Abstract
SET domain lysine methyltransferases (KMTs) catalyze the site- and state-specific methylation of lysine residues in histone and non-histone substrates. These modifications play fundamental roles in transcriptional regulation, heterochromatin formation, X chromosome inactivation and DNA damage response, and have been implicated in the epigenetic regulation of cell identity and fate. The substrate and product specificities of SET domain KMTs are pivotal to eliciting these effects due to the distinct functions associated with site and state-specific protein lysine methylation. Here, we review advances in understanding the molecular basis of these specificities gained through structural and biochemical studies of the human methyltransferases Mixed Lineage Leukemia 1 (MLL1, also known as KMT2A) and SET7/9 (KMT7). We conclude by exploring the broader implications of these findings on the biological functions of protein lysine methylation by SET domain KMTs.
Key words: SET domain, lysine methylation, transcription, S-Adenosylmethionine, crystal structure, substrate specificity, product specificity
Introduction
The genomic DNA of eukaryotes is packaged into a nucleoprotein complex termed chromatin that permits efficient compaction of the DNA within the nucleus. The fundamental repeating unit of chromatin is the nucleosome core particle, which is composed of two copies of each of the four core histones around which is wound approximately 150 base pairs of DNA.1 Covalent modifications of histones and DNA have been implicated in governing chromatin assembly, thereby regulating genomic processes that require access to the DNA template. Histones are subject to numerous modifications including acetylation, arginine methylation, lysine methylation, phosphorylation, ubiquitination and sumoylation.2 Of these modifications, lysine methylation has received considerable attention due to its involvement in transcriptional regulation, DNA damage response, recombination and cell cycle control.3 The major sites of lysine methylation occur in histones H3 and H4 and in the linker histone H1b and have variable effects on gene regulation that are frequently context-dependent. Methylation of Lys-4, Lys-36 and Lys-79 in histone H3 (H3K4, H3K36 and H3K79) is generally enriched in transcriptionally active genes, whereas methylation of H3K9, H3K27, H4K20 and H1bK26 frequently demarcate silent chromatin.4 The signals encoded by these modifications are transduced by effector proteins with binding modules that recognize methyllysines in a sequence-specific manner.5
In 2000, the first histone-specific KMTs, mammalian SUV39H1 and its Schizosaccharomyces pombe homolog, Clr4, were reported, representing a milestone in the field of histone lysine methylation.6 These enzymes are members of a conserved family of S-adenosylmethionine (AdoMet)-dependent KMTs that possess a catalytic SET domain. Subsequent studies isolated numerous SET domain KMTs that display distinct specificities for lysine residues in histones.7–9 In addition, SET domain KMTs display product specificity that is defined as their ability to catalyze mono-, di- or trimethylation of the lysine ε-amine group. Product specificity introduces an additional hierarchy in methyllysine signaling due to the ability of methyllysine binding proteins to discriminate among different degrees of methylation in recognizing their targets. Insights into the molecular basis of the substrate and product specificities of monomeric SET domain KMTs have been gained through numerous structural and functional studies.7–9 While these findings are generally applicable to the SET domain family, certain KMTs, such as SET1/MLL and EZH2 (KMT6), are only functional within large heteromeric complexes whose subunits have a pivotal role in defining their respective substrate and product specificities.10,11 The molecular basis by which these subunits regulate the activity and specificity of SET1/MLL and EZH2 is unresolved and remains an active area of investigation.
Over the past several years, numerous non-histone substrates of SET domain KMTs have been identified, implicating new biological functions for these enzymes.12 For example, the regulatory domain of the tumor suppressor p53 is site-specifically methylated by several KMTs, including SET7/9,13 PR-SET7/SET8 (KMT5A),14 SMYD2 (KMT3C),15 and G9A (KMT1c) and its homolog GLP (KMT1D).16 Other proteins which are subject to site-specific methylation by KMTs include the TFIID-associated factor TAF10,17 HIV TAT,18,19 the yeast ribosomal proteins Rpl12ab, Rpl23ab and Rpl42ab,20–23 DNA methyltransferases I (DNMT1),24–27 the histone acetyltransferase PCAF,28 the retinoblastoma protein (pRb),29–31 the vascular endothelial growth factor receptor,32 and the transcription factors NFκB33–36 estrogen receptor α (ERα),37 androgen receptor (AR),38,39 and E2F1,40 and STAT3.41 The increasing awareness that protein lysine methylation is a widespread modification represents an emerging and exciting area of KMT biology.
In this review, we survey current advances in understanding the substrate and product specificities of SET domain KMTs. Specifically, we focus on recent structural and functional studies of the H3K4-specific methyltransferase MLL1 and on a series of articles reporting novel substrates of human SET7/9. Together, these studies offer new insights into the specificities and biological functions of SET domain KMTs.
MLL1-H3K4 Specificity and Complex Assembly
H3K4 trimethylation is frequently associated with the promoters of actively transcribed genes.42 This modification can function to recruit chromatin remodeling and modifying complexes as well as the basal transcription machinery.5,43,44 Among the enzymes that catalyze this modification are the SET1 and MLL (KMT2) methyltransferases that are conserved from yeast through humans.11 The Saccharomyces cerevisiae genome encodes a single H3K4 methyltransferase, Set1, whereas humans possess at least six homologs: SET1a, SET1b and MLL1-4.45 Unlike many SET domain enzymes, SET1 and MLL KMTs display very weak activity toward H3K4 and require additional subunits to attain maximal activity.46 In budding yeast, Set1 has been isolated in a large macromolecular complex termed COMPASS that is composed of seven subunits, whereas six distinct COMPASS-like complexes have been isolated in humans.11 Despite their variable composition, the human SET1 and MLL complexes share four common subunits, WDR5, RbBP5, ASH2L and DPY30, which form a core complex abbreviated WRAD. Of these complexes, MLL1 has garnered the most attention due to its importance in regulating Hox gene expression during hematopoietic development and its involvement in leukemia.11,47,48 Furthermore, genetic rearrangements of the MLL1 gene yield in-frame translocations to multiple proteins that results in aberrant gene regulation and leukemogenesis.11,47,48 Due to its relevance to cancer, MLL1 has emerged as an archetype for understanding the structure and specificity of the SET1/MLL family of KMTs.
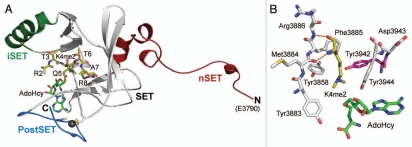
To elucidate its specificity for H3K4, Southall et al. determined the crystal structure of the MLL1 catalytic domain in complex with the product S-adenosylhomocysteine (AdoHcy) as well as a ternary complex with AdoHcy and a dimethylated H3K4 (H3K4me2) peptide.49 The structure of the MLL1 catalytic domain adopts a convoluted β-sheet fold that is conserved in the SET domain family8 and possesses a PostSET motif that contributes three cysteines to a four cysteine zinc cluster adjacent to the substrate binding clefts (Fig. 1A). In the ternary complex, the H3K4me2 peptide is bound in a wide cleft defined by the inserted SET (iSET) helix and the PostSET motif. An alignment of this structure with the ternary complex of Neurospora crassa DIM-5 bound to AdoHcy and an H3K9 peptide substrate illustrates substantial differences in the conformations of the iSET motif in their respective catalytic domains. In DIM-5, the iSET motif is oriented in close proximity to the PostSET motif, forming a narrow, well-defined lysine binding channel that constrains the motion of the K9 side chain to promote its methylation.50 In contrast to this closed conformation, the iSET motif is shifted away from the PostSET motif in MLL1, resulting in an open active site that is unable to optimally orient the K4 side chain for methylation. Based on this observation, Southall et al. hypothesized that the interaction of the WRAD subunits with the iSET motif of MLL1 may induce the closure of the active site (Fig. 1A and B), thereby stimulating H3K4 methylation.49 Consistent with this hypothesis, alanine substitutions of the surface-exposed residues Gln-3867 and Arg-3871 in the iSET helix diminished the activity of the MLL1 complex but did not appreciably affect the activity of the MLL1 catalytic domain itself when compared to the wild-type complex and wild-type catalytic domain. These findings led them to conclude that the interactions between the MLL1 iSET motif and the WRAD subunits may be critical to inducing the closure of the active site to stimulate H3K4 methylation by the core complex. It should be noted that the significance of the open conformation of the MLL1 active site remains uncertain, as a recent analysis of the MLL1 crystal structure suggests that this conformation may be a consequence of contacts with a neighbor molecule in the crystal lattice.51
Figure 1.
Crystal structure of the catalytic domain of human MLL1 bound to AdoHcy and a histone H3 dimethylated Lys-4 (H3K4me2) peptide at 2.2 Å resolution. (A) Ribbon diagram of the catalytic domain of MLL1 with the N-terminal SET (nSET; red), SET (gray), inserted SET (iSET; green) and PostSET (blue) domains denoted (PDB accession code 2W5Z). The PostSET Zn atom (gray sphere) and its coordinating cysteine residues are also illustrated. The H3K4me2 peptide and the product cofactor AdoHcy are depicted in stick representation with gold and green carbon atoms, respectively. The N- and C-termini of the MLL1 catalytic domain are also denoted. Several of the residues in the H3K4me2 peptide adopt alternative conformation. For clarity, these residues are rendered in one conformation. (B) Active site of MLL1 bound to K4me2 and AdoHcy. Residues in the enzyme and ligands are colored as in (A), and hydrogen bonds are shown as orange dashed lines. The Phe/Tyr switch residue Tyr-3942 is highlighted in magenta.
In addition, Southall et al. characterized the product specificity of MLL1 toward H3K4. In the absence of the WRAD complex, MLL1 was shown to methylate H3K4 and H3K4me1 peptides and also exhibited weak activity using an H3K4me2 peptide.49 The extent to which the WRAD subunits augment MLL1 activity was then examined. Incubation of MLL1 with WDR5, which binds to a WDR5-interacting (Win) motif in MLL1 that is N-terminal to the SET domain,52–54 did not appreciably alter H3K4 methyltransferase activity. Conversely, the addition of RbBP5 or WDR5-RbBP5 heterodimer stimulated the activity of MLL1 approximately five-fold, whereas incubation of MLL1 with an ASH2L-DPY30 complex resulted in an approximately three-fold increase in H3K4 methylation.49 The addition of the WRAD complex synergistically stimulated H3K4 methylation nearly 20-fold, in agreement with prior studies showing that core subunits are essential for optimal H3K4 methylation.55
In a parallel study, Patel et al. investigated the assembly and architecture of the MLL1-WRAD complex using a combination of biochemical and biophysical approaches.52 An analysis of the pair wise interactions among MLL and WRAD subunits by analytical ultracentrifugation demonstrated that these proteins associate with low micromolar to nanomolar binding affinities and assemble in the following order: MLL1 ↔ WDR5 ↔ RbBP5 ↔ ASH2L ↔ (DPY30)2 (ASH2L associates with a DPY30 dimer). The association of MLL1 with WDR5 is consistent with prior studies demonstrating that WDR5 specifically binds to a WDR5 interacting (Win) motif that precedes the N-terminal SET (nSET) region of the MLL1 catalytic domain (Fig. 1A).52–54,56
In addition to examining MLL1 complex assembly, Patel et al. investigated the product specificities and kinetic reaction mechanism of the MLL1 catalytic domain and the core complex containing MLL1 and WRAD.52 They reported that the MLL1 catalytic domain monomethylates an H3K4 peptide, whereas the MLL1-WRAD complex dimethylates this peptide with no appreciable levels of trimethylated product detected using a mass spectrometry-based assay. Further analysis revealed that the MLL1-WRAD complex dimethylates H3K4 via a distributive mechanism in which the H3K4me1 intermediate is released into solution prior to its conversion to H3K4me2. To ascertain whether residue(s) in the active site of MLL1 regulate its product specificity, a phenylalanine mutation of the Phe/Tyr switch residue Tyr-3942 was generated. This position within the SET domain active site has been implicated in regulating the product specificity of KMTs.8 The Y3942F mutation in MLL1 altered its specificity from a mono- to a trimethyltransferase,52 consistent with a homologous Y1052F mutation in yeast Set1 that displays enhanced H3K4 trimethylation.57 Collectively, the altered specificity of the Y3942F mutant coupled with the distributive kinetic mechanism of the MLL1-WRAD complex, led Patel et al. to conclude that a second methyltransferase activity exists within the complex that catalyzes H3K4 dimethylation.52 In agreement with this hypothesis, they reported that the WRAD complex possesses intrinsic KMT activity and can dimethylate H3K4 in the presence MLL1. Assays of the individual WRAD proteins showed no enzymatic activity, indicating that the intact WRAD complex is required for H3K4 methylation, perhaps through forming an active site at the interface of the subunits. This finding is surprising given that that previous studies failed to detect methyltransferase activity in WRAD.55 Moreover, a Saccharomyces cerevisiae set1Δ strain exhibits a global loss in H3K4 mono-, di- and trimethylation in vivo,58 implying that the homologous WRAD complex in yeast does not possess intrinsic methyltransferase activity.
In comparing the studies by Southall et al. and Patel et al. there are several differences in their results and conclusions that merit discussion. Most notable is the observation by Patel et al. that MLL1 catalytic domain catalyzes H3K4 monomethylation and that the WRAD complex is responsible for H3K4 dimethylation.52 In contrast, Southall et al. demonstrated that the MLL1 catalytic domain can methylate H3K4 and H3K4me1 peptides to mono- and dimethylated states, respectively, in the absence of WRAD.49 The reasons for the discrepancies in MLL1 product specificity may reflect variations in the assay conditions in each study, such as differences in substrate and enzyme concentrations, variations in the histone H3 peptide length, and differences in the size of the MLL1 constructs assayed.49,52 It is also worth noting that the product specificities measured with peptides may not reflect the specificity of MLL1-WRAD for H3K4 in the context of full length histone or nucleosomal substrates. Additionally, there are marked differences in the reported magnitudes of stimulation of MLL1 by the WRAD complex in each study, with Southall et al. having measured a ∼20-fold increase in activation, whereas Patel et al. observed a 600-fold enhancement in H3K4 methylation. These variations may be due to the length of the MLL1 constructs employed in the respective studies. The MLL1 construct (residues 3,745–3,969) assayed by Patel et al. contains the Win motif (amino acids 3,762–3,767) that was previously shown to be critical for association with WDR5 and proper complex assembly and activation.52–54,56 However, the crystallized MLL1 construct (residues 3,785–3,969) lacks the Win motif (Fig. 1A), providing a possible explanation for its diminished stimulation by the WRAD complex.49
The variations in MLL1 activation by the WRAD complex also highlight differences in the reported complex assembly. The ultracentrifugation data presented by Patel et al. imply a linear assembly of the MLL1-WRAD complex with the interactions between MLL1 and WRAD mediated by the binding of WDR5 to the Win motif in MLL. However, the KMT assays performed by Southall et al. show that RbBP5 and ASH2L/DPY30 modestly stimulate MLL1's activity in the absence of WDR5.49 The latter observations are consistent with the proposed interactions between the iSET helix of the MLL1 SET domain and the WRAD subunits that may induce the closure of the active site to enhance H3K4 methylation. Collectively, these findings illustrate the complexity of the interactions between MLL1 and the WRAD subunits, which are not only required for complex integrity but are also critical in stimulating MLL1 methyltransferase activity. The resolution to these discrepancies will require more detailed structural and functional characterization of the MLL1-WRAD core complex to elucidate the interactions among the subunits and the mechanism by which the MLL1 SET domain is activated.
Recently, two independent groups have investigated the methyltransferase activity of the WRAD complex.59,60 Patel et al. reported that WDR5, RbBP5 and ASH2L (WRA) are necessary and sufficient to reconstitute this activity, whereas DPY30 increases the catalytic efficiency of the complex by reducing the KM values for AdoMet and the histone H3 peptide substrate.60 The activities of WRA and WRAD are relatively low, requiring micromolar concentrations of the complexes to measure H3K4 methylation using a radiometric assay. Consistent with previous studies in reference 53–56, methylation of nucleosomal H3K4 by MLL1 was greatly enhanced in the presence of the WRAD complex and required interactions between the MLL1 Win motif and WDR5.60 Finally, Patel et al. noted that the methyltransferase activity of the WRAD complex is Zn-dependent, based on the observation that its activity is inhibited by metal chelators and is restored by the addition of excess Zn(II).60 This inhibition may be due to the chelation of the Zn(II) ions in the PHD domain of ASH2L, which would presumably destabilize the structure of ASH2L and the WRAD complex.
Cao et al. also investigated the methyltransferase activity of the WRAD complex.59 In contrast to previous studies in reference 52 and 60, they reported that basal activity resides in the ASH2L-RbBP5 heterodimer. This activity was detected using radiometric assays with tritiated AdoMet and required a three week exposure to visualize by fluorography. The ASH2L-RbBP5 complex was shown to methylate an unmodified H3K4 peptide, whereas the catalytic domain of MLL1 methylated unmodified H3K4 and, to a lesser extent, H3K4me1 peptides in radiometric assays. Similarly, Southall et al. demonstrated that the MLL1 catalytic domain can catalyze mono- and dimethylation of H3K4, although MLL1 displayed comparable activity toward H3K4 and H3K4me1 peptides in their assays.49 Cross-linking studies using ultraviolet radiation and radiolabeled-AdoMet revealed that both MLL1 and ASH2L interact with AdoMet and that cross-linking of ASH2L to the cofactor is sensitive to mutations in the SET domain of MLL1, specifically Y3858F and Y3942A.59 Finally, Cao et al. observed that the WRA complex and, to a lesser extent, the ASH2L-RbBP5 heterodimer stimulated MLL1 activity in a concentration-dependent manner. Stimulation of MLL1 activity required a higher concentration of the ASH2L-RbBP5 complex relative to that of WRA complex. These findings may in part explain some of the discrepancies in the activity and specificity of the MLL1 complex in earlier reports in reference 49 and 52. Ultimately, further studies are needed to address the interrelationship between the activities and product specificities of MLL1 and the WRAD complex.
Identification of Non-Histone Substrates of SET7/9
Similar to MLL1 and SET1, human SET7/9 was initially isolated as an H3K4-specific KMT61,62 and was later shown to catalyze monomethylation of this site.63,64 Although classified as an H3K4-specific enzyme, this KMT cannot efficiently methylate nucleosomal substrates compared to H3K4 peptides or full length histone H3,61,62 suggesting that it may possess other substrates beside histone H3. Subsequently, multiple non-histone substrates of SET7/9 have been identified and characterized in vitro and in vivo, including TAF10,17 p53,13 ERα,37 DNMT1,25,27 pRb,29,65 TAT,18 PCAF,28 E2F1,40 AR,38,39 STAT3,41 and the p65/RelA subunit of NFκB.33,35 Methylation of these proteins by SET7/9 has been reported to have differential effects on their stabilities, functions or interactions with binding partners as discussed below.
One function attributed to SET7/9-mediated methylation is to regulate protein:protein or protein:nucleic acid interactions. The first example of this function was reported for the TBP-associated TAF10 that is methylated by SET7/9 at Lys-189 in the loop 2 motif of its histone fold domain.17 This modification has been shown to enhance the affinity of TAF10 for RNA polymerase II, stimulating the transcription of a subset of TAF10-dependent genes. Similarly, Gaughan et al. have reported that SET7/9 methylates AR at Lys-362 in the hinge domain linking the DNA binding and ligand binding domains of the receptor.38 Methylation of Lys-362 induces interactions between the N-terminal and C-terminal domains of AR, promoting the transcription of androgen-responsive genes. However, Ko et al. has recently reported that SET7/9 methylates Lys-360 as opposed to Lys-362 in AR, stimulating transcriptional activation.39 The discrepancies in the methylation site(s) in AR remain unclear and need to be resolved. In contrast to TAF10 and AR, methylation of STAT3 at Lys-140 by SET7/9 promotes the dissociation of the transcription factor from promoter elements, downregulating the transcription of a subset of STAT3-dependent genes.41 Methylation of Lys-140 appears to be dependent on the phosphorylation of Tyr-705 and Ser-727 in STAT3 and can be reversed by the lysine demethylase LSD1 (KDM1). However, the mechanism by which Lys-140 methylation in STAT3 stimulates its dissociation form promoters is currently unknown.
SET7/9 has also been reported to coactivate HIV gene expression. Studies by Pagans et al. have shown that SET7/9 associates with the TAR RNA step-loop in the HIV promoter and interacts with the HIV transactivator TAT and positive transcriptional elongation factor b (P-TEFb).18 Further, SET7/9 monomethylates TAT at Lys-51, a residue that is predicted to interact with the TAR RNA. Pagans et al. have proposed that this modification, coupled with the interactions between SET7/9 and the TAT•P-TEFb complex, stabilize the association of the latter with the TAR RNA to stimulate HIV transcription. Thus, these findings reveal a novel function for SET7/9 in HIV pathogenesis.
In addition to mediating macromolecular interactions, SET7/9 methylation has been implicated in governing protein stability and turnover. The first example of this type of regulation was reported for the tumor suppressor p53, which is methylated by SET7/9 at Lys-372 in its regulatory domain, increasing its half-life and transactivation potential.13 Subsequent studies revealed that SET7/9 methylates this site in response to DNA damage, initiating cell cycle checkpoints and stimulating the acetylation of neighboring lysine residues in the regulatory domain of p53 by the mammalian acetyltransferases p300, CBP and TIP60.66,67 In turn, this acetylation precludes ubiquitin-mediated degradation of p53 by the proteasome, thus explaining the molecular basis by which Lys-372 methylation extends the half-life of p53. Similarly, SET7/9 has been shown to stabilize ERα through methylation of Lys-302 in the hinge motif of the receptor.37 Methylation at this site also promotes the recruitment of ERα to estrogen responsive elements in target genes through a localization mechanism that is not well understood. Interestingly, SET7/9 methylates AR and ERα at the same site within their respective hinge motifs, but the effects of these modifications differ and thus appear to be context-dependent.37,38
While the previous examples illustrate that lysine methylation can enhance protein stability, methylation of other substrates by SET7/9 can result in their degradation. Wang et al. have reported that methylation of Lys-1096 in mouse DNMT1 (Lys-1094 in human DNMT1) increases its turnover in vivo and that LSD1 can reverse this modification to promote DNMT1 stability and DNA methylation.27 In a separate study, Estève et al. have shown that SET7/9 methylates human DNMT1 at Lys-142, inducing ubiquitination and degradation by the proteasome.25 More recent studies have revealed that phosphorylation of Ser-143 in DNMT1 by the AKT1 kinase inhibits methylation at Lys-142 by SET7/9, promoting DNMT1 stability during S phase of the cell cycle.24 Although the reported sites of methylation in human and mouse DNMT1 differ, the resulting effects of these modifications in diminishing DNMT1 stability are similar and reveal a novel mechanism by which lysine methylation can influence DNA methylation. The relationship between SET7/9 methylation and protein degradation is not limited to DNMT1. Kontaki et al. have reported that SET7/9 methylates the cell cycle factor E2F1 at Lys-185, resulting in its ubiquitination and proteasomal degradation.40 In addition, Lys-185 was found to inhibit phosphorylation and acetylation of E2F1, modifications that are associated with the stabilization of the protein in vivo. Collectively, these findings illustrate that the methylation of target proteins by SET7/9 can have distinct effects on their stability that is frequently dependent on the context of other covalent modifications, such as acetylation, phosphorylation and ubiquitination.
In contrast to the preceding proteins, other substrates can be methylated at multiple lysines by SET7/9. For example, SET7/9 can methylate six lysine residues in the lysine acetyltransferase PCAF in vitro, of which Lys-78 and Lys-89 appear to be the preferred methylation sites in the full-length protein.28 Western blot and immunofluorescence analysis confirmed that Lys-89 in PCAF is monomethylated in vivo. Although the function(s) associated with PCAF methylation is presently unresolved, it is conceivable that these modifications may have roles in modulating its stability or its interactions with protein substrates or subunits within PCAF-containing complexes. The tumor suppressor pRb is also subject to multiple methylation by SET7/9 within its C-terminal region. Munro et al. have reported that methylation of Lys-873 serves to recruit heterochromatin protein 1 (HP1) to repress pRb target genes and cell cycle progression.65 Furthermore, they proposed that Lys-873me1 is recognized by the methyllysine-binding chromodomain of HP1, which may recruit other heterochromatin proteins, such as the H3K9-specific KMT SUV39H1 that has previously been shown to interact with pRb to promote transcriptional silencing.30 More recently, Carr et al. have shown that SET7/9 methylates Lys-810 in pRb, which inhibits phosphorylation of the neighboring Ser-807 and Ser-811 by cyclin-dependent kinases (CDKs).29 This finding reveals that Lys-810 methylation by SET7/9 antagonizes CDK-dependent phosphorylation of pRb, thus maintaining the tumor suppressor in a hypo-phosphorylated state that inhibits cell division.
Another substrate that has been shown to be multiply methylated is the RelA/p65 subunit of NFκB, a transcription regulatory complex associated with inflammatory response, autoimmune diseases, cancer and diabetes.33,35 Yang et al. have reported that SET7/9 methylates Lys-314 and Lys-315 in the p65/RelA subunit of NFκB, which represses the expression of NFκB target genes due to ubiquitination and proteasomal degradation of RelA.35 In a subsequent study, they showed that Lys-310 acetylation prolongs the half-life of RelA by antagonizing Lys-314/315 methylation, implying a negative interplay between lysine acetylation and methylation that governs the stability of RelA in vivo.68 In contrast, Ea and Baltimore have reported that SET7/9 methylates RelA at Lys-37 and that this modification stimulates transactivation by NFκB through enhancing DNA binding by RelA.33 Taken together, these results suggest that SET7/9 has opposing functions in NFκB regulation and that methylation at Lys-37 versus Lys-314/315 must be tightly controlled to elicit the appropriate signaling output through the NFκB pathway.
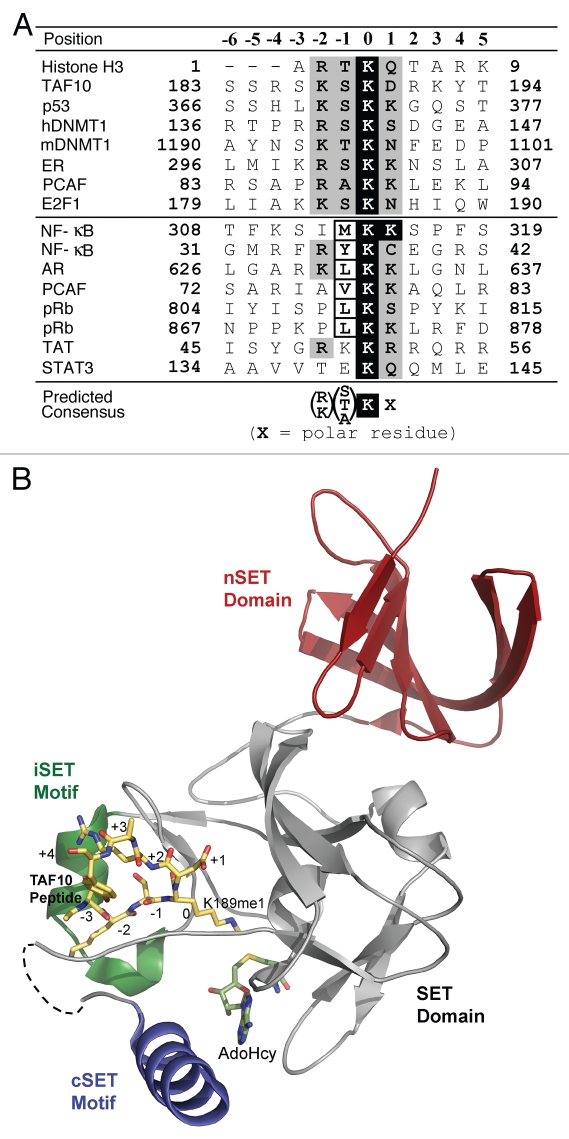
The discovery that SET7/9 methylates multiple proteins has spurred efforts to devise models for its specificity that will facilitate de novo prediction of new substrates and functions for the enzyme. Initial efforts to predict its specificity were based upon sequence alignments of the methylation sites histone H3, TAF10, p53, ERα and DNMT1, representing the first group of substrates identified for SET7/9.13,25,37,69 Together, these analyses led to a predicted consensus recognition motif for SET7/9: (KR)-(STA)-K-X, where K is the methylation site and X is a polar residue (Fig. 2A). Structural studies of the catalytic domain of SET7/9 bound to histone H3, p53, TAF10 and ERα peptides reveal that the residues in the −2, −1 and +1 positions of the methylation site engage in homologous interactions within the enzyme's protein substrate binding cleft, illustrating its preference for basic, small and polar amino acids, respectively, at these positions (Fig. 2B).13,37,63,69 A recent peptide array-based analysis of SET7/9 specificity has expanded the original consensus motif, proposing that the enzyme recognizes: (GRHKPST)-(K > R)-(S > KYARTPN)-K-(QN)-(AQGMSPTYV).70 This revised motif illustrates that SET7/9 displays a broader methylation site specificity than suggested in previous structure-function studies.13,25,37,69
Figure 2.
Substrate specificity of SET7/9. (A) Sequence alignment of the methylation sites of SET7/9 substrates. The methylated lysine is highlighted by white text on a black background and flanking residues that are conserved in at least half of the sequences are depicted with a gray background. The numbers at the top of the alignment denote the relative position of the residues in the methylation sites with respect to the target lysine (position 0). The predicted consensus sequence is shown at the bottom of the alignment and is based on the upper eight methylation sites. (B) The structure of the catalytic domain of human SET7/9 bound to a monomethylated TAF10 peptide (yellow carbon atoms) and AdoHcy (green carbons) at 1.3 Å resolution (PDB accession code 2F69). Positions of the consensus sequence in the TAF10 peptide are indicated by integers. The SET, the nSET, iSET and cSET domains in SET7/9 are colored gray, red, green and blue respectively. The dashed line represents the connectivity of the backbone for amino acids that were not resolved in the crystal structure.
Intriguingly, the more recently identified substrates of SET7/9 possess methylation sites that are strikingly dissimilar from the expanded consensus motif.70 Alignments of the methylation sites in AR, PCAF (Lys-72), pRb (Lys-804 and Lys-867), NFκB RelA (Lys-308 and Lys-309), and STAT3 illustrate that these sequences diverge from the consensus motif at either the −2 or −1 positions, which are directly involved interactions in the enzyme's protein substrate binding cleft (Fig. 2A and B). Further, these sites lack any obvious sequence homology amongst themselves, with the exception of a hydrophobic residue in the −1 position in certain substrates. These findings raise questions concerning the mechanism by which SET7/9 recognizes and methylates these proteins, particularly given that many SET domain KMTs display a high degree of site specificity within their respective substrates. One explanation for the apparent broad specificity of SET7/9 is that interactions distal to its substrate binding cleft may be responsible for the recognition of substrates with methylation sites that diverge from the predicted motif. One candidate for these distal interactions is N-terminal domain of the enzyme. Crystallographic studies have shown that this domain is composed of several MORN motifs that adopt an extended β-sheet structure.71 Recent studies by Pagan et al. have demonstrated that the N-terminal domain of SET7/9 (residues 1–109) binds HIV-1 TAR RNA,18 illustrating that this domain can mediate macromolecular interactions. It is conceivable that the N-terminal domain of SET7/9 may also promote substrate binding through docking of the methylation site of the substrate with the C-terminal SET domain. The non-SET domain KMT PrmA utilizes a similar mechanism to methylate Lys-3, Lys-39 and the N-terminal α-amine of the ribosomal protein L11.72 PrmA possesses an N-terminal domain that grasps L11 protein and facilitates the docking of the different methylation sites into the active site of the enzyme's C-terminal catalytic domain. It remains to be seen whether SET7/9 utilizes a similar binding mode for recognizing and methylating substrates, particularly those possessing methylation sites that diverge form the predict consensus motif. Ultimately, the answers to these questions will require more detailed structural studies of SET7/9 that investigate its plasticity in recognizing sites with diverse sequences and whether distal interactions with enzyme's N-terminal domain participates in substrate recognition.
Summary
Recent studies of SET7/9 and the SET1/MLL family have offered new insights into the substrate and product specificities of SET domain KMTs and their regulation within heteromeric complexes. These studies also illustrate the conceptual challenges associated with understanding the molecular mechanisms of substrate and product specificity. With respect to the SET1/MLL KMTs, the activity and product specificities of these enzymes are dependent on the subunits with which they associate to form catalytically competent methyltransferase complexes. Similarly, the specificity of mammalian EZH2 for H3K27 or H1bK26 is defined by the different Polycomb Repressive Complexes in which the enzyme resides.73,74 Although many SET domain KMTs are active as individual proteins, it is conceivable that these enzymes also associate with subunits that modulate their specificities or provide targeted recruitment to specific intracellular regions to promote site-specific methylation. With respect to MLL1, two independent studies have begun to elucidate the structural basis of complex assembly, illustrating the interactions that mediate contact between a short sequence of RbBP5 and the WD40 repeat domains of WDR5.75,76 In addition, recent structural studies have shown that the N-terminal domain of ASH2L possesses a winged helix DNA binding motif that facilitates recruitment of the MLL1 complex to the β globin and Hox gene loci.77,78 These structures have laid the groundwork for further characterization of the assembly and function of the MLL and SET1 complexes.
Another aspect that has emerged from the biochemical characterization of the MLL1 complex is the novel methyltransferase activity of the WRAD complex.52,59,60 Classification of the domain architectures of the WRAD subunits using the protein domain database SMART79 reveals that they possess WD40, PHD and SPRY domains but lack identifiable methyltransferase catalytic motifs. This sequence analysis implies that the methyltransferase activity of WRAD may reside in the regions of the WRAD subunits that are devoid of classified domains. Alternatively, studies by Cao et al. indicate that the methyltransferase active site is situated at an interface between the ASH2L SPRY domain and residues 330–356 of RbBP5.59 Further studies are required to define the exact location, structure and mechanism of the novel lysine methyltransferase activity reported for the WRAD complex. Finally, the physiological relevance of the WRAD complex's methyltransferase activity needs to be resolved, particularly given its low enzymatic activity.
The identification of multiple substrates of SET7/9 has increased our awareness that lysine methylation is a widespread covalent modification. Dhayalan et al. have very recently reported nine new non-histone substrates of SET7/9, including AKA6, CENPC1, CULLIN1, IRF1, MeCP2, MINT, PPARBP, TTK and ZH8, highlighting the participation of this KMT in multiple signaling pathways.70 In addition to SET7/9, other SET domain KMTs, such as SMYD2,15,31 SMYD3,32 PR-SET7/SET8,14 NSD1,34 SETD6,36 SETDB1 and its homolog SETDB2,19 and G9A and GLP16,80 have been shown to methylate various non-histone substrates, further illustrating that lysine methylation is not confined to histones. Together, these findings imply that other KMTs presently classified as histone-specific possess additional substrates and functions that await characterization. Another open question is whether non-histone proteins are subject dynamic methylation by KMTs and lysine demethylases (KDMs), as has been demonstrated for histone lysine methylation.3 Evidence for dynamic methylation is beginning to emerge based on the findings that LSD1 can reverse methylation at Lys-370 in p53,81 Lys-1094 in human DNMT1,27 Lys-185 in E2F1,40 and Lys-140 in STAT3.41 The identification of alternative substrates of the JmjC family of KDMs has lagged behind that of LSD1, although a recent study has shown that JHDM1A (also known as FBXL11 and KDM2A) can demethylate NFκB at Lys-218 and Lys-221, the sites of methylation by NSD1.34 These findings suggest that other JmjC enzymes may also demethylate non-histone substrates and thus have functions that extend beyond chromatin modification. In summary, these results underscore the concept that lysine methylation is dynamic in nature and has broad functions in intracellular signaling pathways.
Acknowledgments
We thank Ali Shilatifard, Daniel Bochar, Jean-François Couture and Scott Horowitz for reading the review and providing useful comments. R.C.T. is supported by a grant from the National Institutes of Health (GM073839).
References
- 1.Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–196. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 4.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 5.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 7.Couture JF, Trievel RC. Histone-modifying enzymes: encrypting an enigmatic epigenetic code. Curr Opin Struct Biol. 2006;16:753–760. doi: 10.1016/j.sbi.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 14.Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 16.Huang J, Dorsey J, Chuikov S, Zhang XY, Jenuwein T, Reinberg D, et al. G9a and Glp Methylate Lysine 373 in the Tumor Suppressor p53. J Biol Chem. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 18.Pagans S, Kauder SE, Kaehlcke K, Sakane N, Schroeder S, Dormeyer W, et al. The Cellular lysine methyltransferase Set7/9-KMT7 binds HIV-1 TAR RNA, monomethylates the viral transactivator Tat, and enhances HIV transcription. Cell Host Microbe. 2010;7:234–244. doi: 10.1016/j.chom.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Duyne R, Easley R, Wu W, Berro R, Pedati C, Klase Z, et al. Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology. 2008;5:40. doi: 10.1186/1742-4690-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porras-Yakushi TR, Whitelegge JP, Clarke S. A novel SET domain methyltransferase in yeast: Rkm2-dependent trimethylation of ribosomal protein L12ab at lysine 10. J Biol Chem. 2006;281:35835–35845. doi: 10.1074/jbc.M606578200. [DOI] [PubMed] [Google Scholar]
- 21.Porras-Yakushi TR, Whitelegge JP, Clarke S. Yeast ribosomal/cytochrome c SET domain methyltransferase subfamily: identification of Rpl23ab methylation sites and recognition motifs. J Biol Chem. 2007;282:12368–12376. doi: 10.1074/jbc.M611896200. [DOI] [PubMed] [Google Scholar]
- 22.Porras-Yakushi TR, Whitelegge JP, Miranda TB, Clarke S. A novel SET domain methyltransferase modifies ribosomal protein Rpl23ab in yeast. J Biol Chem. 2005;280:34590–34598. doi: 10.1074/jbc.M507672200. [DOI] [PubMed] [Google Scholar]
- 23.Webb KJ, Laganowsky A, Whitelegge JP, Clarke SG. Identification of two SET domain proteins required for methylation of lysine residues in yeast ribosomal protein Rpl42ab. J Biol Chem. 2008;283:35561–35568. doi: 10.1074/jbc.M806006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esteve PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, et al. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol. 2011;18:42–48. doi: 10.1038/nsmb.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pradhan S, Chin HG, Esteve PO, Jacobsen SE. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics. 2009;4 doi: 10.4161/epi.4.6.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nature Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 28.Masatsugu T, Yamamoto K. Multiple lysine methylation of PCAF by Set9 methyltransferase. Biochem Biophys Res Commun. 2009;381:22–26. doi: 10.1016/j.bbrc.2009.01.185. [DOI] [PubMed] [Google Scholar]
- 29.Carr SM, Munro S, Kessler B, Oppermann U, La Thangue NB. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. EMBO J. 2011;30:317–327. doi: 10.1038/emboj.2010.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 31.Saddic LA, West LE, Aslanian A, Yates JR, Rubin SM, Gozani O, et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010 doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunizaki M, Hamamoto R, Silva FP, Yamaguchi K, Nagayasu T, Shibuya M, et al. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 2007;67:10759–10765. doi: 10.1158/0008-5472.CAN-07-1132. [DOI] [PubMed] [Google Scholar]
- 33.Ea CK, Baltimore D. Regulation of NFkappaB activity through lysine monomethylation of p65. Proc Natl Acad Sci USA. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, et al. Regulation of NFkappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF. Negative regulation of NFkappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28:1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, et al. Lysine methylation of the NFkappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NFkappaB signaling. Nat Immunol. 2011;12:29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, et al. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaughan L, Stockley J, Wang N, McCracken SR, Treumann A, Armstrong K, et al. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ko S, Ahn J, Song CS, Kim S, Knapczyk-Stwora K, Chatterjee B. Lysine methylation and functional modulation of androgen receptor by set9 methyltransferase. Mol Endocrinol. 2011;25:433–444. doi: 10.1210/me.2010-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kontaki H, Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol Cell. 2010;39:152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 43.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 45.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 47.Dou Y, Hess JL. Mechanisms of transcriptional regulation by MLL and its disruption in acute leukemia. Int J Hematol. 2008;87:10–18. doi: 10.1007/s12185-007-0009-8. [DOI] [PubMed] [Google Scholar]
- 48.Liedtke M, Cleary ML. Therapeutic targeting of MLL. Blood. 2009;113:6061–6068. doi: 10.1182/blood-2008-12-197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell. 2009;33:181–191. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cosgrove MS, Patel A. Mixed lineage leukemia: a structure-function perspective of the MLL1 protein. Febs J. 2010;277:1832–1842. doi: 10.1111/j.1742-4658.2010.07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the Mechanism of Multiple Lysine Methylation by the Human Mixed Lineage Leukemia Protein-1 (MLL1) Core Complex. J Biol Chem. 2009;284:24242–24256. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J Biol Chem. 2008;283:32162–32175. doi: 10.1074/jbc.M806317200. [DOI] [PubMed] [Google Scholar]
- 54.Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem. 2008;283:35258–35264. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 56.Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J Biol Chem. 2008;283:32158–32161. doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi YH, Lee JS, Swanson SK, Saraf A, Florens L, Washburn MP, et al. Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol Cell Biol. 2009;29:3478–3486. doi: 10.1128/MCB.00013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, et al. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 59.Cao F, Chen Y, Cierpicki T, Liu Y, Basrur V, Lei M, et al. An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. Plos One. 2010;5:14102. doi: 10.1371/journal.pone.0014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A novel non-SET domain multi-subunit methyltransferase required for sequential nucleosomal histone H3 methylation by the mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2011;286:3359–3369. doi: 10.1074/jbc.M110.174524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 63.Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, et al. Structural basis for the product specificity of histone lysine methyltransferases. Molecular Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munro S, Khaire N, Inche A, Carr S, La Thangue NB. Lysine methylation regulates the pRb tumour suppressor protein. Oncogene. 2010;29:2357–2367. doi: 10.1038/onc.2009.511. [DOI] [PubMed] [Google Scholar]
- 66.Ivanov GS, Ivanova T, Kurash J, Ivanov A, Chuikov S, Gizatullin F, et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol. 2007;27:6756–6769. doi: 10.1128/MCB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, et al. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 68.Yang XD, Tajkhorshid E, Chen LF. Functional interplay between acetylation and methylation of the RelA subunit of NFkappaB. Mol Cell Biol. 2010;30:2170–2180. doi: 10.1128/MCB.01343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Couture JF, Collazo E, Hauk G, Trievel RC. Structural basis for the methylation site specificity of SET7/9. Nat Struct Mol Biol. 2006;13:140–146. doi: 10.1038/nsmb1045. [DOI] [PubMed] [Google Scholar]
- 70.Dhayalan A, Kudithipudi S, Rathert P, Jeltsch A. Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem Biol. 2011;18:111–120. doi: 10.1016/j.chembiol.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 71.Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, Blackburn GM, et al. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- 72.Demirci H, Gregory ST, Dahlberg AE, Jogl G. Multiple-site trimethylation of ribosomal protein L11 by the PrmA methyltransferase. Structure. 2008;16:1059–1066. doi: 10.1016/j.str.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 74.Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Avdic V, Zhang P, Lanouette S, Groulx A, Tremblay V, Brunzelle J, et al. Structural and biochemical insights into MLL1 core complex assembly. Structure. 2011;19:101–108. doi: 10.1016/j.str.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 76.Odho Z, Southall SM, Wilson JR. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J Biol Chem. 2010;285:32967–32976. doi: 10.1074/jbc.M110.159921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Y, Wan B, Wang KC, Cao F, Yang Y, Protacio A, et al. Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding. EMBO Rep. 2011 doi: 10.1038/embor.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sarvan S, Avdic V, Tremblay V, Chaturvedi CP, Zhang P, Lanouette S, et al. Crystal structure of the trithorax group protein ASH2L reveals a forkhead-like DNA binding domain. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, et al. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]