 6.Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, et al. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monzo K, et al. Fragile X mental retardation protein controls trailer hitch expression and cleavage furrow formation in Drosophila embryos. Proc Natl Acad Sci USA. 2006;103:18160–18165. doi: 10.1073/pnas.0606508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bechara EG, et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 2009;7:e16. doi: 10.1371/journal.pbio.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiler IJ, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zalfa F, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estes PS, O’Shea M, Clasen S, Zarnescu DC. Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons. Mol Cell Neurosci. 2008;39:170–179. doi: 10.1016/j.mcn.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 15.Pepper AS, Beerman RW, Bhogal B, Jongens TA. Argonaute2 suppresses Drosophila fragile X expression preventing neurogenesis and oogenesis defects. PLoS One. 2009;4:e7618. doi: 10.1371/journal.pone.0007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin P, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 17.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu XL, Li Y, Wang F, Gao FB. The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila. J Neurosci. 2008;28:11883–11889. doi: 10.1523/JNEUROSCI.4114-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palladino MJ, Keegan LP, O’Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 23.Tonkin LA, et al. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 25.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang YQ, et al. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 27.Veraksa A, Bauer A, Artavanis-Tsakonas S. Analyzing protein complexes in Drosophila with tandem affinity purification-mass spectrometry. Dev Dyn. 2005;232:827–834. doi: 10.1002/dvdy.20272. [DOI] [PubMed] [Google Scholar]
- 28.Tsai A, Carstens RP. An optimized protocol for protein purification in cultured mammalian cells using a tandem affinity purification approach. Nat Protoc. 2006;1:2820–2827. doi: 10.1038/nprot.2006.371. [DOI] [PubMed] [Google Scholar]
- 29.Palladino MJ, Keegan LP, O’Connell MA, Reenan RA. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA. 2000;6:1004–1018. doi: 10.1017/s1355838200000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jepson JE, et al. Engineered alterations in RNA editing modulate complex behavior in Drosophila: regulatory diversity of adenosine deaminase acting on RNA (ADAR) targets. J Biol Chem. 2011;286:8325–8337. doi: 10.1074/jbc.M110.186817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 32.Feng Y, et al. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamanini F, et al. Differential expression of FMR1, FXR1 and FXR2 proteins in human brain and testis. Hum Mol Genet. 1997;6:1315–1322. doi: 10.1093/hmg/6.8.1315. [DOI] [PubMed] [Google Scholar]
- 34.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 35.Koh YH, Gramates LS, Budnik V. Drosophila larval neuromuscular junction: molecular components and mechanisms underlying synaptic plasticity. Microsc Res Tech. 2000;49:14–25. doi: 10.1002/(SICI)1097-0029(20000401)49:1<14::AID-JEMT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 36.Gatto CL, Broadie K. Temporal requirements of the fragile X mental retardation protein in the regulation of synaptic structure. Development. 2008;135:2637–2648. doi: 10.1242/dev.022244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keegan LP, et al. Tuning of RNA editing by ADAR is required in Drosophila. EMBO J. 2005;24:2183–2193. doi: 10.1038/sj.emboj.7600691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallo A, Keegan LP, Ring GM, O’Connell MA. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J. 2003;22:3421–3430. doi: 10.1093/emboj/cdg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerjee P, et al. Substitution of critical isoleucines in the KH domains of Drosophila fragile X protein results in partial loss-of-function phenotypes. Genetics. 2007;175:1241–1250. doi: 10.1534/genetics.106.068908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darnell JC, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 41.Darnell JC, et al. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zang JB, et al. A mouse model of the human Fragile X syndrome I304N mutation. PLoS Genet. 2009;5:e1000758. doi: 10.1371/journal.pgen.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dickman DK, Lu Z, Meinertzhagen IA, Schwarz TL. Altered synaptic development and active zone spacing in endocytosis mutants. Curr Biol. 2006;16:591–598. doi: 10.1016/j.cub.2006.02.058. [DOI] [PubMed] [Google Scholar]
- 45.Aravamudan B, Fergestad T, Davis WS, Rodesch CK, Broadie K. Drosophila UNC-13 is essential for synaptic transmission. Nat Neurosci. 1999;2:965–971. doi: 10.1038/14764. [DOI] [PubMed] [Google Scholar]
- 46.Zhang B, et al. Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron. 1998;21:1465–1475. doi: 10.1016/s0896-6273(00)80664-9. [DOI] [PubMed] [Google Scholar]
- 47.Dockendorff TC, et al. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–984. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 48.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 49.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 50.Zeng F, et al. A protocol for PAIR: PNA-assisted identification of RNA binding proteins in living cells. Nat Protoc. 2006;1:920–927. doi: 10.1038/nprot.2006.81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.