Abstract
Genetic loci displaying environmentally responsive epigenetic marks, termed metastable epialleles, offer a solution to the paradox presented by genetically identical yet phenotypically distinct individuals. The murine viable yellow agouti (Avy) metastable epiallele exhibits stochastic DNA methylation and histone modifications associated with coat color variation in isogenic individuals. The distribution of Avy variable expressivity shifts following maternal nutritional and environmental exposures. To characterize additional murine metastable epialleles, we utilized genome-wide expression arrays (N = 10 male individuals, 3 tissues per individual) and identified candidates displaying large variability in gene expression among individuals (Vi = inter-individual variance), concomitant with a low variability in gene expression across tissues from the three germ layers (Vt = inter-tissue variance), two features characteristic of the Avy metastable epiallele. The CpG island in the promoter of Dnajb1 and two contraoriented ERV class II repeats in Glcci1 were validated to display underlying stochasticity in methylation patterns common to metastable epialleles. Furthermore, liver DNA methylation in mice exposed in utero to 50 mg bisphenol A (BPA)/kg diet (N = 91) or a control diet (N = 79) confirmed environmental lability at validated candidate genes. Significant effects of exposure on mean CpG methylation were observed at the Glcci1 Repeat 1 locus (p < 0.0001). Significant effects of BPA also were observed at the first and fifth CpG sites studied in Glcci1 Repeat 2 (p < 0.0001 and p = 0.004, respectively). BPA did not affect methylation in the promoter of Dnajb1 (p = 0.59). The characterization of metastable epialleles in humans is crucial for the development of novel screening and therapeutic targets for human disease prevention.
Key words: epigenetics, metastable epiallele, viable yellow agouti, environmental epigenomics
Introduction
Within human, mouse and other animal species, individuals considered genetically identical, such as monozygotic twins and inbred strains, often display phenotypic discordance in various traits and disease susceptibility, even after controlling for the environment. Epigenetic programming has been proposed to play an important role in the diverse phenotypes of genetically identical individuals. A handful of murine metastable epialleles have been identified (Avy, AxinFu, CabpIAP) in which the activity of a contraoriented intracisternal A particle (IAP) retrotransposon controls expression of an adjacent gene.1–5 Variable expressivity results from stochastic DNA methylation of the 5′ long terminal repeat (LTR) of the IAP, producing genetically identical individuals with varying phenotypes. The distribution of variable expressivity has been shifted at these metastable epialleles following maternal exposure to nutritional and environmental factors.6–10
The Avy metastable epiallele displays stochastic methylation profiles7,11 and histone modification patterns12 that are associated with individual variation in coat color and adult-onset obesity. Interestingly, Avy methylation profiles are similar within tissues derived from the three germ layers (brain representing ectodermal; kidney representing mesodermal; and liver representing endodermal origin); thus, metastable epiallele epigenetic profiles are likely set prior to germ layer differentiation.7–9 To aid in the genome-wide characterization of developmentally-labile metastable loci, we previously introduced the “Agouti Expression Fingerprint” concept, defined by large variability in gene expression among individuals (Vi = inter-individual variation), concomitant with a low variability in gene expression among tissues from the three germ layers (Vt = inter-tissue variation).13 Genes expressing this fingerprint, therefore, will be identified by a large Vi:Vt ratio or R value. Consequently, genome-wide expression arrays should be useful in detecting metastable epialleles in both inbred mice and monozygotic twins where genetic variation is presumably eliminated.
Herein, we report the development of an expression microarray approach to predict candidate metastable epialleles in the mouse genome, identify and validate candidate metastable loci, and discuss the application of this approach to the identification of metastable loci in the human genome. This methodological approach preferentially identifies genes whose expression patterns are epigenetically established prior to embryonic stem cell differentiation, and does not inform the identification of genes that undergo epigenetic drift with age.14 The epigenome is particularly vulnerable to environmental perturbations during embryogenesis because the elaborate DNA methylation patterning and chromatin structure required for normal growth is established early in development.15,16 Thus, the identification of epigenomic targets such as metastable epialleles that are particularly vulnerable to dysregulation during early development will facilitate the understanding of the developmental origins of health and disease (DOHaD).17
Results
Agouti, housekeeping and tissue-specific gene expression.
Expression data from tissues of the three germ layers (liver, kidney, brain) demonstrated high variance in agouti RNA levels among isogenic animals (Table 1, rows) coupled with low variance among tissue types in individual animals (Table 1, columns). Agouti expression levels in liver, kidney and brain were approximately 3- to 4-fold higher in yellow, slightly mottled and mottled animals compared to pseudoagouti animals. Agouti expression levels in the liver, kidney and brain were about 1.5- to 2-fold higher in heavily mottled animals compared to pseudoagouti animals. The inter-tissue expression levels exhibited low variability, with the exception of yellow and slightly mottled animals, which exhibited higher expression levels in kidney, compared to liver and brain.
Table 1.
Mean agouti (A) expression levels across individuals and tissue type (N = 2 animals per cell)
| Yellow | Slightly mottled | Mottled | Heavily mottled | Pseudoagouti | |
| Liver | 294 | 280 | 307 | 179 | 101 |
| Kidney | 688 | 402 | 349 | 199 | 91 |
| Brain | 339 | 319 | 244 | 152 | 97 |
The R value, defined as the ratio of inter-individual (Vi) to inter-tissue (Vt) variance in gene expression, for the Agouti (A) gene was 1.97, indicating that average expression variance between isogenic animals is ∼2-fold higher than average expression variance among the three germ layers. Overall, these data serve as proof of concept for the Agouti Fingerprint, defined as high inter-individual variation (Vi) in Agouti expression coupled with low inter-tissue expression variation (Vt). Accordingly, expression levels for housekeeping genes such as beta actin (Actb) should be similar across individuals and tissue type. The R value for Actb was 1.03, indicating similar inter-individual and inter-tissue variance, as expected. The R value for albumin (Alb1), which normally exhibits post-natal liver-specific expression,18 was 0.05, indicating low variance across individuals concomitant with high variance across tissues. Indeed, Alb1 expression signals in the liver were over 25,000 units compared to background level in brain and kidney.
Candidate Agouti Fingerprint genes.
Using Affymetrix expression array data, we queried the entire mouse genome for candidate metastable epialleles that display the Agouti Fingerprint. Approximately 100 of the greater than 40,000 transcripts (0.25%) on the mouse array displayed an expression pattern characterized as high inter-individual variation coupled with low inter-tissue variation (R value >1.5). This set of transcripts represents our candidate list of genes potentially modifiable by early environmental exposures via epigenetic mechanisms.
The eight annotated genes with the highest R values (Table 2) included D site albumin promoter binding protein (Dbp), agouti (A, proof of principle control), Dnaj homolog subfamily B member 1 (Dnajb1), translation initiation factor epsilon subunit (Eif2b5), T-complex protein 1 subunit eta (Cct7), glucocorticoid induced transcript 1 (Glcci1), ribosome biogenesis regulatory protein (Rrs1) and nuclear distribution protein C (Nudc). Table 2 lists the chromosomal location for these candidate metastable epialleles, and indicates the presence of a CpG island within 1kb of the putative promoter or the existence of LTR and non-LTR retrotransposon repeat elements (LINEs, long interspersed nuclear elements and SINEs, short interspersed nuclear elements). The top R value gene, Dbp, was not included for further analysis because of its confounding circadian expression pattern.19,20 Although tissues used in this study were collected during daylight hours, collection time was not standardized. Two of the remaining top candidates, Dnajb1 and Glcci1, were selected for further analysis based on genetic characteristics thought to be predictive of DNA stochasticity: contraoriented ERV class II repeats in the latter and a promoter CpG island devoid of a retroelement in the former. As the Dnajb1 candidate metastable epiallele lacks repetitive elements often associated with known metastable epialleles, we focused on the promoter as a source of stochastic methylation.
Table 2.
High R-value gene names and characteristics
| Gene and gene symbol | Chrom.location | Variation ratio (R-value) | CpG Island at putative promoter | Repeat elements |
| D site albumin promoter (Dbp)* | 7B4 | 2.04 | Yes | None |
| Agouti (A)** | 2H2 | 1.97 | No | SINEs LINEs LTRs^^ |
| Dnaj homolog subfamily B member 1 (Dnajb1) | 8C3 | 1.67 | Yes | None |
| Translation initiation factor eIF-2B epsilon subunit (Eif2b5) | 16B1 | 1.65 | Yes | SINEs LINEs LTRs |
| T-complex protein subunit eta (Cct7) | 6D1 | 1.62 | Yes | SINEs LTRs |
| Glucocorticoid induced transcript 1 (Glcci1) | 6A1 | 1.61 | Yes | SINEs LINEs LTRs^^ |
| Ribosome biogenesis regulatory protein (Rrs1) | 1A2 | 1.58 | Yes | SINEs |
| Nuclear distribution protein C (Nudc) | 4D3 | 1.56 | Yes | SINEs LTRs |
Expression controlled by circadian-controlled histone modifications.
Gene responsible for coat color variation in Avy mice (Agouti Fingerprint gene).
LTR ERV class II (IAP-like repeats) predicted.
We performed unsupervised hierarchical clustering analysis to identify gene clusters associated with sample characteristics, including animal coat color and sample tissue type. Clustering was not observed based on coat color phenotype. Clustering, however, was observed based on sample tissue of origin; 6,625 out of 45,101 probes segregated the samples into three major branches depicting brain, liver and kidney. As expected, 7 out of 8 genes with high R values (Table 2) did not display transcripts within this tight tissue-specific clustering tree. Of the 10 probes for Glcci1 on the array, 2 were associated with tissue cluster and 8 were not.
Methylation analysis of candidate loci: methylation patterns characteristic of the Agouti Fingerprint.
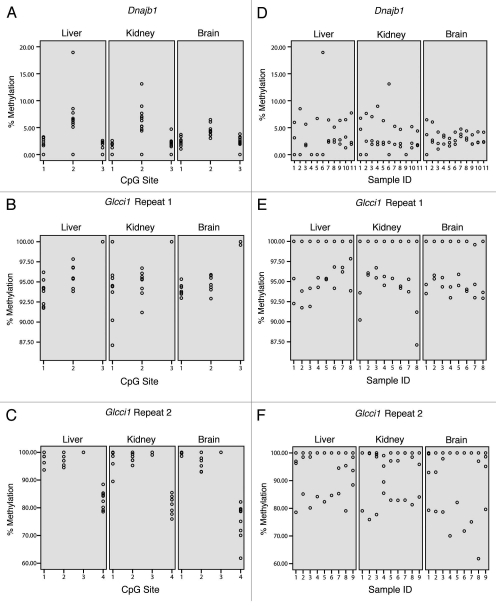
The candidate metastable epiallele, Dnajb1, is the only top ranked candidate gene that contains a CpG promoter island (972 bp, 67% GC content) in the absence of a retrotransposon repeat element (Table 2). Bisulfite sequencing of the promoter of Dnajb1 revealed low to moderate levels of methylation at three sites within this CpG island, analyzed in liver, kidney and brain. The Dnajb1 candidate metastable locus exhibited a wide range of methylation values across individuals (Fig. 1A). The inter-individual range of percent methylation was 0 to 18.95 in liver, 0 to 13.11 in kidney and 0 to 6.46 in brain (Table 3). Mean inter-individual variance in percent methylation was 8.76 in liver, 3.73 in kidney and 0.85 in brain (Table 3). Quantitative PCR analysis of Dnajb1 showed patterns in expression variance that mirror those seen in methylation variance: large inter-individual fold-change variance in liver RNA levels (variance = 57.7), with smaller expression variance in kidney (variance = 0.12) and brain (variance = 0.86) RNA levels. Site-specific inter-individual methylation ranges and variances are reported in Supplemental Table 1. Conversely, Dnajb1 displayed a narrower range of methylation within tissues of individuals (Fig. 1D). The inter-tissue range across individuals was 0 to 3.7 at site 1 and 0 to 4.71 at site 3. A wider-range of methylation was observed, however, at site 2 (0 to 18.95). Mean inter-tissue variance in percent methylation was 0.88 at site 1, 9.48 at site 2 and 0.92 at site 3.
Figure 1.
Candidate metastable epialleles, Dnajb1 and Glcci1 exhibit high inter-individual variation and low inter-tissue variation. Candidate loci exhibit a wide range of methylation values across individuals (left-hand column). A narrower range within individuals (right-hand column) can be observed by comparing values for each individual in each tissue type.
Table 3.
Validation of candidate metastable epialleles
| Locus | Tissue | Minimum methylation | Maximum methylation | Mean methylation ± SEM | Range | Average variance |
| Liver | 0 | 18.95 | 3.27 ± 0.67 | 18.95 | 8.76 | |
| Dnajb1 (N = 11) | Kidney | 0 | 13.11 | 3.21 ± 0.52 | 13.11 | 3.73 |
| Brain | 0 | 6.46 | 3.04 ± 0.23 | 6.46 | 0.85 | |
| Glcci1 Repeat 1 (N = 10) | Liver | 91.74 | 100 | 96.27 ± 0.54 | 8.26 | 1.44 |
| Kidney | 87.11 | 100 | 96.33 ± 0.63 | 12.89 | 4.87 | |
| Brain | 92.92 | 100 | 95.96 ± 0.58 | 7.08 | 1.49 | |
| Glcci1 Repeat 2 (N = 9) | Liver | 78.56 | 100 | 95.00 ± 1.22 | 21.44 | 5.19 |
| Kidney | 75.94 | 100 | 94.52 ± 1.35 | 24.06 | 6.37 | |
| Brain | 61.80 | 100 | 93.10 ± 1.84 | 38.20 | 6.70 |
Inter-individual ranges and variances of percent methylation at three candidate loci, Dnajb1, Glcci1 Repeat 1, and Glcci1 Repeat 2 are characteristic of the Agouti Expression Fingerprint.
The Glcci1 candidate metastable epiallele was the only gene to exhibit endogenous retrovirus (ERV) class II LTR repeats analogous to those observed in the Avy metastable epiallele (Table 2). Nine ERV class II LTR repeats are found within the Glcci1 gene, four of which are contraoriented with respect to Glcci1 transcription. Since three of the identified murine metastable epialleles (Avy, AxinFu, CabpIAP) are associated with contraoriented IAP insertions,1,2,5 we chose to focus methylation analysis on the two contraoriented ERV class II repeats closest to the Glcci1 promoter. Both repeats exhibited high levels of methylation at all sites studied (3 sites at Glcci1 Repeat 1, 6 sites at Glcci1 Repeat 2) in liver, kidney and brain. The repeat loci in Glcci1 displayed a wide range of methylation across individuals (Fig. 1B and C). The range of percent methylation at Repeat 1 was 91.74 to 100 in liver, 87.11 to 100 in kidney and 92.92 to 100 in brain; at Repeat 2, ranges were 78.56 to 100 in liver, 75.94 to 100 in kidney and 61.80 to 100 in brain (Table 3). Mean variance in percent methylation at Repeat 1 was 1.44 in liver, 4.87 in kidney and 1.49 in brain; at Repeat 2, mean variance was 5.19 in liver, 6.37 in kidney and 6.70 in brain (Table 3). Quantitative PCR analysis of Glcci1 expression revealed large inter-individual fold-change variance in liver RNA levels (variance = 147.6), with smaller expression variance in kidney (variance = 0.24) and brain (variance = 4.4), reflective of variance in methylation in these tissues. Site-specific methylation ranges and variances are reported in Supplemental Table 1. On the other hand, the repeat loci in Glcci1 exhibit a narrower range within tissues of individuals (Fig. 1E and F). The inter-tissue range across individuals at Repeat 1 was 87.11 to 100 at site 1, 91.19 to 97.84 at site 2 and 99.29 to 100 at site 3; at Repeat 2, values were 89.51 to 100 at site 1, 92.93 to 100 at site 2, 99.04 to 100 at site 3 and 70.04 to 88.44 at site 4. Mean inter-tissue variance in percent methylation at Repeat 1 was 3.33 at site 1, 2.62 at site 2 and 0.02 at site 3; at Repeat 2, values were 5.39 at site 1, 6.82 at site 2, 0.03 at site 3 and 21.84 at site 4.
Methylation analysis of candidate loci: environmentally labile methylation.
In order to assess the environmental lability of our candidate metastable epialleles, we assessed DNA methylation changes in offspring exposed in utero to bisphenol-A (BPA), a representative endocrine active compound, as described previously in reference 9. We evaluated liver tissue methylation levels in 79 control offspring (32 a/a and 47 Avy/a) and 91 BPA-exposed offspring (19 a/a and 72 Avy/a; Table 4). Methylation was not associated with genotype, coat color or sex in any of the analyses. Significant effects of BPA exposure on mean CpG methylation at the Dnajb1 locus were not observed (p = 0.59). No significant effects of BPA exposure were seen at any of the three CpG sites assayed (p = 0.25, p = 0.67 and p = 0.76), respectively. Quantitative PCR gene expression analysis on a subset of offspring available for liver tissue RNA isolation (N = 17 control and N = 18 BPA exposed offspring) revealed marginally significant differences in relative expression of Dnajb1 (Control fold change = 0.58 and BPA fold change = 0.50; p = 0.07).
Table 4.
Metastable epialleles are environmentally labile in response to maternal BPA exposure
| Locus | Test of fixed effects (p-value) | Least squares means (Control ± SEM, BPA-exposed ± SEM) | Test of effect slices (p-value) | |
| Dnajb1 | 10.59 | |||
| Site 1 | 0.88 ± 0.10, 1.03 ± 0.09 | 0.25 | ||
| Site 2 | 2.80 ± 0.14, 2.88 ± 0.12 | 0.67 | ||
| Site 3 | 1.31 ± 0.14, 1.25 ± 0.13 | 0.76 | ||
| Glcci1 Repeat 1 | <0.0001 | |||
| Site 1 | 91.68 ± 0.30, 92.47 ± 0.28 | 0.05 | ||
| Site 2 | 92.42 ± 0.28, 94.25 ± 0.25 | <0.0001 | ||
| Site 3 | 99.70 ± 0.26, 98.94 ± 0.23 | 0.49 | ||
| Glcci1 Repeat 2 | 0.45 | |||
| Site 1 | 94.39 ± 0.54, 97.84 ± 0.49 | <0.0001 | ||
| Site 2 | 95.54 ± 0.34, 95.50 ± 0.31 | 0.92 | ||
| Site 3 | 99.60 ± 0.17, 99.68 ± 0.15 | 0.73 | ||
| Site 4 | 83.84 ± 0.43, 83.52 ± 0.39 | 0.59 | ||
| Site 5 | 86.75 ± 0.37, 85.28 ± 0.34 | 0.004 | ||
| Site 6 | 79.04 ± 0.41, 78.53 ± 0.37 | 0.36 |
Repeated measures ANOVA with individual CpG sites defined as repeated measures. The effect of BPA exposure (N = 91 exposed and N = 79 control offspring) on mean methylation was assessed via tests of fixed effects and site-specific toxicant effects via tests of effect slices for each CpG site.
On the other hand, significant effects of BPA exposure on mean CpG methylation were observed at the Glcci1 Repeat 1 locus (p < 0.0001; Table 4). Significant effects of BPA exposure were seen at the first two CpG sites assayed (p = 0.05 and p < 0.0001, respectively); no significant effect was seen at the third CpG site (p = 0.49). Although significant effects of BPA exposure on mean CpG methylation at the Glcci1 Repeat 2 locus were not observed (p = 0.45), significant effects of BPA exposure were seen at the first and fifth CpG sites studied (p < 0.0001 and p = 0.004, respectively). The remaining four CpG sites studied exhibited no significant effects. Site-specific effect sizes due to exposure at all three loci are shown in Table 4. Quantitative PCR analysis in the subset of available tissues for RNA analysis failed to reveal differences (p = 0.80) in Glcci1 gene expression levels in BPA exposed offspring (N = 18; fold change = 0.73) compared to controls (N = 16; fold change = 0.85).
Discussion
To date, the discovery of metastable epialleles in the mouse, such as Avy and AxinFu, was solely dependent on directly observed phenotypes (coat color and tail-kink, respectively); whereas, CabpIAP was identified in the mouse with the use of a bioinformatics approach.5 Furthermore, currently available epigenome-wide DNA assay technologies may fail to identify metastable epialleles due to the highly repetitive content of their regulatory and coding regions.21,22 For example, a recent analysis characterizing the transcriptome of mouse embryonic stem cells aligned 74.5% of Illumina deep-sequencing reads to the mouse genome,23 suggesting that some proportion of the 25.5% of unaligned sequence reads were located in highly repetitive sequence regions.
We utilized genome-wide murine expression arrays to develop methodologies for identifying candidate metastable epialleles in the mouse genome. Utilizing this approach we identified a relatively modest number of candidate loci (∼100 transcripts with an R value >1.5), suggesting that the vast majority of the genome is insensitive to epigenetic stochasticity reflective of metastable epialleles. Initiating this analysis in isogenic mice allowed us to control for genetic and environmental heterogeneity, a luxury not readily available within human analyses. Using the microarray approach, we were able to confirm previous studies by Kaput et al.24 and Wolff et al.25 that the inter-individual variation in Agouti RNA levels corresponds to coat color, with yellow animals displaying 3- to 6-fold enrichment of agouti compared to pseudoagouti animals, as measured by Affymetrix arrays. This is an important internal control or “proof of concept” to confirm that expression arrays can detect the Agouti Fingerprint, characterized as high ratio of inter-individual (Vi) to inter-tissue (Vt) variance (R value) in gene expression.
We report on the identification and characteristics of 6 novel candidate metastable epialleles (Table 2). Using unsupervised hierarchical clustering, our top candidate metastable epialleles did not cluster by coat color, suggesting that individual metastable loci have unique and independent underlying stochasticity. Furthermore, our candidate metastable epialleles were not among those probes that clustered by tissue type, confirming low expression variability across tissues from the three germ layers. Two of the candidate metastable epialleles were further analyzed for variable methylation in either the promoter region CpG island (Dnajb1) or within the 5′ end of two contraoriented LTR ERV class II repeats (Glcci1). The candidate metastable epiallele Dnajb1 is a heat shock protein whose increased expression has been associated with bipolar disorder and schizophrenia in humans.26,27 The Glcci1 candidate metastable epiallele is a protein-coding gene expressed in multiple murine tissues. Its function is relatively unknown; however, it has been shown to be an early marker of apoptosis and can be induced by steroid hormones.28
Since metastable epialleles are prone to a high degree of stochastic change, they are characterized by inter-individual variability in DNA methylation, even in the absence of environmental exposure. Our analysis specifically filtered for genes with low inter-tissue expression variance and high inter-individual expression variance, with the goal of identifying loci subject to epigenetic stochasticity early in development. The three loci investigated all exhibited greater inter-individual range and variance than inter-tissue range and variance in methylation profiles. Quantitative gene expression analysis at the two candidate loci also reflected large inter-individual range and variance in liver RNA levels. It is interesting to note that stochasticity appeared to be both site- and tissue-specific. For example, although methylation of Glcci1 Repeat 1 CpG site 1 appeared variable in all three tissue types, variance was higher in kidney (11.67) than in liver or brain (2.15 and 3.15, respectively). A similar pattern in variances was observed at Glcci1 Repeat 2 CpG site 1 (12.47 in kidney, 4.97 in liver and 0.21 in brain) and at Glcci1 Repeat 2 CpG site 2 (8.94 in brain, 4.87 in liver and 2.99 in kidney). Site specificity in CpG methylation at metastable epialleles is not unprecedented, as investigations of the Avy metastable epiallele have shown individual sites to be more informative than others.8 Thus, the use of technologies, such as pyrosequencing that can resolve individual CpG sites is particularly important in investigating metastable epialleles.29,30 Analogous variation in tissue and individual methylation patterns was recently observed at putative human candidate metastable epialleles by Waterland et al.31
As the distribution of methylation has been shifted at known murine metastable epialleles following maternal exposure to nutritional and environmental factors,6–10 we investigated whether our candidate genes display environmental lability in response to in utero BPA exposure. Site-specific differences in BPA exposed vs. control groups were seen at two of three CpG sites analyzed in Glcci1 Repeat 1 and at two of six CpG sites analyzed in Glcci1 Repeat 2. Moreover, an overall increase in mean methylation comparing exposed to controls was observed in Glcci1 Repeat 1. Significant effect sizes, however, were subtle; environmental exposure was associated with only 1–3% methylation change at CpG sites with statistical significance. Further, these subtle differences in methylation were not confirmed by altered expression levels of Glcci1 in a small subset of the samples used for methylation analysis, although this lack of confirmation may be due to small sample size. In contrast, the Avy and CabpIAP loci have been reported to exhibit approximately 12% and 13% change in mean tail tissue methylation, respectively, on exposure to BPA.9 These data suggest that metastable epialleles may fall on a continuum of environmental responsiveness. Further analysis with expanded sample sizes will be needed to assess whether BPA-induced changes in DNA methylation correlate to altered expression profiles at the candidate metastable epialleles, as well as whether these molecular changes are associated with phenotype.
The data presented here represent validation of a novel method for identification of metastable epialleles in mice and confirmation of both the inter-tissue and inter-individual variance in methylation as well as the environmental responsiveness of such loci. The further characterization of epigenetically labile metastable epialleles in humans also will be crucial. Although humans do not have a metastable Agouti homologue, the Agouti Fingerprint pattern may still be extremely useful in identifying developmentally labile genes subject to stochastic DNA methylation profiles established prior to stem cell differentiation. The detection of a genome-wide set of environmentally-labile metastable epialleles in humans will allow us to establish their role in human health, and to also better determine the value of the mouse as a toxicological model for assessing human risk from agents that elicit their biological effect primarily by altering the epigenome.
Methods
Animals and tissues.
For expression microarray studies, liver, kidney and brain tissue from 10 male Avy/a mice (2 per each of the 5 coat color classes) were collected at time of weaning and coat color determination (d22) and flash frozen in liquid nitrogen prior to storage at −80°C. Animals were housed in polycarbonate-free cages and provided ad libitum access to their respective diets and water. Mice were obtained from a colony that has been maintained with sibling mating and forced heterozygosity for the Avy allele for over 220 generations, resulting in a genetically invariant background. These mice were originally obtained from a colony at the Oak Ridge National Laboratory, in which the Avy mutation arose spontaneously in the C3H/HeJ strain, as previously described in reference 7. Animals used in this study were maintained in accordance with the Guidelines for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, 1996) and were treated humanely and with regard for alleviation of suffering. The study protocol was approved by the University of Michigan Committee on Use and Care of Animals and the Duke University Institutional Animal Care and Use Committee.
RNA isolation for microarray.
Total RNA was isolated from frozen brain, kidney and liver tissues by homogenization in RNA-Stat 60 (Tel-Test, Friendswood, TX), and subsequent processing was performed as recommended by the manufacturer. RNA quality and yield was checked on an agarose gel and with an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA) before submission to Expression Analysis (Durham, NC).
Microarray hybridization.
Affymetrix GeneChip Mouse Genome 2.0 arrays (Santa Clara, CA) were processed by Expression Analysis. Briefly, double stranded cDNA was generated from 10 µg of the RNA template. Biotin-labeled cDNA transcripts were synthesized in vitro, fragmented and hybridized to the array chip followed by washing and scanning for bound fluorescent tags. Raw and MAS5 normalized hybridization signal data were returned to the investigators for further analysis.
Expression data analysis.
MAS5 hybridization signal data were analyzed using Microsoft Excel (Redmond, WA). Genes with average expression signals less than 30 units (background) were excluded from the analysis. For each gene on the array, a tissue specific inter-individual expression level variance was calculated. The three tissue specific inter-individual variances were averaged to obtain a mean inter-individual variance (Vi). For each individual, an inter-tissue expression level variance was also calculated. The inter-tissue variance was averaged across individuals to obtain an average inter-tissue variance (Vt). For each gene, a ratio (R value) or Vi to Vt was determined, and genes with high R values (>1.5) were classified as potential Agouti Fingerprint gene candidates. Individual CEL files and a combined Excel file have been uploaded to GEO (Accession Number: GSE28559). Unsupervised hierarchical clustering analysis was performed using the centroid linkage algorithm to identify gene clusters associated with sample characteristics. Probes were filtered out if standard deviation value was less than 1.
CpG island and repeat identification.
CpG islands of candidate genes were identified using GrailEXP (http://compbio.ornl.gov/Grail-1.3) under default parameters. RepeatMasker (http://repeatmasker.org/cgi-bin/WEBRepeatMasker) was used to identify simple and complex repeats, including the LTR endogenous retrovirus-like (ERV) class II repeats (the repeat classification of murine IAP elements).
Candidate gene validation and assessment of BPA-induced changes in methylation.
To validate candidate metastable epialleles for variable methylation patterns reflective of the Agouti Fingerprint, banked liver, kidney and brain tissue from d22 Avy/a and a/a (N = 10 for Glcci1 Repeat 1; N = 9 for Glcci1 Repeat 2; N = 11 for Dnajb1) were utilized. To assess whether BPA, a representative environmental chemical, induces methylation changes in candidate metastable loci, banked liver tissue of d22 mice exposed in utero and during lactation to either a control AIN-93G diet (diet 95092 with 7% corn oil substituted for 7% soybean oil; Harlan Teklad, Madison, WI) (N = 79) or the control diet supplemented with 50 mg BPA/kg diet (N = 91), as described previously in reference 9. All diet ingredients were supplied by Harlan Teklad except BPA, which was supplied by Sigma. For these efforts, DNA isolation, bisulfite conversion and pyrosequencing were performed, as detailed below.
DNA isolation and methylation analysis.
Total DNA was isolated from the mouse tissues using buffer ATL, proteinase K and RNase A (Qiagen Inc., Valencia, CA), followed by phenol-chloroform extraction and ethanol precipitation. DNA quality and concentration was assessed using a ND1000 spectrophotometer (NanoDrop Technology, Wilmington, DL).
Quantitative DNA methylation patterns at candidate loci were analyzed via sodium bisulfite modification followed by pyrosequencing.30 Bisulfite treatment of genomic DNA converts unmethylated cytosine to uracil, which is read as a thymidine during polymerase chain reaction (PCR). Methylated cytosines are protected from bisulfite conversion and thus remain unchanged, resulting in genome-wide methylation-dependent differences in DNA sequence that can be interrogated by PCR and quantified via sequencing. Approximately 1 ug of DNA was bisulfite converted using the EpiTect Bisulfite Kit (Qiagen Inc., Valencia, CA) with clean-up automation on the QIAcube® purification system.
Following conversion, a 25 µL PCR was carried out using approximately 50 ng of bisulfite-converted DNA, HotStarTaq master mix (Qiagen) and forward (1 pmol) and reverse (0.5 pmol) primers. PCR and sequencing primers for candidate loci are available in Supplemental Table 2. Reverse primers were biotin-labeled in order to purify the final PCR product by Sepharose beads. The PCR product was checked for amplicon size by gel electrophoresis and pyrosequenced on a PyroMark MD (Qiagen), using 0.4 µM sequencing primer. Sequences to analyze are also provided in Supplemental Table 2. The percentage of cells methylated was quantified using PyroMark software, which computes the degree of methylation as %5-methylated cytosines (%5mC) over the sum of methylated and unmethylated cytosines. To test for complete conversion following bisulfite treatment, the Pyro Q-CpG Software was set to incorporate internal quality controls of C nucleotides into the dispensation order for non-CpG cytosines in the original sequence that should have been fully converted to Ts. Samples for candidate gene validation were sequenced once; samples for environmental lability testing were run in duplicate and the average of the two replicates was used in the statistical analysis. The three CpG sites studied in the promoter of the Dnajb1 locus are located in region 86131987–86132024 of GenBank accession number NC_000074. The three CpG sites studied in the first contraoriented ERV class II repeat analyzed at the Glcci1 locus are located in region 8536167–8536545 and the six CpG sites studied in the second contraoriented ERV class II repeat are located in region 8566843–8567147 of GenBank accession number NC_000072.
RNA isolation and qPCR for expression validation.
Total RNA was purified from mice used for methylation analyses for both validation (N = 12 each; liver, kidney, brain) and environmental lability (N = 18 BPA exposed, N = 17 control; liver tissue only) via an adapted protocol for the RNeasy® Mini Kit (Qiagen, Valencia, CA), in conjunction with the QIAcube® automated nucleic acid purification system (Qiagen, Valencia, CA). Briefly, approximately 10–15 mg of tissue from liver, kidney or brain was homogenized using three 30 second bursts at 30 Hz in a TissueLyser® II (Qiagen, Valencia, CA). Crude lysates were processed on a QIAcube® automated nucleic acid purification system. Total RNA quality and concentration were assessed using a ND1000 spectrophotometer (NanoDrop Technology, Wilmington, DL). For each sample, a volume equivalent to 1 µg of total RNA was used with the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) to generate DNA complementary to the messenger RNA.
Primers for real-time PCR of metastable epiallele candidates Glcci1 and Dnjab1 and of housekeeping gene beta actin were designed using NCBI Primer BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome). Both primer pairs were designed to have a Tm = 59°C ± 1°C, contain ≤3 Gs or Cs within 5 bases of the 3′ end, and produce a product between 50–150 bp in length. Primers for Glcci1 (expected product 108 bp) were designed to cross an exon-exon junction; as all samples were assayed for both Dnajb1 (expected product 55 bp) and Glcci1, any sample DNA contamination would have been apparent. Each real-time PCR amplification was performed as a 25 µL reaction using iQ™ SYBR® Green Supermix (Bio-Rad, Hercules, CA), 2 µL of undiluted cDNA and a 100 nM final concentration of each primer. Reactions were carried out in triplicate on a CFX96™ Real-Time System (Bio-Rad, Hercules, CA) and included no template control reactions for each primer set. Calibrator samples were run in duplicate for beta actin, Glcci1 and Dnajb1. PCR conditions were 95°C for 3 minutes, 40 cycles of 95°C for 10 seconds, 55°C for 30 seconds and 95°C for 10 seconds. This was followed by a melt curve analysis of 65°C–95°C in 0.5°C increments. Following real-time PCR and melt curve analysis, products were run on a 1.5% agarose gel to verify appropriate product size.
CFX Manager™ Version 1.0 software (Bio-Rad, Hercules, CA) was used to calculate the average threshold cycle (C(t)) for each set of triplicate reactions. C(t) values for each sample for both Glcci1 and Dnajb1 were normalized to the reference gene beta actin and calibrated to account for inter-plate variation, as previously described in reference 32. Data is described as fold change in expression (2−ΔΔC(t)) on a linear scale.
Statistics for variable methylation patterns.
Variable methylation patterns at candidate metastable epialleles Glcci1 and Dnajb1 were validated by calculating the range of and variance in percentage of cells methylated at each CpG site at each of three loci (Glcci1 Repeat 1, Glcci1 Repeat 2, Dnajb1) in three tissue types (liver, kidney, brain). Inter-individual and inter-tissue ranges were calculated by computing the range in percent methylation at each site in each tissue across individuals in the former and at each site in all individuals across tissue in the latter. Mean inter-individual and inter-tissue variances were calculated by computing the average of variances at each site in a given tissue in the former and at each site in a given individual in the latter. Site-specific inter-individual variance was calculated by computing the variance in methylation at a given site within a tissue type at a given locus.
The ability of BPA to induce methylation changes in the same epialleles was assessed using repeated measures ANOVA (PROC MIXED), Bonferroni-corrected for multiple comparisons, with individual CpG sites defined as repeated measures. Thus, this analysis accounted for the lack of independence of multiple sites at a locus within a single animal. Since we observed an interaction between BPA exposure and individual CpG sites (that is, methylation at individual CpG sites were affected differentially by BPA exposure), we tested the effect of BPA for each CpG site using the ‘slice’ option in PROC MIXED. The ‘slice’ option specifies effects by which to separate interaction LSMEANS effects, effectively comparing each individual site in exposed relative to unexposed groups, accounting for correlation of sites within a locus within a subject. Thus, the effect of BPA exposure on mean methylation was assessed via tests of fixed effects and site-specific BPA effects via tests of effect slices for each CpG site. Normality of percent methylation at each locus was assessed visually using histograms and normal probability plots. No more than five potential outliers were observed at each locus; analysis was run both with and without these values, with no appreciable change. The final analysis excluded these few outliers, as the results were more conservative. Fold changes in expression in both BPA and control groups were analyzed for both candidate genes using Wilcoxon rank sum tests, due to non-normality of the data. All analyses were carried out using SAS version 9.2 software (Cary, NC).
Acknowledgments
This work was supported by NIH grant ES017524 and the University of Michigan NIEHS P30 Core Center P30 ES017885. Support for MSN was provided by NIEHS Institutional Training Grant T32 ES007062. The authors thank Christopher Faulk for his critical reading of the manuscript, as well as Ken Guire and Joe Kazemi from UM CSCAR (Center for Statistical Consultation and Research) for assistance with statistical analysis and figure.
Abbreviations
- Avy
viable yellow agouti
- BPA
bisphenol A
- ERV
endogenous retrovirus
- IAP
intracisternal A particle
- LTR
long terminal repeat
- LINEs
long interspersed nuclear elements
- SINEs
short interspersed nuclear elements
- Vi
inter-individual variance
- Vt
inter-tissue variance
Supplementary Material
References
- 1.Duhl D, Vrieling H, Miller K, Wolff G, Barsh G. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 2.Vasicek T, Zeng L, Guan X, Zhang T, Costantini F, Tilghman S. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics. 1997;147:777–786. doi: 10.1093/genetics/147.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruvinsky A, Flood W, Costantini F. Developmental mosaicism may explain spontaneous reappearance of the Axin(Fu) mutation in mice. Genesis. 2001;29:49–54. doi: 10.1002/1526-968x(200102)29:2<49::aid-gene1004>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 5.Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucl Acids Res. 2004;32:5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:2393–2400. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 7.Waterland R, Jirtle R. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolinoy DC, Wiedman J, Waterland R, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaminen-Ahola N, Ahola A, Maga M, Mallitt KA, Fahey P, Cox T, et al. Maternal Ethanol Consumption Alters the Epigenotype and the Phenotype of Offspring in a Mouse Model. PLoS Genet. 2010;6:1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2006;2:399–405. doi: 10.1371/journal.pgen.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolinoy D, Weinhouse C, Jones T, Rozek L, Jirtle R. Variable histone modifications at the A (vy) metastable epiallele. Epigenetics. 2010;5:637–644. doi: 10.4161/epi.5.7.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolinoy D, Das R, Weidman J, Jirtle R. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res. 2007;61:30–37. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- 14.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterland R, Jirtle R. Early nutrition, epigenetic changes at transposons and imprinted genes and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barker D. The developmental origins of adult disease. J Am Coll Nut. 2004;23:588–595. doi: 10.1080/07315724.2004.10719428. [DOI] [PubMed] [Google Scholar]
- 18.Chojkier M. Inhibition of albumin synthesis in chronic diseases: molecular mechanisms. J Clin Gastroenterol. 2005;39:143–146. doi: 10.1097/01.mcg.0000155514.17715.39. [DOI] [PubMed] [Google Scholar]
- 19.Ripperger J, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 20.Wuarin J, Schibler U. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell. 1990;63:1257–1266. doi: 10.1016/0092-8674(90)90421-a. [DOI] [PubMed] [Google Scholar]
- 21.Bertone P, Trifonov V, Rozowsky JS, Schubert F, Emanuelsson O, Karro J, et al. Design optimization methods for genomic DNA tiling arrays. Genome Res. 2006;16:271–281. doi: 10.1101/gr.4452906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graf S, Nielsen FGG, Kurtz S, Huynen MA, Birney E, Stunnenberg H, et al. Optimized design and assessment of whole genome tiling arrays. Bioinformatics. 2007;23:195–204. doi: 10.1093/bioinformatics/btm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenkranz R, Borodina T, Lehrach H, Himmelbauer H. Characterizing the mouse ES cell transcriptome with Illumina sequencing. Genomics. 2008;94:187–194. doi: 10.1016/j.ygeno.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 24.Kaput J, Klein K, Reyes E, Kibbe W, Cooney C, Jovanovic B, et al. Identification of genes contributing to the obese yellow Avy phenotype: caloric restriction, genotype, diet x genotype interactions. Physiol Genomics. 2004;18:316–324. doi: 10.1152/physiolgenomics.00065.2003. [DOI] [PubMed] [Google Scholar]
- 25.Wolff G, Stanley J, Ferguson M, Simpson P, Ronis M, Badger T. Agouti signaling protein stimulates cell division in “viable yellow” (A(vy)/a) mouse liver. Exp Bio Med. 2007;232:1326–1329. doi: 10.3181/0704-BC-95. [DOI] [PubMed] [Google Scholar]
- 26.Iwamoto K, Bundo M, Washizuka S, Kakiuchi C, Kato T. Expression of HSPF1 and LIM in the lymphoblastoid cells derived from patients with bipolar disorder and schizophrenia. J Hum Genet. 2004;49:227–231. doi: 10.1007/s10038-004-0136-5. [DOI] [PubMed] [Google Scholar]
- 27.Uchiyama Y, Takeda N, Mori M, Terada K. Heat shock protein 40/DjB1 is required for thermotolerance in early phase. J Biochem. 2006;140:805–812. doi: 10.1093/jb/mvj212. [DOI] [PubMed] [Google Scholar]
- 28.Chapman M, Askew D, Kuscuoglu U, Miesfeld R. Transcriptional control of steroid-regulated apoptosis in murine thymoma cells. Mol Endocrinol. 1996;10:967–978. doi: 10.1210/mend.10.8.8843413. [DOI] [PubMed] [Google Scholar]
- 29.Shen L, Waterland R. Methods of DNA methylation analysis. Curr Opin Clin Nutr Metab Care. 2007;10:576–581. doi: 10.1097/MCO.0b013e3282bf6f43. [DOI] [PubMed] [Google Scholar]
- 30.Colella S, Shen L, Baggerly K. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechnique. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 31.Waterland R, Kellermayer R, Laritsky E, Rayco-Solon P, Harris R, Travisano M, et al. Season of conception in rural gambia affects DNA methylation at putative human metastable epialleles. PLoS Genetics. 2010;6:1001252. doi: 10.1371/journal.pgen.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Applied_Biosystems, author. Guide to performing relative quantitation of gene expression using real-time quantitative PCR. 2008 Document Part Number 4371095 Rev B. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.