Abstract
In a recent publication,1 we identified a novel F-box protein, encoded by fates-shifted (fsd), that plays a role in targeting Bcd for ubiquitination and degradation. Our analysis of mutant Drosophila embryos suggests that Bcd protein degradation is important for proper gradient formation and developmental fate specification. Here we describe further experiments that lead to an estimate of Bcd half-life, <15 min, in embryos during the time of gradient formation. We use our findings to evaluate different models of Bcd gradient formation. With this new estimate, we simulate the Bcd gradient formation process in our own biologically realistic 2-D model. Finally, we discuss the role of Bcd-encoded positional information in controlling the positioning and precision of developmental decisions.
Key words: morphogen gradient, Bicoid, half-life, diffusion constant, steady state, positional information, robustness, Drosophila, fates-shifted, ubiquitination
Models of Bcd Gradient Formation
Bcd is a morphogenetic protein that forms a concentration gradient along the anterior-posterior (A-P) axis in early Drosophila embryos.2 It controls embryonic patterning by activating its target genes in a concentration-dependent manner.3–7 Despite extensive studies, it currently remains controversial how the Bcd concentration gradient is formed. There are two broad, contrasting models. A simple diffusion model8 has been widely used to explain the exponential gradient of Bcd,9–11 where Bcd protein is synthesized at the anterior, diffuses and decays throughout the embryo (hence also referred to as the SDD model). The diffusion model produces a steady state exponential profile of the Bcd concentration: B = Ae-x/λ, where A is the amplitude, x is distance from the anterior and λ is the length constant.9 However, since bcd mRNA, the source for Bcd production, is not restricted to a single point in the actual embryo,12,13 the idealized version of this model is inadequate for Bcd. In fact, based on the observed redistribution of bcd mRNA, a contrasting model was proposed recently in reference 14. In this model, the process of Bcd gradient formation and its final shape are dictated by the redistribution process of bcd mRNA.14,15 We refer to this model as the mRNA-dictated-gradient model. Since the measured bcd mRNA profile differs from the exponential Bcd protein profile,14 the pure form of the proposed mRNA-dictated-gradient model also appears inadequate for explaining fully how the Bcd protein gradient is formed.12,16
In addition to these two contrasting models, several other models have also been proposed for Bcd.17–19 Since Bcd diffusion remains a component of all these models, they as a group are more related to the diffusion model than the mRNA-dictated-gradient model. Each of these models was proposed to explain specific properties of the Bcd gradient system. For example, an intriguing property of the Bcd gradient is the stability of its nuclear concentrations as a function of developmental time.20 While the nuclear number undergoes an exponential increase after each nuclear division, the profiles of nuclear Bcd concentration remain relatively stable at the interphase of nuclear cycles 10–14. This observation led to the proposal of a nuclear trapping model,17 where Bcd protein is reversibly trapped by the nucleus to allow the formation of a stable nuclear Bcd concentration gradient. The nuclear trapping model proposes that Bcd does not decay during the period of gradient formation.17 Another model, a pre-steady-state model,18 was proposed to explain the precision of the expression patterns of Bcd target genes among different embryos. This model proposes that the positional information provided by the Bcd gradient is decoded before the gradient reaches its steady state. It also requires Bcd to be a stable protein with a half-life comparable to the proposed decoding time (60∼90 min).18
An Estimate of Bcd Half-Life to Evaluate Different Models
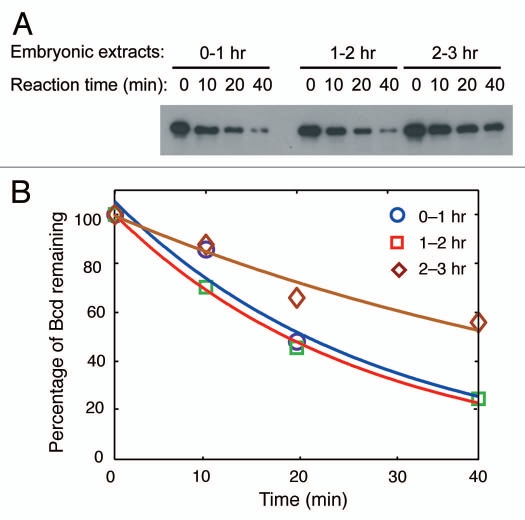
An important finding reported in our recent study is that perturbed Bcd degradation, in embryos from fsd females (referred to as fsd embryos), led to an altered Bcd gradient profile.1 These results suggest that Bcd degradation is important for the gradient formation process. To evaluate different models of Bcd gradient formation, it is critical to have an estimate of the Bcd half-life, t1/2, in embryos at the time of Bcd gradient formation. Such a value is currently unavailable. Based on the kinetics of the disappearance of Bcd after cellularization,2 it was estimated that Bcd t1/2 in cellularized embryos is <30 min. This value is for embryos that are actively “clearing up” the Bcd gradient that is no longer needed. In our reported study in reference 1, we used 0–3 hr embryonic extracts to assay Bcd degradation. These extracts reflect, collectively, the properties of embryos that are both undergoing Bcd gradient formation and actively clearing up the Bcd gradient (i.e., before and after cellularization, respectively). To gain further insights into Bcd degradation properties in embryos as a function of developmental time, we generated extracts from staged 0–1, 1–2 and 2–3 hr embryos, with all experiments performed side-by-side to allow direct comparisons. Our Bcd degradation assays in these extracts revealed an estimated t1/2 of 19.7, 18.9 and 43.6 min, respectively (Fig. 1). These results show that embryos undergoing the process of Bcd gradient formation (0–1 and 1–2 hr) actually have higher Bcd degradation activities than later embryos (2–3 hr). Since the 2–3 hr embryos contain those that are actively clearing up the Bcd gradient, we used the previous estimate obtained from such embryos2 to calibrate our biochemical data. Using such calibration, we estimate that Bcd t1/2 in embryos at the time of Bcd gradient formation is <15 min. While we are aware of the inherent limitations of biochemical assays for quantifying biophysical properties inside an embryo, this value represents the best estimate currently available.
Figure 1.
Bcd degradation and estimation of Bcd half-life. (A) Extracts generated from 0–1, 1–2 and 2–3 hr w1118 embryos were used to assay Bcd degradation.1 As shown previously in reference 1, Bcd degradation in embryonic extracts is inhibited by MG132, indicating that proteasome-dependent activities, as opposed to some non-specific activities, are responsible for Bcd degradation. All experiments shown here were performed side-by-side. As discussed previously in reference 1, data from experiments that are not done side-by-side cannot, and should not, be compared with each other. (B) The plot shows the percentage of Bcd protein remaining at different time points of the degradation reaction. Solid lines represent exponential fitting to experimental data. (For further details, see text and ref. 1).
Our estimated Bcd t1/2 of <15 min has important implications for the mechanisms of Bcd gradient formation. First, this value is not consistent with the assumptions made in the nuclear trapping and pre-steady-state models,17,18 where Bcd is proposed not to decay or to decay slowly during the gradient formation process. Our reported finding that Bcd degradation is important for gradient formation1 is also inconsistent with the mRNA-dictated-gradient model because, according to this model, the final shape of the protein gradient is determined exclusively by the bcd mRNA gradient, rather than Bcd protein degradation and diffusion.12,14–16 Second, our estimated Bcd t1/2 sheds light on an outstanding controversy over the diffusion constant D of Bcd in the cytoplasm of embryos. While an earlier study20 suggested a D value of ∼0.3 µm2s−1, a more recent study in reference 21 revealed a value that is >20 times larger, ∼7.4 µm2s−1. In a simple diffusion model,8,9 the length constant λ of the steady state exponential profile is a function of the diffusion constant D and decay rate ω, λ2 = D/ω. Under this framework, our estimated half-life of Bcd (t1/2 = 15 min corresponds to ω = 0.0008 s−1) and a consensus λ estimate of ∼100 µm are more consistent with a large D value. We note that, since our estimated Bcd t1/2 represents an upper bound, a formal possibility exists that the actual D value for Bcd could be even larger than the reported ∼7.4 µm2s−1.
Bcd Gradient Formation Simulated in a Biologically Realistic Model
We have developed recently a biologically realistic 2-D model for the Bcd gradient formation process.12,22 This model is different from the simple diffusion model because it has incorporated several key biological features relevant to the Bcd gradient system, including the location and amount of bcd mRNA and the exponential increase in nuclear numbers after each nuclear division (reviewed in ref. 12). Using this model and our newly estimated Bcd t1/2, we conducted simulation studies to evaluate Bcd gradient properties (for details, see Fig. 2 legend). Figure 2A shows the simulated profiles of nuclear Bcd concentration, [Bn], at nuclear cycles 10–14, which exhibit an experimentally observed stability.20 As further discussed in reference 12, an interaction between Bcd and the genome within the nucleus in our model plays an important role in maintaining the stability of nuclear Bcd concentrations during the time when the nuclear number is increasing exponentially (see also ref. 17). Figure 2B shows that our simulated [Bn] profile and experimentally measured Bcd profile,23 both at early nuclear cycle 14, exhibit strong resemblance to each other. These and additional results (below) show that our biologically realistic model can recapitulate key properties of the Bcd gradient. For example, it also readily explains the differences in Bcd gradient profiles caused by the geometric asymmetry between the dorsal and ventral sides of the embryo.22 In addition, it recapitulates the scaling properties of the Bcd gradient23 by simply assuming a correlation between the Bcd production rate and embryo volume.12 This hypothesized correlation was recently observed experimentally13 through quantitative measurements of bcd mRNA in large and small embryos from selected Drosophila lines.24 Since our model is based on Bcd diffusion and degradation while incorporating relevant, biologically realistic features of the embryo, our results suggest that the diffusion model—despite the inadequacies of its idealized form in fully capturing Bcd gradient properties—remains a useful framework in guiding our thinking and analyses of the Bcd gradient formation process.
Figure 2.
Simulated nuclear Bcd gradient profiles. (A) Nuclear Bcd concentration [Bn], in arbitrary units, within the cortical layer of a simulated embryo was obtained in a biologically realistic 2-D model.12 Here the simulated [Bn] profiles as a function of fractional embryo length x/L are shown for nuclear cycles 10 to 14. These simulated [Bn] profiles, as seen experimentally,20 differ by <10%, with a calculated g value of −0.09 (for details see ref. 12). This simulation was performed using ω = 0.0008 s−1 and D = 8 µm2s−1. All other parameters used here were the same as in the main model described in reference 12, except the center coordinate of the bcd mRNA sphere (55 µm in current work). (B) Comparison between simulated [Bn] profile and the experimentally measured mean Bcd profile (with standard deviation shown).23 Both profiles are from early nuclear cycle 14 and each has a length constant λ of ∼100 µm.
Interpretation of Bcd-Encoded Positional Information
One of the fundamental questions regarding the actions of morphogens relates to the precision of the positional information provided by a morphogen gradient and the extent to which such information can influence the precision of developmental decisions.25,26 For the Bcd gradient profile, embryo-to-embryo variations were initially reported to be very large, suggesting that the positional information provided by the Bcd gradient is imprecise.9,27,28 These findings prompted the proposal of different models that can correct embryo-to-embryo fluctuations in Bcd-encoded positional information.18,29–33 However, more recent studies revealed that the Bcd gradient profile is highly precise among different embryos.23,34,35 But whether a precise Bcd gradient is important for precise patterning remains controversial. It was suggested that, based on thermal perturbations, a precise Bcd gradient is not required for precise patterning.36 Our own studies based on genetic perturbations have led to a contrasting proposal.23,35 In two separate cases where mutant embryos exhibit increased variability in the hunchback (hb) expression boundary, we were able to trace the origin of such variability directly to perturbed Bcd gradient properties.23,35 To our knowledge, these results represent the only experimental evidence that a precise Bcd gradient is necessary for precise developmental decisions. Our finding1 that the shift in Bcd-encoded positional information in fsd embryos matches the shift in hb expression boundary further underscores a direct and dominant role of Bcd in instructing hb expression (see also ref. 22). Exactly how the positional information provided by the Bcd gradient feeds into precise patterning decisions is a subject of intense theoretical investigations.34,35,37–39 In a recent experimental study,40 we investigated the role of Bcd in the actual transcriptional events of its target genes in developing embryos. Our results suggest that Bcd acts as a direct and sustained input for these transcriptional events. In addition, a comparison between the noise in Bcd-dependent transcriptional events and the noise in Bcd-dependent transcriptional products provides a first experimental demonstration of the effect of time/space averaging in reducing the output noise.40
Recent studies of the role of the terminal system in the expression of Bcd target genes have led to the proposal of a morphogen network model.41–44 This model emphasizes the integration of maternal inputs of both the terminal system and the Bcd gradient; cross-regulation between the zygotic products of gap genes is also a hallmark feature of the proposed gene regulatory network models.18,33,45,46 Understanding mechanistically how genes respond to distinct Bcd concentration thresholds remains an important and challenging problem.41–43,47,48 Meanwhile, how hb is expressed in response to Bcd has attracted most extensive studies in the field.9,22,23,33–35,46,49,50 Unlike genes that are expressed near the head region, the hb expression boundary does not appear to be influenced strongly by the terminal system.41 Thus, the positioning and precision of the hb expression boundary provides a best readout of Bcd-encoded positional information and properties intrinsic to the Bcd gradient system. In addition, the hb expression boundary is located near the middle of the embryo, where the scaling properties can influence the patterning landscape along the entire A-P axis.51 Importantly, the scaling properties of hb can also be traced directly to the scaling properties of the Bcd gradient.12,13,23 These and other results have led us to propose in reference 23 that the Bcd gradient itself is a robust system (see also refs. 34 and 49). Understanding the precise mechanisms of Bcd-activated hb transcription will further expand our knowledge of not only how Bcd works in particular, but also how morphogens work in general.
Acknowledgments
This work was supported in part by grants from NIH and NSF (to Jun Ma) and an AHA postdoctoral fellowship (to Feng He).
Extra View to: Liu J, Ma J. Fates-shifted is an F-box protein that targets Bicoid for degradation and regulates developmental fate determination in Drosophila embryos. Nature Cell Biol. 2011;13:22–29. doi: 10.1038/ncb2141.
References
- 1.Liu J, Ma J. Fates-shifted is an F-box protein that targets Bicoid for degradation and regulates developmental fate determination in Drosophila embryos. Nat Cell Biol. 2011;13:22–29. doi: 10.1038/ncb2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Driever W, Nüsslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 3.Driever W, Nüsslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration dependent manner. Cell. 1988;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- 4.Struhl G, Struhl K, Macdonald P. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989;57:1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- 5.Driever W, Thoma G, Nüsslein-Volhard C. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding site for the bicoid morphogen. Nature. 1989;340:363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- 6.Driever W, Ma J, Nusslein-Volhard C, Ptashne M. Rescue of bicoid mutant Drosophila embryos by Bicoid fusion proteins containing heterologous activating sequences. Nature. 1989;342:149–154. doi: 10.1038/342149a0. [DOI] [PubMed] [Google Scholar]
- 7.Ephrussi A, St Johnston D. Seeing is believing. The bicoid morphogen gradient matures. Cell. 2004;116:143–152. doi: 10.1016/s0092-8674(04)00037-6. [DOI] [PubMed] [Google Scholar]
- 8.Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969;25:1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 9.Houchmandzadeh B, Wieschaus E, Leibler S. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature. 2002;415:798–802. doi: 10.1038/415798a. [DOI] [PubMed] [Google Scholar]
- 10.Wartlick O, Kicheva A, Gonzalez-Gaitan M. Morphogen gradient formation. Cold Spring Harb Perspect Biol. 2009;1:1255. doi: 10.1101/cshperspect.a001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimm O, Coppey M, Wieschaus E. Modelling the Bicoid gradient. Development. 2010;137:2253–2264. doi: 10.1242/dev.032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng J, Wang W, Lu LJ, Ma J. A two-dimensional simulation model of the Bicoid gradient in Drosophila. PLoS ONE. 2010;5:10275. doi: 10.1371/journal.pone.0010275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung D, Miles C, Kreitman M, Ma J. Scaling of the Bicoid morphogen gradient by a volume-dependent production rate. Development. 2011 doi: 10.1242/dev.064402. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spirov A, Fahmy K, Schneider M, Frei E, Noll M, Baumgartner S. Formation of the bicoid morphogen gradient: an mRNA gradient dictates the protein gradient. Development. 2009;136:605–614. doi: 10.1242/dev.031195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dilao R, Muraro D. mRNA diffusion explains protein gradients in Drosophila early development. Journal of theoretical biology. 2010;264:847–853. doi: 10.1016/j.jtbi.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Little SC, Tkacik G, Kneeland TB, Wieschaus EF, Gregor T. The formation of the bicoid morphogen gradient requires protein movement from anteriorly localized mRNA. PLoS biology. 2011;9:1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coppey M, Berezhkovskii AM, Kim Y, Boettiger AN, Shvartsman SY. Modeling the bicoid gradient: diffusion and reversible nuclear trapping of a stable protein. Developmental biology. 2007;312:623–630. doi: 10.1016/j.ydbio.2007.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergmann S, Sandler O, Sberro H, Shnider S, Schejter E, Shilo BZ, et al. Pre-steady-state decoding of the Bicoid morphogen gradient. PLoS biology. 2007;5:46. doi: 10.1371/journal.pbio.0050046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecht I, Rappel WJ, Levine H. Determining the scale of the Bicoid morphogen gradient. Proc Natl Acad Sci USA. 2009;106:1710–1715. doi: 10.1073/pnas.0807655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007;130:141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abu-Arish A, Porcher A, Czerwonka A, Dostatni N, Fradin C. High mobility of bicoid captured by fluorescence correlation spectroscopy: implication for the rapid establishment of its gradient. Biophys J. 2010;99:33–35. doi: 10.1016/j.bpj.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He F, Wen Y, Cheung D, Deng J, Lu LJ, Jiao R, et al. Distance measurements via the morphogen gradient of Bicoid in Drosophila embryos. BMC Dev Biol. 2010;10:80. doi: 10.1186/1471-213X-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He F, Wen Y, Deng J, Lin X, Lu J, Jiao R, et al. Probing intrinsic properties of a robust morphogen gradient in Drosophila. Dev Cell. 2008;15:558–567. doi: 10.1016/j.devcel.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miles CM, Lott SE, Luengo Hendriks CL, Ludwig MZ, Manu, Williams CL, et al. Artificial selection on egg size perturbs early pattern formation in Drosophila melanogaster. Evolution. 2010;65:33–42. doi: 10.1111/j.1558-5646.2010.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaeger J, Martinez-Arias A. Getting the measure of positional information. PLoS biology. 2009;7:81. doi: 10.1371/journal.pbio.1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolpert L. Positional information and patterning revisited. J Theor Biol. 2011;269:359–365. doi: 10.1016/j.jtbi.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 27.Spirov AV, Holloway DM. Making the body plan: Precision in the genetic hierarchy of Drosophila embryo segmentation. In Silico Biol. 2002;3:9. [PubMed] [Google Scholar]
- 28.Holloway DM, Harrison LG, Kosman D, Vanario-Alonso CE, Spirov AV. Analysis of pattern precision shows that Drosophila segmentation develops substantial independence from gradients of maternal gene products. Dev Dyn. 2006;235:2949–2960. doi: 10.1002/dvdy.20940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howard M, Ten Wolde PR. Finding the center reliably: robust patterns of developmental gene expression. Physical Rev Lett. 2005;95:208103. doi: 10.1103/PhysRevLett.95.208103. [DOI] [PubMed] [Google Scholar]
- 30.Houchmandzadeh B, Wieschaus E, Leibler S. Precision domain specification in the developing Drosophila embryo. Pysical Rev E. 2005;72:061920. doi: 10.1103/PhysRevE.72.061920. [DOI] [PubMed] [Google Scholar]
- 31.Aegerter-Wilmsen T, Aegerter CM, Bisseling T. Model for the robust establishment of precise proportions in the early Drosophila embryo. Journal of theoretical biology. 2005;234:13–19. doi: 10.1016/j.jtbi.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 32.McHale P, Rappel WJ, Levine H. Embryonic pattern scaling achieved by oppositely directed morphogen gradients. Phys Biol. 2006;3:107–120. doi: 10.1088/1478-3975/3/2/003. [DOI] [PubMed] [Google Scholar]
- 33.Manu, Surkova S, Spirov AV, Gursky VV, Janssens H, Kim AR, et al. Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation. PLoS biology. 2009;7:1000049. doi: 10.1371/journal.pbio.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He F, Saunders T, Wen Y, Cheung D, Jiao R, ten Wolde P, et al. Shaping a morphogen gradient for positional precision. Biophys J. 2010;99:697–707. doi: 10.1016/j.bpj.2010.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucchetta EM, Vincent ME, Ismagilov RF. A precise Bicoid gradient is nonessential during cycles 11–13 for precise patterning in the Drosophila blastoderm. PLoS One. 2008;3:3651. doi: 10.1371/journal.pone.0003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tostevin F, ten Wolde PR, Howard M. Fundamental limits to position determination by concentration gradients. PLoS Computational Biol. 2007;3:78. doi: 10.1371/journal.pcbi.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okabe-Oho Y, Murakami H, Oho S, Sasai M. Stable, precise and reproducible patterning of bicoid and hunchback molecules in the early Drosophila embryo. PLoS Computational Biol. 2009;5:1000486. doi: 10.1371/journal.pcbi.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erdmann T, Howard M, ten Wolde PR. The role of spatial averaging in the precision of gene expression patterns. Phys Rev Lett. 2009;103:258101. doi: 10.1103/PhysRevLett.103.258101. [DOI] [PubMed] [Google Scholar]
- 40.He F, Ren J, Wang W, Ma J. A Multiscale investigation of bicoid-dependent transcriptional events in Drosophila embryos. PLoS ONE. 2011;6:e19122. doi: 10.1371/journal.pone.0019122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochoa-Espinosa A, Yu D, Tsirigos A, Struffi P, Small S. Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc Natl Acad Sci USA. 2009;106:3823–3828. doi: 10.1073/pnas.0807878105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Löhr U, Chung HR, Beller M, Jackle H. Antagonistic action of Bicoid and the repressor Capicua determines the spatial limits of Drosophila head gene expression domains. Proc Natl Acad Sci USA. 2009;106:21695–21700. doi: 10.1073/pnas.0910225106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Löhr U, Chung HR, Beller M, Jackle H. Bicoid: Morphogen function revisited. Fly (Austin) 2010;4:236–240. doi: 10.4161/fly.4.3.11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porcher A, Dostatni N. The bicoid morphogen system. Curr Biol. 2010;20:249–254. doi: 10.1016/j.cub.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 45.Jaeger J, Surkova S, Blagov M, Janssens H, Kosman D, Kozlov KN, et al. Dynamic control of positional information in the early Drosophila embryo. Nature. 2004;430:368–371. doi: 10.1038/nature02678. [DOI] [PubMed] [Google Scholar]
- 46.Holloway DM, Lopes FJ, da Fontoura Costa L, Travencolo BA, Golyandina N, Usevich K, et al. Gene expression noise in spatial patterning: hunchback promoter structure affects noise amplitude and distribution in Drosophila segmentation. PLoS Computational Biol. 2011;7:1001069. doi: 10.1371/journal.pcbi.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu D, Zhao C, Ma J. Enhancer sequences influence the role of the amino terminal domain of Bicoid in transcription. Mol Cell Biol. 2003;23:4439–4448. doi: 10.1128/MCB.23.13.4439-4448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ochoa-Espinosa A, Yucel G, Kaplan L, Pare A, Pura N, Oberstein A, et al. The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila. Proc Natl Acad Sci USA. 2005;102:4960–4965. doi: 10.1073/pnas.0500373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crauk O, Dostatni N. Bicoid determines sharp and precise target gene expression in the Drosophila embryo. Curr Biol. 2005;15:1888–1898. doi: 10.1016/j.cub.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 50.Porcher A, Abu-Arish A, Huart S, Roelens B, Fradin C, Dostatni N. The time to measure positional information: maternal hunchback is required for the synchrony of the Bicoid transcriptional response at the onset of zygotic transcription. Development. 2010;137:2795–2804. doi: 10.1242/dev.051300. [DOI] [PubMed] [Google Scholar]
- 51.de Lachapelle AM, Bergmann S. Precision and scaling in morphogen gradient read-out. Mol Syst Biol. 2010;6:351. doi: 10.1038/msb.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]