Abstract
Transcriptional coregulators control the activity of many transcription factors and are thought to have wide ranging effects on gene expression patterns. We show here that muscle-specific nuclear receptor corepressor 1 (NCoR1) knockout mice have rather selective phenotypic changes, characterized by enhanced exercise endurance due to an increase of both muscle mass and of mitochondrial number and activity. The activation of selected transcription factors that control muscle function, such as MEF2, PPARβ/δ and ERRs, underpinned these phenotypic alterations. NCoR1 levels are decreased in conditions that require fat oxidation resetting transcriptional programs to boost oxidative metabolism. The capacity of NCoR1 to modulate oxidative metabolism may be conserved as the knockdown of gei-8, the sole C.elegans NCoR homolog, also robustly increased muscle mitochondria and respiration. Collectively, our data suggest that NCoR1 plays an adaptive role in muscle physiology and that interference with NCoR1 action could be used to improve muscle function.
Keywords: NCoR, ERR, MEF2, Muscle, Mitochondria, PPARβ/δ
Introduction
Transcription factors are key mediators in homeostatic circuits, as they process environmental signals into transcriptional changes (Desvergne et al., 2006; Francis et al., 2003). Transcriptional coregulators have recently emerged as equally important modulators of such adaptive transcriptional responses. The fact that the activity of coactivators and corepressors is tightly regulated through the spatial and temporal control of their expression and activity levels opens hence another avenue to adapt transcription to environmental cues (Feige and Auwerx, 2007; Rosenfeld et al., 2006; Smith and O'Malley, 2004; Spiegelman and Heinrich, 2004). Interestingly, many of these coregulators do not operate in isolation, but are part of large multi-protein complexes, that integrate complex signaling pathways. The convergence of an elaborate coregulator network on the peroxisome proliferator-activated receptor (PPAR) coactivator α (PGC)-1α illustrates this principle well, as its activity depends on several other coregulators, including the steroid receptor coactivators, NR interacting protein 1 or RIP140, CREB binding protein, p300, protein arginine methyltransferase 1, general control of amino acid synthesis 5, and SIRT1 (Fernandez-Marcos and Auwerx, 2011; Handschin and Spiegelman, 2006).
The corepressor (NCoR1) and the silencing mediator for retinoid and thyroid hormone receptor (SMRT or NCoR2) are also acting as cofactor scaffolding platforms. NCoR1 and SMRT hardwire corepressor pathways that incorporate several deacetylases [including class I (HDAC3), class II (HDAC4, 5, 7, and 9) and class III (SIRT1) HDACs], transducin beta-like 1 (TBL1) and TBLR1, two highly related F box/WD40-containing factors, and the G-protein-pathway suppressor 2 [reviewed in (Perissi et al., 2010)]. Since germline NCoR1-/- and SMRT-/- mice are embryonically lethal (Jepsen et al., 2000; Jepsen et al., 2007), information on the role of these proteins in adult physiology is limited. Studies of mice with mutations in the NR interaction domains (RIDs) 1 and 2 of SMRT (SMRTmRID), which solely disrupts its interaction with NRs, indicated that lethality of SMRT-/- mice is caused by non-NR transcription factors (Nofsinger et al., 2008). Work in 3T3-L1 cells in which NCoR1 or SMRT expression was reduced by RNA interference, demonstrated that they repress adipogenesis by inhibiting PPARγ (Yu et al., 2005). In line with this, adipogenesis was enhanced in mouse embryonic fibroblasts (MEFs) from SMRTmRID mice (Nofsinger et al., 2008). Interestingly, SIRT1 is also part of the NCoR1/SMRT complex and contributes to the inhibition of PPARγ (Picard et al., 2004).
Contrary to adipose tissue, the function of NCoR1/SMRT in skeletal muscle has not yet been established. We here report the generation and characterization of muscle-specific NCoR1-/- (NCoR1skm-/-) mice, which displayed a remarkable enhanced exercise capacity. This was the result of increased muscle mass and a muscle fiber type shift towards more oxidative fibers, coordinated by the induction of genes involved in mitochondrial biogenesis and function, ensuing from the activation of PPARβ/δ, ERR, and MEF2. Worms with a muscle-selective knockdown of gei-8, the sole C.elegans NCoR homolog, also had improved mitochondrial activity. These data combined with the specific reduction in the expression levels of NCoR1, but not SMRT, in situations of enhanced fat oxidation, establish NCoR1 as a key physiological regulator of muscle mass and function.
Results
NCoR1skm-/- mice have increased muscle mass
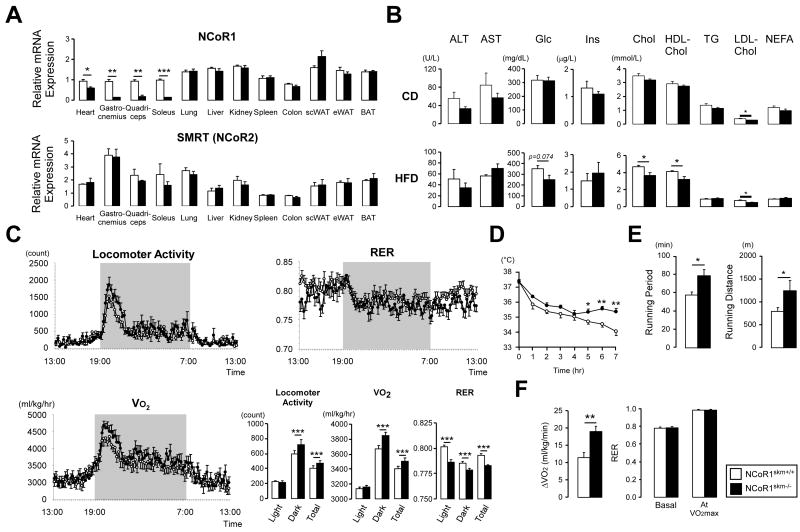
Given the embryonic lethality of germline NCoR1-/- mice [(Jepsen et al., 2000), Suppl. Table 1], we generated a floxed NCoR1 mouse line in which exon 11 of the NCoR1 gene (Horlein et al., 1995) was flanked with LoxP sites, priming it for subsequent deletion using the Cre-LoxP system. These mice, bearing floxed NCoR1 L2 alleles, were then bred with a skeletal muscle (skm)-specific Cre driver (human α-skeletal actin promoter) (Miniou et al., 1999) to yield NCoR1skm-/- and NCoR1skm+/+ mice (Suppl. Fig.1). As expected, NCoR1 mRNA expression was significantly decreased in soleus, gastrocnemius and quadriceps and modestly reduced in the heart muscle of NCoR1skm-/- mice, but not altered in other tissues (Figure 1A). No compensatory induction of the related co-repressor SMRT/NCoR2 (Chen and Evans, 1995) was observed (Figure 1A). We also tried to determine NCoR1 protein levels in muscle, but failed to detect the endogenous protein with the currently available NCoR1 antibodies.
Figure 1. Validation and metabolic phenotypes of NCoR1skm-/- mice.
(A) mRNA levels of NCoR1 and Smrt in different tissues were determined by qRT-PCR. Values were normalized to 36B4. (n=8-10/group). (B) Biochemical analysis of the plasma from NCoR1skm+/+ and skm-/- mice after 6 hr fasting (n=8) either on chow diet (CD) (top) or HFD (bottom). (C) Circadian activity, measured as the total locomotor activity, and energy expenditure was evaluated by the measurement of oxygen consumption (VO2), and by the calculation of the respiratory exchange ratio (RER) over a 24 hr period after 12 wks of HFD. The bar graphs represent the average for each group (n = 12). (D) Body temperature was measured for 7 hr in mice exposed to 4°C after 18-wks of HFD (n=7, 8). (E and F) Exercise experiments, measuring running time and distance till exhaustion (endurance exercise) (E) and the increment of VO2max, during exercise, and RER levels at basal and VO2max condition (F) were performed after 14- and 17-wk HFD.
Data are expressed as mean ± SEM.
See also Suppl. Figs. 1-3 and Suppl. Tables 1-3.
NCoR1skm-/- mice were indistinguishable from NCoR1skm+/+ mice upon visual inspection and no gross organ anomalies were revealed upon autopsy. The relative mass of the soleus muscle was higher, whereas the mass of the gastrocnemius showed a trend towards an increase, which did not reach statistical significance (Suppl. Fig. 2A-B). The soleus was also more intensely red and there were larger sections with reddish color in the gastrocnemius in NCoR1skm-/- mice (Suppl. Fig. 2C). Body weight evolution and food intake of male NCoR1skm-/- and NCoR1skm+/+ mice after weaning was comparable both on chow diet (CD) and on high fat diet (HFD) (Suppl. Fig. 2A and data not shown). On CD, carbohydrate and lipid profiles were similar, except for LDL cholesterol, which was reduced in NCoR1skm-/- mice (Figure 1B). In addition to the lower LDL cholesterol on CD, total and HDL cholesterol levels were also reduced in NCoR1skm-/- mice on HFD (Figure 1B). Furthermore, glucose edged down (p=0.074) in the wake of similar insulin levels on HFD. The slightly reduced area under the curve in intraperitoneal glucose tolerance test (IPGTT; Suppl. Fig. 2D) and the delayed recovery from hypoglycemia during intraperitoneal insulin tolerance test (IPITT; Suppl. Fig. 2E) in mutant mice on HFD, may suggest a discrete improvement in insulin sensitivity but without a clear impact on glucose tolerance.
Enhanced exercise performance in NCoR1skm-/- mice
We next evaluated energy expenditure by indirect calorimetry and actiometry in CD and HFD fed mice (Figure 1C and Suppl. Fig. 2F). Total locomotor activity was significantly higher in NCoR1skm-/- mice. Consistent with this, O2 consumption (VO2) was increased under both CD and HFD. Interestingly, the NCoR1skm-/- mice displayed a marked decrease in the respiratory exchange ratio (RER) on a HFD (Figure 1C), indicating an enhanced use of fat as main energy source. NCoR1skm-/- mice were also more cold tolerant, as they maintained their body temperature better when exposed to 4°C (Figure 1D).
Exercise performance was strikingly improved in NCoR1skm-/- mice (Figure 1E-F, and Suppl. Fig. 3A-D). In endurance exercise, NCoR1skm-/- mice ran for a significantly longer time and distance before exhaustion (Figure 1E, and Suppl. Fig. 3A-B). The increase of the VO2 values (ΔVO2) during exercise and the maximal ability to utilize oxygen during exercise (VO2max), which critically determines the endurance performance of skeletal muscle, was slightly higher in NCoR1skm-/- mice on both CD (Suppl. Fig. 3D and data not shown) and HFD (Figure 1F). Despite the moderate reduction in NCoR1 mRNA levels in cardiac muscle of NCoR1skm-/- mice, heart rate, blood pressure, cardiac morphology and function were not changed (Suppl. Fig. 3E-G and Suppl. Table 2).
NCoR1skm-/- muscle demonstrates increased oxidative capacity
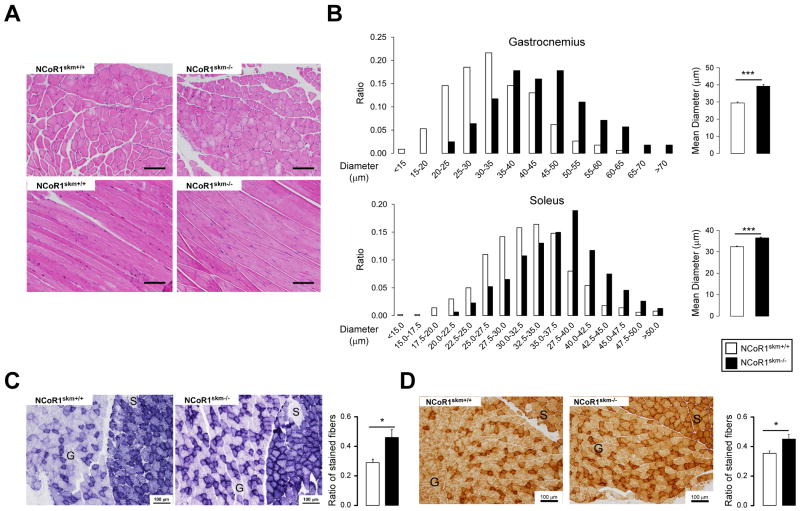
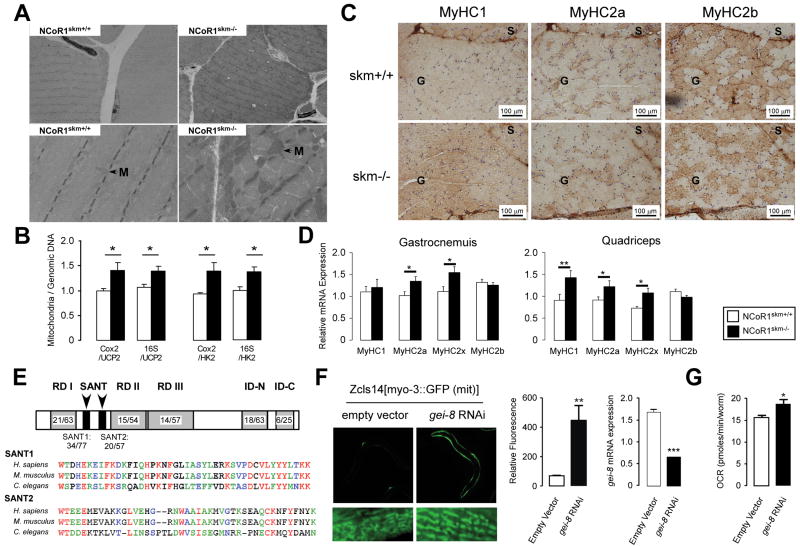
The enhanced exercise capacity, associated with the increase in overall muscle mass and change in muscle appearance, led us to examine muscle morphology. Upon staining muscles with hematoxylin/eosin or toluidine blue, not only the diameter of single muscle fibers was larger, but also the connective tissue between the muscle bundles was less abundant in NCoR1skm-/- mice (Figure 2A-B and Suppl. Fig. 4A). The increased number of intensely stained fibers upon succinate dehydrogenase (SDH) and cytochrome oxidase (COX) (Figure 2C-D) staining, further testified of increased mitochondrial activity in the NCoR1skm-/- gastrocnemius. Two mitochondrial DNA markers, cyclooxygenase 2 (Cox2) and 16S ribosomal RNA, normalized by genomic DNA markers [uncoupling protein 2 (Ucp2) and hexokinase 2 (Hk2)] were both significantly higher in NCoR1skm-/- muscle, indicative of increased mitochondrial content (Figure 3B). This observation was also underscored by electron microscopy, which revealed more abundant and larger mitochondria with normal structure (Figure 3A). Immunohistochemical analysis of the myosin heavy chain (MyHC) isoforms (Schiaffino et al., 1989) demonstrated a decreased number of the more glycolytic MyHC2b fibers, with a concomitant increase in the number of more oxidative MyHC2x and 2a fibers in the NCoR1skm-/- gastrocnemius (Figure 3C). This observation was consolidated by analysis of MyHC isoform mRNAs, which indicated an increased expression of the mRNAs of MyHC2x and 2a (more oxidative fibers) compared to that of MyHC2b (more glycolytic) in both NCoR1skm-/- gastrocnemius and quadriceps (Figure 3D). In quadriceps, but not gastrocnemius, the expression of MyHC1 mRNA was also increased. Finally, staining of platelet-endothelial cell adhesion molecule (PECAM)-1, an endothelial cell marker of angiogenesis and tissue vascularization, which contributes to enhanced myocellular aerobic capacity, also increased in NCoR1skm-/- muscle (Suppl. Fig. 5C).
Figure 2. Histological analyses of the muscles of control and NCoR1skm-/- mice.
(A) Histological analysis of gastrocnemius sections stained with H&E. (B) Distribution and mean diameter of muscle fibers in gastrocnemius and soleus. (C and D) Histological analysis of gastrocnemius and soleus sections by succinate dehydrogenase (C) and cytochrome C oxidase staining (D). (C). S, soleus; G, gastrocnemius. The ratio of the stained fibers is indicated in the graph.
Data are expressed as mean ± SEM.See also Suppl. Fig. 4.
Figure 3. Histological analyses of the muscles of mice and C. elegans.
Transmission electron microscopy of non-oxidative fibers of NCoR1skm+/+ and skm-/- gastrocnemius. M, mitochondria. (B) Relative mitochondrial DNA content (Cox2 or 16S) in gastrocnemius was measured and normalized by genomic DNA content (Ucp2 and Hk) (n=6). (C) Representative MyHC1, 2a, and 2b immunohistochemical detection on serial sections of the soleus and gastrocnemius. (D) Subtypes of MyHCs in gastrocnemius and quadriceps were analyzed by qRT-PCR in NCoR1skm+/+ and skm-/- mice (n=6). (E) Indentity/similarity (%) in the sequences of gei-8 with mammalian NCoR1. Multiple alignment of SANT domains of NCoR1 homologs. Color identifies similarity. (F) Representative pictures of the effects of RNAi-mediated knockdown of gei-8 on mitochondrial morphology and number in a C.elegans strain carrying a mitochondrial GFP-reporter driven by the muscle-specific myo-3 promoter (left panel). Quantification of the mitochondrial induction by fluorescence upon gei-8 knockdown (middle panel), and of the efficacy of the RNAi-mediated gei-8 knockdown by qRT-PCR analysis (right panel). (G) Muscle-specific RNAi inhibition of gei-8 enhances respiration in C.elegans.
Data are expressed as mean ± SEM.
See also Suppl. Fig. 3 and Suppl. Tables 2 and 3.
The control of muscle mitochondria by NCoR1 is conserved in C.elegans
To investigate whether the effects of NCoR1 deficiency are evolutionary conserved, we took advantage of the power of C.elegans genetics. A protein blast search indicated that gei-8 (GEX interacting protein family member 8) is the only putative NCoR1 homolog in the C.elegans genome. Further analysis showed that the total amino acid sequence of gei-8 is 43% homologous to mouse NCoR1 and contained conserved SANT (switching-defective protein 3 (Swi3), adaptor 2 (Ada2), nuclear receptor co-repressor (N-CoR), transcription factor (TF) IIIB)) domains (34% identical/77% similar for SANT1; 20% identical/57% similar for SANT2) (Figure 3E). Other important functional domains (Repressor Domain (RD) and nuclear receptor Interaction Domain (ID)) were also conserved (Figure 3E). Upon the robust gei-8 knockdown in worms expressing a mitochondrial GFP-reporter driven by the muscle-specific myo-3 promoter, a striking enlargement of the mitochondria was observed in body wall muscle (Figure 3F). This result is not due to an indirect effect on transcriptional activity through the myo-3 promoter because no increase in GFP expression is observed with another strain carrying the pmyo-3∷GFP reporter (Supp. Fig. 4B). We also measured O2 consumption in NR350 transgenic worms fed with gei-8 dsRNA. NR350 worms lack rde-1, an essential component of the RNAi machinery encoding a member of the PIWI/STING/Argonaute family, in all tissues except the body wall muscle in which the wild-type rde-1 gene has been rescued using the hlh-1 promoter (Durieux et al., 2011). Consistent with the effects observed in the mouse, also the muscle-specific knockdown of gei-8 enhanced O2 consumption in these NR350 worms (Figure 3G), suggesting that the function of gei-8 to control mitochondrial metabolism is conserved through evolution.
NCoR1 negatively correlates with key mitochondrial and myogenic genes
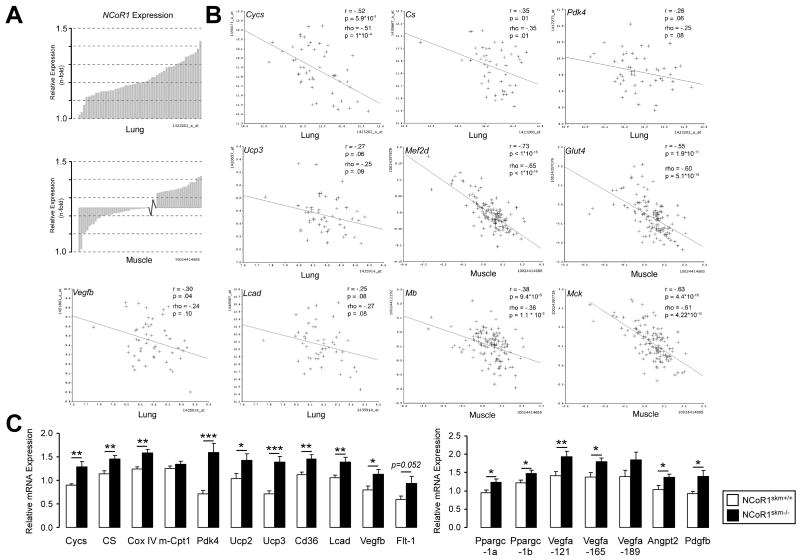
After establishing these striking mitochondrial effects of NCoR1 in mice and worms, we exploited a complementary systems genetics approach to evaluate NCoR1s molecular coexpression partners in the mouse (Argmann et al., 2005; Houtkooper et al., 2010). Expression of NCoR1 in two panels of genetically heterogeneous mice, made by intercrossing C57BL/6 with C3H/HeJ (the BXH F2 cross), or C57BL/6 with DBA/2J (the BXD genetic reference population) mice, varied ±1.5-fold between cases in both lung and muscle (Figure 4A). A large number of transcripts covaried significantly with NCoR1 in the different mice lines belonging either to the BXH cross or BXD strains. Most distinctively, only a fraction of these covariates were negative, which was against the dogma expected for a corepressor such as NCoR1s (Suppl. Tables 4 and 5). In skeletal muscle from the BXH cross (n=124 females), strong covariates of NCoR1 include myocyte-specific enhancer factor 2D (Mef2d), myoglobin (Mb), muscle creatine kinase (Mck), and glucose transporter type 4 (Glut4) (van Nas et al., 2010). A similar analysis of lung tissue from the BXD cross (n=51 strains) includes genes such as cytochrome c (Cycs), citrate synthase (Cs), pyruvate dehydrogenase kinase 4 (Pdk4), uncoupling protein 3 (Ucp3), vascular endothelial growth factor b (Vegfb), and long chain acyl-CoA dehydrogenase (Lcad) (Schughart and Williams, unpublished data) (Figure 4B and Suppl. Table 4 and 5). This analysis significantly extends the number of NCoR1 targets and covariates, with several of them being consistent with increased mass and mitochondrial biogenesis observed in NCoR1skm-/- muscle.
Figure 4. Identification of NcoR1-correlated genes.
(A) Expression of NCoR1 mRNA in lung tissue of the different BXD strains (upper) and in muscle tissue from an F2 intercross between C57BL/6J and C3H/HeJ (lower panel). Natural expression variation across the animals is ∼1.5 fold in each tissue. (B) Pearson's r and Spearman's rank correlation coefficient, rho, were calculated with corresponding p values for the mRNA covariation between NCoR1 and genes involved in oxidative phosphorylation (Cycs, Cs, and Pdk4), mitochondrial uncoupling (Ucp3), fatty acid metabolism (Lcad), angiogenesis (Vegfb), glucose uptake (Glut4), and myogenesis (Mef2d, Mb, Mck). The tissue from which data were generated is indicated. (C) Gene expression analysis by qRT-PCR in NCoR1skm+/+ and skm-/- gastrocnemius (n=10).
Data are expressed as mean ± SEM.See also Suppl. Fig. 5 and Suppl. Tables 3-5.
This initial set of NCoR1 covariates (Figure 4B) was then included together with other potential candidates for qRT-PCR analysis in mixed fiber muscle, including the gastrocnemius and quadriceps (Figure 4C, Suppl. Fig. 5A, data not shown). Whereas the mRNAs of most relevant NRs were unchanged, mRNA levels of PGC-1α and β (Ppargc1a and 1b) increased. Several genes involved in mitochondrial function, including those encoding for proteins involved in TCA cycle and oxidative phosphorylation [Cs, cytochrome c oxidase subunit IV (CoxIV), Pdk4], uncoupling [Ucp2 and Ucp3], fatty acid uptake and metabolism [Cd36 and Lcad], were robustly induced in NCoR1skm-/- muscle. In addition, mRNA levels of Vegfb and its receptor Flt1, which regulates trans-endothelial fatty acid transport (Hagberg et al., 2010), were also induced. Interestingly, the expression of hypoxia inducible factor (Hif) 1α and of its targets, glucose transporter 1 (Glut1), fibroblast growth factor (Fgf) and Fgf-receptor 2 (Fgfr2), were unchanged (Suppl. Fig. 5A), whereas all three Vegfa isoforms, i.e. Vegfa-121, -165, and -189, were induced in NCoR1-/- quadriceps, gastrocnemius, and soleus (Figure 4C, Suppl. Fig. 5B). Together with this increase in Vegfa, both Angpt2 and Pdgfb mRNA levels were induced (Figure 4C), suggesting that myocellular aerobic capacity is facilitated by a HIF1α-independent angiogenic pathway in NCoR1skm-/- mice (Arany et al., 2008).
Enhanced PPARβ/δ and/or ERR function in NCoR1skm-/- muscle
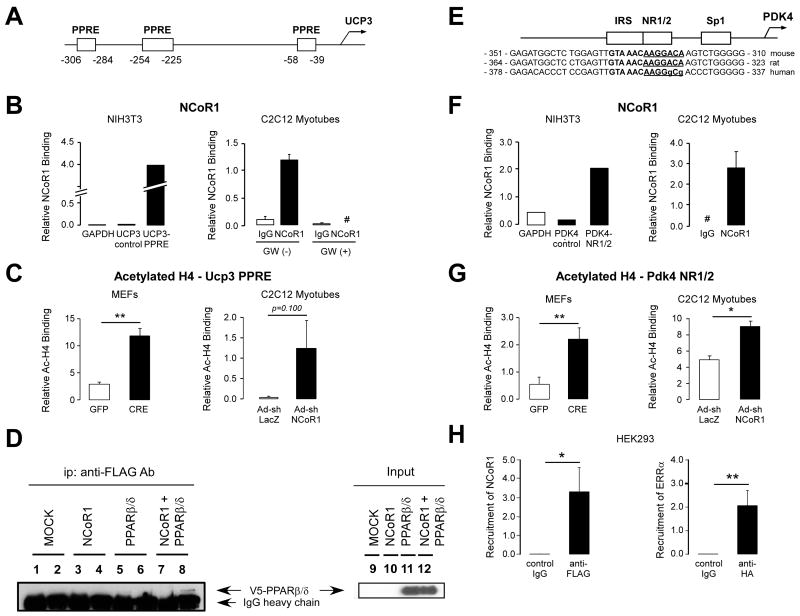
Several genes whose expression is changed in the absence of NCoR1 are PPARβ/δ and/or ERR targets (Figure 4). Since the expression of PPARβ/δ and/or ERR was unchanged in NCoR1skm-/- mice (Suppl. Fig. 5A), a direct effect of NCoR1 on the expression of these targets through the activation of these NRs was expected. As cases in point to demonstrate the recruitment of NCoR1 to these genes, we selected the mouse Ucp3 and Pdk4 promoters, which contain three PPAR responsive elements (PPREs) (Figure 5A) and extended NR half-sites (NR1/2), known to bind members of the ERR subfamily (Figure 5E) (Zhang et al., 2006), respectively. We first used NIH-3T3 cells in which an epitope-tagged version of NCoR1 (NCoR1-FLAG) was expressed. The two PPREs adjacent to the Ucp3 transcription start site recruited NCoR1 more efficiently, compared to two control sequences in the Gapdh and Ucp3 promoter that lack PPREs (Figure 5B, left). Likewise, NCoR1 bound avidly to the mouse Pdk4 promoter NR1/2 site in transfected NIH-3T3 cells (Figure 5F, left). Although there is a two nucleotide difference in NR1/2 site of the human Pdk4 promoter (Figure 5E), NCoR1 and ERRα were also recruited to this site in human HEK293 cells (Figure 5H).
Figure 5. Increased PPARβ/δ and ERR activity in NCoR1skm/- muscle.
(A, B, E, F, and H) NCoR1 recruitment to the PPREs on mouse Ucp3 promoter and to the ERR-RE on human and mouse Pdk4 promoter determined by ChIP in NIH-3T3 cells transfected with an NCoR1-FLAG vector or in C2C12 myotubes. A schematic of the promoters of the Ucp3 (A) and Pdk4 (E) genes and the sequence alignment of the mouse, rat and human Pdk4 promoter is also shown to highlight the conservation of the NR1/2 (or ERR-RE) (E). Boxes indicate putative PPREs in the Ucp3 and the NR1/2, IRS, and Sp1 in the Pdk4 promoter. ChIP experiments for the Ucp3 promoter were performed in C2C12 myotubes both before and 6 hr after addition of a PPARβ/δ agonist (100 nM GW501516). #; not detected. ChIP experiments in HEK293 cells transfected with FLAG-NCoR1 and HA-ERRα vector (H). (C and G) Binding of acetylated histone 4 (H4) to the PPREs on the Ucp3 and to the NR1/2 on the Pdk4 promoters in ChIP assays, using either immortalized NCoR1L2/L2 MEFs, infected with an adenovirus either expressing GFP or Cre recombinase, or C2C12 myotubes infected with the Ad-shNCoR1 virus. Representative data is shown from 3 experiments. (D) Interaction between PPARβ/δ and NCoR1 determined by in vitro co-IP experiments from HEK293 cells, in which NCoR1-FLAG and/or V5-PPARβ/δ are expressed. IP was performed with control IgG (lanes 1, 3, 5, and 7) or anti-FLAG antibody (lanes 2, 4, 6, and 8) and the immunoblot was developed with an anti-V5 antibody. PPARβ/δ co-immunoprecipitated by the anti-FLAG antibody is indicated by an arrow. Input samples are shown in lanes 9 -12.
Data are expressed as mean ± SEM.
See also Suppl. Fig. 6.
We then used a highly specific NCoR1 antibody, which we recently generated (Barish & Evans, unpublished), for ChIP experiments in C2C12 myotubes. Confirming our data in NIH-3T3 cells that express NCoR1-FLAG, endogenous NCoR1 occupied the same Ucp3 PPREs (Figure 5B, right). The recruitment of NCoR1 to the Ucp3 promoter was robustly inhibited by the addition of the selective PPARβ/δ ligand GW501516. Likewise, endogenous NCoR1 was readily detected on the NR1/2 in the Pdk4 promoter in C2C12 myotubes (Figure 5F, right).
Subsequently, we explored whether NCoR1 gene deletion in NCoR1L2/L2 MEFs by means of adenoviral Cre recombination, or NCoR1 gene knockdown in C2C12 myotubes infected by an NCoR1 shRNA adenovirus, modulates histone H4 acetylation on the Ucp3 and Pdk4 promoters. Consistent with NCoR1 binding to these promoters in NIH-3T3 cells and C2C12 myotubes (Figure 5B and F), NCoR1 deletion or silencing induced H4 acetylation of both target promoters in MEFs and C2C12 cells, indicating chromatin opening (Figure 5C and G).
To further consolidate these observations, we analyzed whether NCoR1 interacts directly with PPARβ/δ or ERRα, using nuclear extracts of HEK293 cells, transfected with tagged versions of NCoR1, PPARβ/δ or ERRα. Although a specific association between NCoR1 and PPARβ/δ was evident in these co-IP experiments (Figure 5D, lane 8), we failed to detect a similar interaction between ERRα and NCoR1 (data not shown).
MEF2 is hyperacetylated and activated in the absence of NCoR1
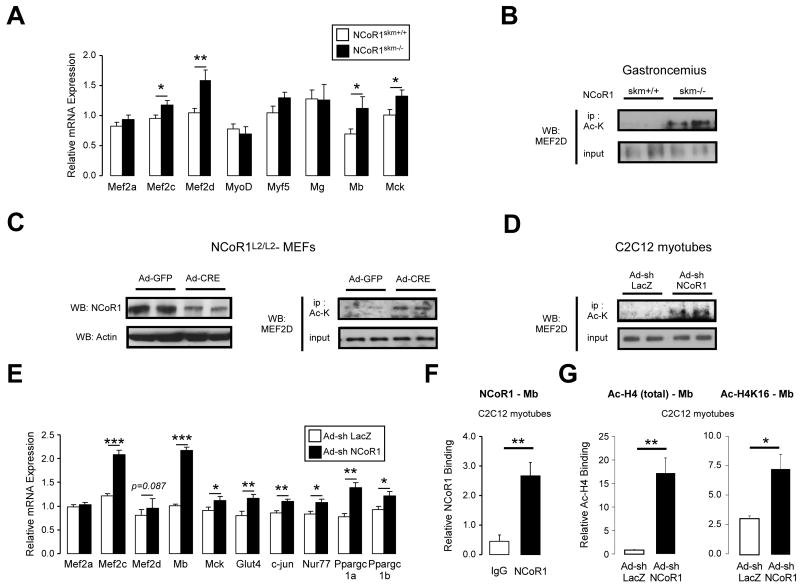
The increased muscle mass observed in NCoR1skm-/- mice indicated that the absence of NCoR1 not only induced oxidative metabolism, but also stimulated myogenesis. In line with this, mRNA levels of two markers of myogenesis, Mb and Mck were increased in NCoR1skm-/- quadriceps (Figure 6A). Amongst several myogenic regulatory factors, only the expression of two Mef2 family members, i.e. Mef2c and Mef2d, negatively correlated with NCoR1 expression in our systems genetics analysis (Figure 4 and Suppl. Fig. 6A). The selective induction of Mef2c and Mef2d mRNA was furthermore confirmed by qRT-PCR of NCoR1skm-/- gastrocnemius and quadriceps, while no changes were found in MyoD, myf5, and myogenin mRNA (Figure 6A, Suppl. Fig. 6E, data not shown).
Figure 6. Enhanced MEF2 activity in NCoR1skm-/- muscle.
(A) Gene expression of myogenesis-related genes was measured by qRT-PCR in NCoR1skm+/+ and skm-/- quadriceps (n=10). (B, C, and D) Acetylation levels of MEF2D were determined by Western blot after immunoprecipitation with an Ac-Lys Ab from gastrocnemius (B), from NCoR1L2/L2-MEFs infected with Ad-GFP or Ad-Cre recombinase (C, right), and from C2C12 myotubes infected with Ad-shLacZ or Ad-shNCoR1 (D). MEF2D expression in total protein extracts was shown in the lower panels. The expression of NCoR1 and actin in NCoR1L2/L2-MEFs was also shown (C, left). (E) MEF2 target mRNAs determined by qRT-PCR in C2C12 myotubes infected with either Ad-shLacZ or Ad-shNCoR1 (n=6). (F) NCoR1 recruitment to the MEF2 site of the mouse Mb promoter determined by ChIP in C2C12 myotubes. (G) Binding of either global acetylated histone 4 (H4) or H4 acetylated on K16 (H4K16) to the MEF2 site of the Mb gene was evaluated by ChIP from C2C12 myotubes infected as in (E). Data are expressed as mean ± SEM.
See also Suppl. Fig. 6 and Suppl. Table 3.
The activity of MEF2 family members is not only controlled by their expression levels, but is also modulated by their acetylation status. MEF2 is acetylated and activated by p300, whereas it is deacetylated by HDAC3 and HDAC4, which are part of the NCoR1 corepressor complex (Ma et al., 2005; Nebbioso et al., 2009). Since the expression of the Mef2d isoform is most prominently correlated with NCoR1 expression, we investigated MEF2D acetylation in gastrocnemius and found that its acetylation levels were enhanced in NCoR1skm-/- mice (Figure 6A-B).
We then compared the acetylation of MEF2D in floxed NCoR1L2/L2 MEFs, infected with an adenovirus expressing either GFP as control or Cre-recombinase to reduce NCoR1 protein expression (Figure 6C, left panel). Whereas MEF2D protein levels were stable in NCoR1-/- MEFs, perhaps due to the more acute nature of the deletion, MEF2D was robustly hyperacetylated when NCoR1 levels were attenuated (Figure 6C, right panel). Likewise, a slight but consistent MEF2D hyperacetylation was observed in C2C12 myotubes infected with Ad-shNCoR1 to knockdown NCoR1 expression (Figure 6D and Suppl. Fig. 6B-D), further underscoring the importance of MEF2D deacetylation by the NCoR1 complex.
Given the induction of MEF2D expression and its hyperacetylation and consistent with our systems genetics analysis (Figure 4B), mRNA levels of the MEF2 targets, Mb and Mck, were robustly induced in NCoR1skm-/- gastrocnemius (Figure 6A). Silencing of NCoR1 in C2C12 myotubes also resulted in a similar induction of several MEF2 target genes, including Mb, Mck, Glut4, c-jun, Nur77, PGC-1α, PGC-1β (Figure 6E and Suppl. Fig. 6B-D). In line with these data, endogenous NCoR1 was readily detected on MEF2 binding sites on these target promoters in C2C12 myotubes, as illustrated for the Mb promoter (Figure 6F). The induction of these MEF2 targets by NCoR1 knock-down was furthermore accompanied by H4K16 and global H4 hyperacetylation on their promoters (e.g. Mb, Glut4, Mck) (Figure 6G and Suppl. Fig. 6F).
NCoR1 levels are regulated in response to physiological stimuli
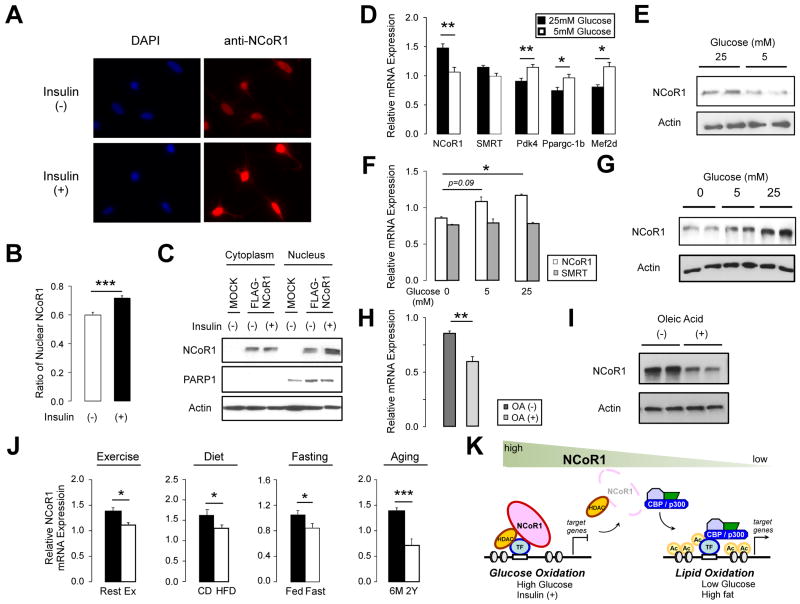
We next investigated whether NCoR1 function could be altered by different physiological stimuli in vitro and in vivo. One hour after stimulation of 293T cells with 1 μM insulin, higher amounts of endogenous NCoR1 (Figure 7A) or transfected FLAG-NCoR1 (Suppl. Fig. 7A) were detected in nuclei as evidenced by immunofluorescence and subcellular fractionation (Figure 7A-C).
Figure 7. Localization and expression of NCoR1 in physiological condition.
(A-C) Localization of NCoR1 protein was determined either by immunofluorescence and quantified (A and B) and by western blot (C). 293T cells grown without (-) or with (+) 1 μM insulin for 1 hr were stained by DAPI or anti-NCoR1 (A). Quantification of nuclear NCoR1 is shown in (B). Nuclear and cytosolic fractions were separated from FLAG-NCoR1 transfected 293T cells after a 1 hr-stimulation without (-) or with 1 μM insulin (+) and protein levels were determined by western blotting (C). (D) mRNA levels of NCoRs and its target genes determined by qRT-PCR in MEFs cultured for 24 hr in 5 or 25 mM glucose (n=6). (E) NCoR1 protein was determined by western blotting from MEFs cultured for 24 hr in 5 or 25 mM glucose. (F) NCoR1 and SMRT mRNA was determined in MEFs cultured for 48 hr in 0, 5, and 25 mM glucose. (G) NCoR1 protein was determined by western blotting from MEFs cultured for 24 hrs in 0, 5, or 25 mM glucose. (H) NCoR1 mRNA expression in MEFs grown for 48 hr in 0 mM glucose with or without 0.03 mM oleic acid (n=4). (I) NCoR1 protein determined by western blotting from MEFs cultured as indicated in (G). (J) NCoR1 mRNA measured by qRT-PCR in muscles of resting mice or 3 hrs after an endurance run (14-wk old; n =5), in mice that were fed for 20 wks either with HFD or CD (28-wk old; n=8), in mice that were fasted or fed for 16 hrs (14-wk old; n=10), and in 6-month or 2-year old mice (n=10). (K) Model schematizing how different levels of NCoR1 controls transcription of muscle genes by controlling the activity of transcription factors (TFs; i.e. PPARβ/δ, ERR, and MEF2).
Data are expressed as mean ± SEM.
See also Suppl. Fig. 7 and Suppl. Table 3.
At the transcriptional level, NCoR1 mRNA changed in response to different concentrations of glucose in the culture media (Figure 7D-E). Growing MEFs in low glucose decreased NCoR1 mRNA (Figure 7D) and protein (Figure 7E) levels, concomitant with the induction of its target genes (Pdk4, Vgefb, Mef2d, etc.). Similar results, i.e. decreased mRNA levels of NCoR1 associated with increased expression of its targets (Pdk4, Ucp2, Ucp3, Vegfb, Mb, Mck, Glut4), were also obtained in glucose-deprived C2C12 myotubes (Suppl. Fig. 7B). Interestingly, the reduction of glucose decreased mRNA levels of NCoR1, but not those of SMRT (Figure 7F). The tight dose-dependent correlation between NCoR1 expression and glucose levels in the culture medium (Figure 7E-F), suggested the possibility that NCoR1 could block the oxidation of lipid substrates when glucose was available. We therefore evaluated NCoR1 mRNA and protein in MEFs cultured in different fatty acid concentrations (Figure 7H-I). The addition of oleic acid (OA) to the medium to force fatty acid oxidation also decreased NCoR1 levels, independently of the glucose concentration (Figure 7H). These effects seemed again specific to NCoR1, as only a small difference in SMRT mRNA levels was observed with OA in the absence of glucose (data not shown). Together, these data indicate that settings that favor fatty acid oxidation (i.e. low glucose, low insulin, and high fatty acid) are all associated with a reduction of NCoR1.
We then test whether different conditions that enhance fatty acid oxidation also modulate NCoR1 mRNA levels in the muscle in vivo. This was indeed the case, as muscle NCoR1, but not SMRT, mRNA levels decreased after exercise (3 hrs after a resistance run), high fat feeding (20 wks of high fat feeding), fasting (after a 16 hr fast), and aging (6-mo vs 2-yr-old mice) (Figure 7J, data not shown). Interestingly, the reduction in NCoR1 mRNA levels match well with the potentiation of lipid oxidation after exercise (Kiens and Richter, 1998; Pilegaard et al., 2000), high fat feeding (Watanabe et al., 2006), fasting (Storlien et al., 2004) and in aged mice (unpublished data R. Houtkooper and J. Auwerx). Also in epidydimal white adipose tissue, HFD feeding reduced specifically NCoR1, and not SMRT, mRNA (Suppl. Fig. 7C). Unlike for the muscle, where we were unable to detect NCoR1 with the available antibodies, NCoR1 protein levels almost disappeared from epidydimal fat upon HFD (Suppl. Fig. 7D). Altogether, these data led us to suggest that NCoR1 is a negative transcriptional regulator of fatty acid oxidation and that a reduction of NCoR1 enables the muscle (and adipose tissue), to deal with lipid substrates more efficiently.
Discussion
The increased muscle mass, associated with a strikingly improved exercise capacity, is the most prominent phenotypic outcome of the muscle-specific NCoR1 gene deletion. The enhanced exercise capacity is associated with a reprogramming of glycolytic to more oxidative muscle fibers and a corresponding stimulation of oxidative mitochondrial metabolism, indicative of an improved intrinsic quality of the muscles. The increased muscle quantity and oxidative profile in NCoR1skm-/- mice also contributes to the slight improvement in metabolic parameters after high fat feeding and to the cold resistance subsequent to shivering thermogenesis (Canto and Auwerx, 2009).
Combined these properties suggest that NCoR1 acts as a master modulator of mitochondrial metabolism in the muscle, a hypothesis bolstered by the fact that the inhibition of the single worm NCoR homolog, gei-8, also robustly boosts muscle oxidative mitochondrial metabolism in C. elegans. The evolutionary conservation of the structure and function of this corepressor makes it tempting to speculate that NCoR may have evolved to facilitate metabolic adaptation of the mitochondria to energy availability, as has been described for other cofactors as SIRT1 (Canto and Auwerx, 2009).
As to how the absence of NCoR1 in the muscle achieves these remarkable effects, it is important to recall that NCoR1 docks histone deacetylases, such as HDAC3 (Alenghat et al., 2008) and SIRT1 (Picard et al., 2004). The use of mice with a point mutation in the NCoR1 deacetylase activation domain, incapacitating its interaction with HDAC3, indicated that this interaction is a nodal point in epigenetic regulation (Alenghat et al., 2008; Feng et al., 2011). The physiological alterations in the NCoR1skm-/- muscle together with the study of the expression correlates of NCoR1 suggested that, rather than the generalized transcriptional activation expected upon ablation of a corepressor platform protein, only a small set of transcriptional pathways was selectively affected.
Several transcription factors control muscle differentiation and development, including MyoD, myogenin, Myf5, Myf6, and MEF2 (Black and Olson, 1998; McKinsey et al., 2001, 2002; Potthoff and Olson, 2007). MEF2 especially caught our attention, as only its expression negatively correlated with NCoR1 (Figure 4B and Suppl. Fig. 10) and was induced in NCoR1skm-/- muscle (Figure 6A). MEF2 is key for muscle development, and also participates in muscle stress-response and remodeling in adulthood, such as occurs during muscle fiber type switch (Potthoff and Olson, 2007; Zhang et al., 2002). MEF2 activity is not only controlled at the level of its expression, but also by a wide range of intracellular signaling pathways and interacting co-regulator molecules [reviewed by (Potthoff and Olson, 2007)]. The histone acetyltransferase s, p300/CBP bind, acetylate, and activate MEF2 (Ma et al., 2005; McKinsey et al., 2001), whereas Class I [HDAC3 (Gregoire et al., 2007)], Class II [HDAC4, 5, 7, and 9 (Haberland et al., 2007; McKinsey et al., 2001)], and class III HDACs [SIRT1 (Zhao et al., 2005)] all are reported to interact with MEF2 and prevent the activation of its target genes (McKinsey et al., 2001). The absence of NCoR1 would hence favor the acetylation and activation of MEF2. The hyperacetylation of MEF2D and histone 4 in NCoR1 deficient MEFs, C2C12 myotubes, and muscles, which translates into the induction of MEF2 targets and the gain of muscle mass in NCoR1skm-/- mice, is fully in line with this idea (Figure 6).
The increased expression of genes related to fatty acid catabolism and mitochondrial respiration, underlie the oxidative mitochondrial changes observed in the NCoR1skm-/- mice. In the muscle, the expression of these gene sets is tightly controlled by NRs belonging to the PPAR and ERR families (Alaynick, 2008). PPARβ/δ, the predominant PPAR isoform in oxidative fibers, regulates oxidative capacity and enhances slow fiber-type function, resulting in improved exercise capacity and metabolic protection (Luquet et al., 2003; Tanaka et al., 2003; Wang et al., 2004). Amongst the ERRs, mainly ERRα and ERRγ seem to be involved in the coordination of muscle energy homeostasis [reviewed in (Giguere, 2008; Villena and Kralli, 2008)]. In line with this, a genome-wide location analysis of ERRα and ERRγ identified binding sites in genes of a large number of mitochondrial proteins (Dufour et al., 2007), and studies in mouse models show that they coordinate many aspects of muscle oxidative metabolism, including endurance capacity (Giguere, 2008; Narkar et al., 2011; Villena and Kralli, 2008). Several features of the NCoR1skm-/- mice are suggestive of the activation of PPARβ/δ and ERR. The demonstration that NCoR1 is recruited to the PPREs in the Ucp3 and the NR1/2 in Pdk4 promoters and that histone 4 is hyperacetylated on these promoters when NCoR1 is absent, suggests that coactivators now activate the transcription of these genes in a fashion unopposed by the NCoR1 corepressor platform (Figure 7K). Given that PGC-1α is a key coactivator of PPARβ/δ and ERR transcriptional programs [reviewed in (Handschin and Spiegelman, 2006)], it is also no surprise that several phenotypic features, ranging from similarities in gene expression patterns over the induction of mitochondrial oxidative metabolism and exercise capacity to HIF1α-independent angiogenesis, are shared between mice that lack NCoR1 or that overexpress PGC-1α (Arany et al., 2008; Lin et al., 2002).
NCoR1 action is tightly regulated by various physiological challenges and this is achieved through, at least, two different mechanisms. First, nuclear levels of NCoR1 are regulated. Our results show how insulin, which stimulates glucose oxidation at the expense of fatty acid oxidation, increases NCoR1 levels in the nucleus, enabling it to subsequently repress lipid oxidation genes. Interestingly, this effect of insulin on nuclear NCoR1 accumulation, is consistent with the positive effect of mTORC1 on nuclear NCoR1 accumulation, recently reported in hepatocytes (Sengupta et al., 2010). The second mechanism involves the modulation of NCoR1 expression. Exposing cells to media with low glucose and/or high fatty acid levels, reduces specifically NCoR1 (but not SMRT) mRNA and protein levels, ultimately de-repressing genes that control oxidative lipid metabolism. Likewise, endurance exercise, fasting, high fat feeding and aging, conditions paired with increased fat oxidation, also are characterized by attenuated muscle NCoR1 mRNA expression. Although we are unable to detect NCoR1 protein in the muscle with the currently available antibodies, our data in adipose tissue unequivocally show that the specific reduction in NCoR1 mRNA observed in that tissue after HFD is matched with a spectacular drop in NCoR1 protein. If the reduced NCoR1 mRNA levels in muscle also translate in corresponding changes in NCoR1 protein, they will prime the muscle for mitochondrial oxidation. These selective effects of NCoR1 to repress muscle fatty acid oxidation, hence suggest that changes in NCoR1 levels adapt transcriptional outcomes to physiological energy needs (Figure 7K).
In conclusion, we demonstrated here that NCoR is an evolutionary conserved negative regulator of both muscle mass and mitochondrial oxidative metabolism in nematode and mammals. In the mouse, NCoR1 achieves these effects through controlling a rather selected set of functional pathways, which are governed by MEF2, PPARβ/δ and the ERRs. The NCoR1skm-/- muscle phenotype furthermore mirrors many features of the stimulation of PGC-1α, a coactivator, whose action is less constrained by the absence of the NCoR1 corepressor scaffold. Our work also provides evidence that NCoR1 expression is regulated in a dynamic fashion and as such could play a role, similar to PGC-1α, in transcriptional adaptation to physiological challenges. Moreover, pharmacological inhibition of NCoR1 and/or its interaction with deacetylases may be a viable approach to improve muscle mass and oxidative metabolism. The fact that the inhibition of HDACs increases muscle cell size supports this concept (Iezzi et al., 2004). It is also tempting to speculate that the beneficial effects of the inhibition of mTORC1 and insulin signaling on health- and life-span, may in part rely on the attenuation of NCoR1 activity and the subsequent induction of oxidative metabolism in the muscle [reviewed in (Houtkooper et al., 2010)].
Experimental Procedures
Animal studies
NCoR1 floxed (NCoR1L2/L2), NCoR1skm+/+ and skm-/- mice were generated at the Mouse Clinical Institute (Strasbourg, France) and phenotyped (Champy et al., 2004; Champy et al., 2008) according to standard procedures that are described in detail in the Suppl. Materials. C. elegans strains, RNAi feeding experiments, GFP expression analysis are described in the Suppl. Materials. C.elegans O2 consumption was measured in 200 two-day old worms using a Seahorse XF24 as described in the Suppl. Materials.
Histological and EM analyses
Staining of muscles with hematoxylin/eosin, immunohistochemical and EM analysis, analysis of enzymatic activity of SDH and COX was carried out as described (Lagouge et al., 2006).
mRNA analysis and identification of NCoR1-correlated genes
The mRNA expression levels were measured in cells and tissues using qRT-PCR (Lagouge et al., 2006) The GeneNetwork program (http://www.genenetwork.org) was used to generate a broad range of NCoR1-correlated genes that may contribute to the phenotype of NCoR1skm-/- mice. Skeletal muscle mRNA expression was analyzed using 124 females from a classic F2 intercross between C57BL/6J and C3H/HeJ [UCLA BHHBF2 Muscle; (van Nas et al., 2010); GEO GSE12795]. NCoR1 (10024414685, 3′ UTR) was compared across all transcripts to find muscle covariates. Lung mRNA in a recombinant inbred intercross between C57BL/6J and DBA/2J was analyzed across 51 strains [HZI BXD Lung M430v2 (Apr08) RMA; (Alberts et al., 2011)]. Four NCoR1 probe sets from this microarray were analyzed (1423200_at, 3′ UTR; 1435914_at, 3′ UTR; 1423202_a_at, exonic and 3′ UTR; 1423201_at, exonic). For all probe sets, the top correlates were calculated (Suppl. Tables 4, 5). Strong or interesting correlates were selected for validation by qRT-PCR.
Cell culture, adenoviral infections, ChIP, co-IP, and Western blot experiments
Experimental details are provided in the Suppl. procedures.
Statistical Analyses
Statistical analyses were performed with a Student's t test for independent samples. Data are expressed as mean ± SEM, and p values smaller than 0.05 were considered as statistically significant. Statistical significance is displayed as * (p < 0.05), ** (p < 0.01), or *** (p < 0.001).
Supplementary Material
Acknowledgments
We acknowledge M. Lazar (University of Pennsylvania, Philadelphia), A. Kralli (Scripps Research Institute, San Diego), J-S. Annicotte and L. Fajas (Institut de Génétique Moléculaire de Montpellier, France), A. Lusis (University of California, Los Angeles), K. Schughart (Helmholtz Zentrum, Hannover, Germany), R. Williams (University of Tennessee Health Science Center, Memphis), and the Caenorhabditis Genetics Center (CGC) for generous sharing of research reagents and data. We thank N. Messadeq (Institut Clinique de la Souris, Strasbourg, France) for EM analysis, and the Center for PhenoGenomics (CPG) at the EPFL for help with mouse phenotyping. This work was supported by the École Polytechnique Fédérale de Lausanne, Swiss National Science Foundation, NIH [DK059820 (JA), DK062434 (RME), 1K08HL092298 (GDB), HD027183 (RME), DK057978 (RME)] the EU ideas program (ERC-2008-AdG-23118), the Helmsley Charitable Trust, the Glenn Foundation and the Howard Hughes Medical Institute (HHMI). HY was supported by an FRM fellowship. RME is an Investigator of the HHMI and the March of Dimes Chair in Molecular and Developmental Biology. JA is the Nestle Chair in Energy Metabolism. We thank the members of the Auwerx lab and R. Williams for discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alaynick WA. Nuclear receptors, mitochondria and lipid metabolism. Mitochondrion. 2008;8:329–337. doi: 10.1016/j.mito.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts R, Lu L, Williams RW, Schughart K. Genome-wide analysis of the mouse lung transcriptome reveals novel molecular gene interaction networks and cell-specific expression signatures. Respir Res. 2011;12:61. doi: 10.1186/1465-9921-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bucan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, Cooper M, Laznik D, Chinsomboon J, Rangwala SM, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- Argmann CA, Chambon P, Auwerx J. Mouse phenogenomics: the fast track to “systems metabolism”. Cell Metab. 2005;2:349–360. doi: 10.1016/j.cmet.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champy MF, Selloum M, Piard L, Zeitler V, Caradec C, Chambon P, Auwerx J. Mouse functional genomics requires standardization of mouse handling and housing conditions. Mamm Genome. 2004;15:768–783. doi: 10.1007/s00335-004-2393-1. [DOI] [PubMed] [Google Scholar]
- Champy MF, Selloum M, Zeitler V, Caradec C, Jung B, Rousseau S, Pouilly L, Sorg T, Auwerx J. Genetic background determines metabolic phenotypes in the mouse. Mamm Genome. 2008;19:318–331. doi: 10.1007/s00335-008-9107-z. [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011;93:884S–890. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis GA, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annu Rev Physiol. 2003;65:261–311. doi: 10.1146/annurev.physiol.65.092101.142528. [DOI] [PubMed] [Google Scholar]
- Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- Gregoire S, Xiao L, Nie J, Zhang X, Xu M, Li J, Wong J, Seto E, Yang XJ. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol Cell Biol. 2007;27:1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Arnold MA, McAnally J, Phan D, Kim Y, Olson EN. Regulation of HDAC9 gene expression by MEF2 establishes a negative-feedback loop in the transcriptional circuitry of muscle differentiation. Mol Cell Biol. 2007;27:518–525. doi: 10.1128/MCB.01415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg CE, Falkevall A, Wang X, Larsson E, Huusko J, Nilsson I, van Meeteren LA, Samen E, Lu L, Vanwildemeersch M, et al. Vascular endothelial growth factor B controls endothelial fatty acid uptake. Nature. 2010;464:917–921. doi: 10.1038/nature08945. [DOI] [PubMed] [Google Scholar]
- Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Williams RW, Auwerx J. Metabolic networks of longevity. Cell. 2010;142:9–14. doi: 10.1016/j.cell.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iezzi S, Di Padova M, Serra C, Caretti G, Simone C, Maklan E, Minetti G, Zhao P, Hoffman EP, Puri PL, et al. Deacetylase inhibitors increase muscle cell size by promoting myoblast recruitment and fusion through induction of follistatin. Dev Cell. 2004;6:673–684. doi: 10.1016/s1534-5807(04)00107-8. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Kiens B, Richter EA. Utilization of skeletal muscle triacylglycerol during postexercise recovery in humans. Am J Physiol. 1998;275:E332–337. doi: 10.1152/ajpendo.1998.275.2.E332. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Luquet S, Lopez-Soriano J, Holst D, Fredenrich A, Melki J, Rassoulzadegan M, Grimaldi PA. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. FASEB J. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- Ma K, Chan JK, Zhu G, Wu Z. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol Cell Biol. 2005;25:3575–3582. doi: 10.1128/MCB.25.9.3575-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- Miniou P, Tiziano D, Frugier T, Roblot N, Le Meur M, Melki J. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res. 1999;27:e27. doi: 10.1093/nar/27.19.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Fan W, Downes M, Yu RT, Jonker JW, Alaynick WA, Banayo E, Karunasiri MS, Lorca S, Evans RM. Exercise and PGC-1alpha-Independent Synchronization of Type I Muscle Metabolism and Vasculature by ERRgamma. Cell Metab. 2011;13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebbioso A, Manzo F, Miceli M, Conte M, Manente L, Baldi A, De Luca A, Rotili D, Valente S, Mai A, et al. Selective class II HDAC inhibitors impair myogenesis by modulating the stability and activity of HDAC-MEF2 complexes. EMBO Rep. 2009;10:776–782. doi: 10.1038/embor.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nofsinger RR, Li P, Hong SH, Jonker JW, Barish GD, Ying H, Cheng SY, Leblanc M, Xu W, Pei L, et al. SMRT repression of nuclear receptors controls the adipogenic set point and metabolic homeostasis. Proc Natl Acad Sci U S A. 2008;105:20021–20026. doi: 10.1073/pnas.0811012105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilegaard H, Ordway GA, Saltin B, Neufer PD. Transcriptional regulation of gene expression in human skeletal muscle during recovery from exercise. Am J Physiol Endocrinol Metab. 2000;279:E806–814. doi: 10.1152/ajpendo.2000.279.4.E806. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Gorza L, Sartore S, Saggin L, Ausoni S, Vianello M, Gundersen K, Lomo T. Three myosin heavy chain isoforms in type 2 skeletal muscle fibres. J Muscle Res Cell Motil. 1989;10:197–205. doi: 10.1007/BF01739810. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- Smith CL, O'Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Storlien L, Oakes ND, Kelley DE. Metabolic flexibility. Proc Nutr Soc. 2004;63:363–368. doi: 10.1079/PNS2004349. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Yamamoto J, Iwasaki S, Asaba H, Hamura H, Ikeda Y, Watanabe M, Magoori K, Ioka RX, Tachibana K, et al. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci U S A. 2003;100:15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nas A, Ingram-Drake L, Sinsheimer JS, Wang SS, Schadt EE, Drake T, Lusis AJ. Expression quantitative trait loci: replication, tissue- and sex-specificity in mice. Genetics. 2010;185:1059–1068. doi: 10.1534/genetics.110.116087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena JA, Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol Metab. 2008;19:269–276. doi: 10.1016/j.tem.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- Yu C, Markan K, Temple KA, Deplewski D, Brady MJ, Cohen RN. The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor gamma transcriptional activity and repress 3T3-L1 adipogenesis. J Biol Chem. 2005;280:13600–13605. doi: 10.1074/jbc.M409468200. [DOI] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma K, Sadana P, Chowdhury F, Gaillard S, Wang F, McDonnell DP, Unterman TG, Elam MB, Park EA. Estrogen-related receptors stimulate pyruvate dehydrogenase kinase isoform 4 gene expression. J Biol Chem. 2006;281:39897–39906. doi: 10.1074/jbc.M608657200. [DOI] [PubMed] [Google Scholar]
- Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.