Abstract
Clostridium difficile infection (CDI) arises in the setting of antibiotic administration where disruption of the normal indigenous gut microbiota leads to susceptibility to C. difficile colonization and colitis. Using a murine model of CDI, we demonstrate that changes in the community structure of the indigenous gut microbiota are associated with the loss of colonization resistance against C. difficile. Several antibiotic regimens were tested in combination for the ability to overcome colonization resistance, including a five antibiotic cocktail consisting of kanamycin, gentamicin, colistin, metronidazole and vancomycin administered in drinking water for three days, a single intraperitoneal dose of clindamycin or 10 days of cefoperazone in drinking water. Following antibiotic treatment animals were challenged with 105 colony forming units of C. difficile strain VPI 10463 via oral gavage. Animals that received the antibiotic cocktail and clindamycin prior to C. difficile challenge followed one of two clinical courses, either becoming clinically ill and moribund within 2–4 days post challenge, or remaining clinically well. Animals that became clinically ill developed histologically severe colitis. These histopathologic findings were significantly less severe in animals that remained clinically well. Analysis of 16s rRNA gene sequences retrieved from gut tissue at necropsy demonstrated that Proteobacteria dominated the gut microbiota in clinically ill animals. In contrast, the gut microbial community of clinically well animals more closely resembled untreated animals, which were dominated by members of the Firmicutes. All animals that received cefoperazone treatment prior to C. difficile challenge were clinically ill and moribund by 2–5 days post challenge in a dose dependent manner. The gut communities in these animals were dominated by C. difficile suggesting that cefoperazone treatment resulted in a greater loss in colonization resistance. Thus, the severity of colitis that arises in this system reflects the interplay between the expansion of C. difficile in the gut community and the ecologic dynamics of the indigenous microbial community as it recovers from antibiotic perturbation. We demonstrate that altering the balance of these two opposing processes alters clinical outcome and thus may lead to novel preventative and therapeutic approaches for CDI.
Keywords: Clostridium difficile, colonization resistance, microbial ecology, antibiotic-associated diarrhea, C. difficile infection
Introduction
The gastrointestinal (GI) tract of mammals is inhabited by a complex microbial community that plays a crucial role in maintaining gut homeostasis.1,2 The GI tract microbiota performs a number of beneficial metabolic functions3 and also aids in the normal development of the mucosal epithelium and maturation of the mucosal immune system.4–7 The indigenous microbiota protects the host from colonization by potentially pathogenic organisms, a function that is termed colonization resistance.8 It has been hypothesized that following the successful colonization by a pathogen, the ultimate pathology depends on the interplay between the host, pathogen and the indigenous microbiota.9 Thus, the resident microbiota can potentially modulate the outcomes of any pathogen/host interaction.
C. difficile is a Gram-positive, toxin-producing bacterium first described in 1935 as a commensal organism in the fecal microbiota of healthy newborn infants.10 It is currently the most common cause of health care-associated diarrhea and colitis and is responsible for significant morbidity and increased healthcare cost.11 Clostridium difficile infection (CDI) is associated with the use of broad-spectrum antibiotic therapy, increasing patient age and hospitalization.12 In recent years, the appearance of an epidemic strain (BI/NAP1/027) with potentially increased virulence has prompted renewed interest in the pathogenesis and epidemiology of this bacterium.11,12 Additionally, it appears that the overall incidence of C. difficile infection has been increasing.13
As C. difficile is not normally a significant component of the GI tract microbiota of adult humans, it is proposed that the indigenous gut microbiota is important in mediating colonization resistance against this pathogenic bacterium.14,15 According to this hypothesis, disruption of the indigenous gut microbiota by the administration of antibiotics results in a decrease in colonization resistance. Furthermore, recurrent CDI appears to occur in the setting where the indigenous microbiota is sufficiently disturbed so that colonization resistance cannot be restored even after cessation of the inciting antibiotics and completion of specific treatment directed against C. difficile.16 We have demonstrated that patients with recurrent C. difficile infection have decreased diversity of the indigenous gut microbiota which may reflect a corresponding defect in colonization resistance.17
A number of animal models have been developed to facilitate the study of C. difficile pathogenesis. The hamster model has been used extensively and it was in this host that Koch's postulates were fulfilled for C. difficile as the causative agent of antibiotic-associated colitis.18 In this model colitis develops after exposure to clindamycin and subsequent C. difficile challenge. However, the resulting disease is severe and lethal within three days after initial infection. This does not represent the usual course and spectrum of CDI in humans, which can range from asymptomatic to severe colitis.13 Furthermore, the limited availability of reagents to study host responses in hamsters has dampened the usefulness of this model. Gnotobiotic mice challenged with C. difficile also develop intestinal disease but this model precludes an examination of the role of indigenous microbiota in mediating colonization resistance.19–21 Thus, the available animal models have limited studies of C. difficile pathogenesis.
It has been reported that treatment of mice with various antibiotics can render the animals susceptible to C. difficile colonization.22 In some cases this can lead to the development of colitis.23,24 In this present study, we utilized antibiotic-treated mice to demonstrate that altering the community structure of the indigenous gut microbiota is associated with both the loss of colonization resistance against C. difficile and differences in the severity of disease. Our results indicate that a better understanding of the role of the indigenous microbiota in CDI could lead to novel and improved mechanisms for prevention and treatment.
Results
Overcoming colonization resistance to C. difficile.
To compare the ability of different antibiotic regimens to overcome colonization resistance against C. difficile six to eight week old C57BL/6 mice were treated with either an antibiotic cocktail (kanamycin, gentamicin, colistin, metronidazole and vancomycin), clindamycin or the combination of both prior to challenge with 1 × 105 CFU of C. difficile (VPI 10463) via oral gavage (Fig. 1A and B). Control animals were challenged with C. difficile in the absence of any antibiotic pretreatment. Colonization was monitored by culture and C. difficile-specific PCR was performed on DNA isolated from stool pellets or from gut tissue harvested at necropsy, which occurred 2 to 4 days post-challenge. Animals were monitored daily for signs of clinical CDI including diarrhea, weight loss and hunched posture. C. difficile was never recovered from animals that were challenged without antibiotic pretreatment. Animals that received the antibiotic cocktail without clindamycin prior to C. difficile challenge were also resistant to colonization. Of the 9 animals that received only a single dose of clindamycin prior to C. difficile challenge, 4 of them shed low amounts of the organism in their feces for the first 2 days following challenge, but the organism was no longer detectable in stool or tissue when the animals were euthanized four days following challenge. All 12 animals that received both the antibiotic cocktail and clindamycin prior to C. difficile challenge shed the organism in their feces throughout the experiment and C. difficile was found in tissue at the time of necropsy. These results indicate that the combination of the antibiotic cocktail and clindamycin is required to completely overcome colonization resistance against C. difficile.
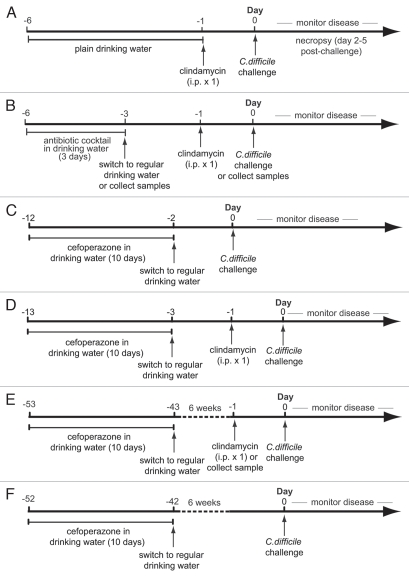
Figure 1.
Experimental designs for alteration of microbiota and C. difficile infection. Wild-type mice were treated with: (A) a single intraperitoneal dose of clindamycin and challenged with 105 CFU of C. difficile (VPI 10463). (B) a 5 antibiotic cocktail in drinking water for 3 days; a 5 antibiotic cocktail in drinking water for 3 days followed by a 2-day period without the drug and a single dose of clindamycin; or a 5 antibiotic cocktail in drinking water for 3 days followed by a 2-day period without the drug and a single dose of clindamycin followed by challenge with 105 CFU of C. difficile one day later. (C) 10 days cefoperazone treatment followed by 2 days off drug with or without C. difficile challenge. (D) 10 days cefoperazone treatment followed by 2 days off drug, a single dose of clindamycin followed by one day recovery with or without C. difficile challenge. (E) 10 days cefoperazone treatment followed by 6 weeks off drug, a single dose of clindamycin followed by one day recovery prior to C. difficile challenge. (F) 10 days cefoperazone treatment followed by 6 weeks off drug then C. difficile challenge.
Clinical disease in C. difficile infected mice.
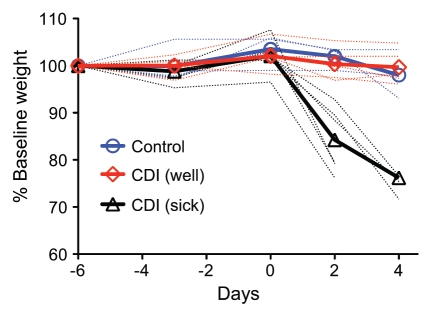
We monitored the development of disease in mice that received both the antibiotic cocktail and clindamycin prior to C. difficile challenge. Five of 12 animals that received both the antibiotic cocktail and clindamycin did not show overt clinical signs of CDI despite remaining colonized with C. difficile. The remaining seven mice exhibited signs of disease including diarrhea, hunched posture and significant (>20% from baseline) weight loss (Fig. 2). One animal was found dead at 2 days post challenge while 6 animals were moribund and euthanized 2 to 4 days post challenge.
Figure 2.
Weight loss in C. difficile infected mice. Weight loss curves for untreated (control) animals (n = 5) and animals treated with the 5 antibiotic cocktail for 3 days (days -6 to -3) then given a single dose of clindamycin after 2 days off antibiotics followed by challenge with C. difficile one day later (n = 11). 6 of 11 mice lost >20% body weight (sick) while one animal died 1–2 days post challenge (not shown). The remaining 5 animals did not lose significant body weight (well) when compared to control animals. Weight loss percentage is based on the starting weight at day -6 with thick lines showing the average for animals in each clinical group and dotted lines showing the data for each individual. CDI-C. difficile infection.
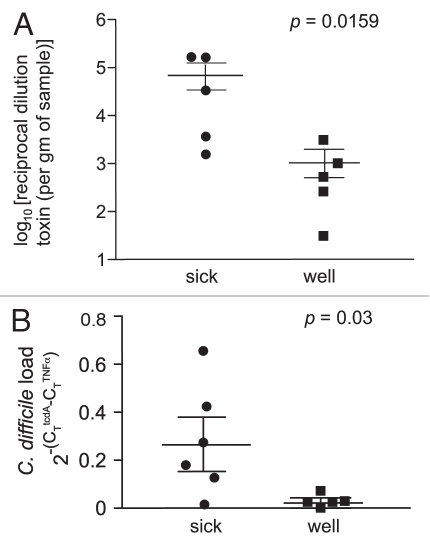
A central feature of the pathogenicity of C. difficile is the production of two large glucosyltransferase toxins encoded by tcdA and tcdB that modify and inactivate the small GTPases Rho, Rac and Cdc42.25 We measured the activity levels of C. difficile toxin in the gut using a tissue culture cytotoxin assay. High levels of C. difficile cytotoxin were detected in samples obtained from animals with severe clinical disease (Fig. 3A). Additionally, quantitative PCR analysis indicates that animals with severe clinical disease had significantly higher numbers of C. difficile in their gut at the time of necropsy compared to those animals that were clinically well (Fig. 3B). These results suggest that the animals that did not develop severe CDI in the multi-antibiotic treatment protocol had the ability to control the population size of colonizing C. difficile and to limit the production of toxin.
Figure 3.
Increased cytotoxin activity and C. difficile load are associated with increased CDI severity. (A) Vero cell tissue culture was used to determine the log10 reciprocal cytotoxin dilution per gram of sample. Each point represents individual animals that were either sick (n = 5) or well (n = 5) after treatment with antibiotics/clindamycin and exposure to C. difficile. Sick animals had increased levels of cytotoxin production compared to animals with less clinical disease. (B) Quantitative PCR was performed on cecal DNA from animals treated with antibiotics/clindamycin and exposed to C. difficile. Values represent the relative abundance of the tcdA gene normalized to the single copy mouse tumor necrosis factorα (TNFα) gene. Sick animals (n = 6) had increased levels of C. difficile compared to well animals (n = 5) with less clinical disease.
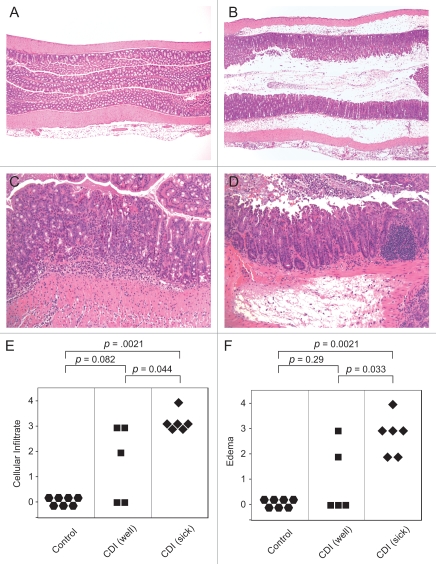
Histopathologic changes were seen in both the cecal and colonic tissue of all animals that received the antibiotic cocktail, clindamycin and C. difficile. Pathologic changes consisted of neutrophilic inflammation in the mucosa and submucosa with varying degrees of submucosal edema (Fig. 4). Of note, animals that were infected and clinically ill had significantly more severe colonic inflammation and particularly, submucosal edema than animals that were infected but remained clinically well (Fig. 4E and F). In the most severely affected animals, there were areas of erosion and in rare cases, ulceration. Occasionally, severely affected animals had luminal exudates comprised of degenerate neutrophils, hemorrhage and sloughed epithelium embedded in a fibrinous matrix suggestive of pseudomembranes (Fig. 4D). Untreated animals or those that received antibiotics without C. difficile challenge had no histological alterations.
Figure 4.
Animals with clinically severe CDI have increased colonic histopathology. (A) Proximal colon of an untreated (control) mouse. HE. Original magnification x40. (B) Colon of a sick antibiotic treated mouse infected with C. difficile showing severe submucosal edema. HE. Original magnification x40. (C) Increased magnification of colon from a well antibiotic treated mouse infected with C. difficile showing moderate neutrophilic mucosal and submucosal inflammation but lacking significant submucosal edema. HE. Original magnification x200. (D) Sick C. difficile infected mouse showing marked submucosal edema in addition to neutrophilic inflammation. There is also a pseudomembrane on the luminal surface consisting of degenerate neutrophils, sloughed epithelial cells and hemorrhage within a fibrinous matrix. HE. Original magnification x200. (E) Categorical cellular infiltration scores of untreated (control) animals, C. difficile infected well animals and C. difficile infected sick animals. (F) Categorical edema scores of untreated (control) animals, C. difficile infected well animals and C. difficile infected sick animals. Comparisons between groups were performed using the non-parametric Krustal Wallis test.
Shifts in microbial ecology associated with antibiotic treatment and C. difficile infection.
We have previously shown that antibiotic administration can decrease the overall mass of bacteria within the gut.26 The administration of the 5 antibiotic cocktail significantly decreased the overall bacterial population by 20-fold when measured immediately after the treatment period (data not shown). However, a single administration of clindamycin did not change the total microbial population size when measured 24 hours after the dose was given. Furthermore, following administration of both the antibiotic cocktail and clindamycin, at the time corresponding to C. difficile challenge, the overall bacterial population size was similar to untreated animals. Therefore, the loss of colonization resistance against C. difficile following antibiotic administration was not directly related to changes in the overall density of the gut microbiota.
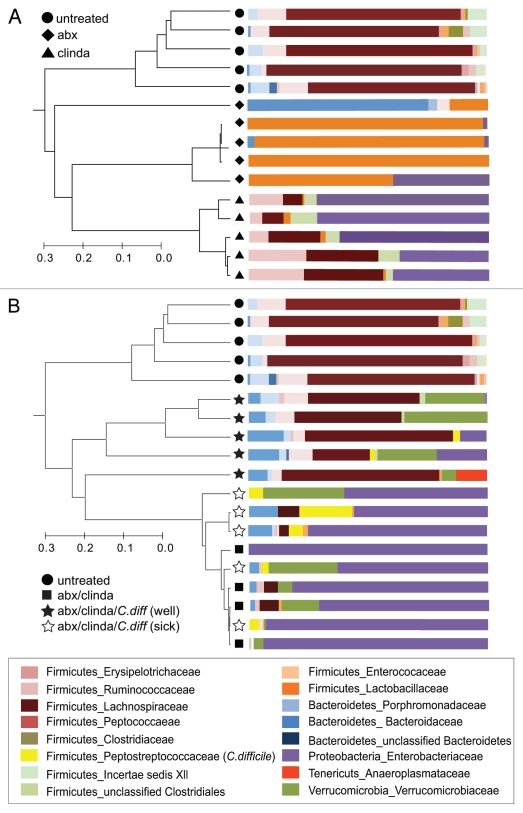
Since the loss of colonization resistance against C. difficile was not associated with an overall decrease in bacterial density we analyzed 16S rRNA gene sequences retrieved from gut tissue at the time of necropsy to examine the specific changes in the community structure of the gut microbiota that resulted from antibiotic treatment and C. difficile infection. In control mice that never received antibiotics, the gut microbial community was dominated by members of the phylum Firmicutes with lower numbers of Bacteroidetes. The administration of either the antibiotic cocktail, clindamycin or both resulted in a significant change in the structure of the gut microbial community (Fig. 5A and B). The administration of the antibiotic cocktail resulted in a shift in the community structure to one dominated by bacteria from the family Lactobacillaceae. Clindamycin treatment alone shifted the community composition to a dominance of Proteobacteria belonging to the family Enterobacteriaceae (Fig. 5A). When clindamycin was administered following treatment with the antibiotic cocktail, there was again a predominance of Proteobacteria in the gut community (Fig. 5B).
Figure 5.
Shifts in microbial community structure and composition associated with antibiotic administration and C. difficile infection. The community structure of the gut microbiota was determined by 16s rRNA gene clone library construction. (A) The microbial communities in animals that were treated with the antibiotic cocktail (abx) or with clindamycin alone (clinda) were altered from that seen in untreated controls. The antibiotic cocktail alone resulted in the appearance of significant numbers of lactobacilli, whereas clindamycin administration was associated with an increase in Proteobacteria. (B) The microbial communities in animals that received the combination of the antibiotic cocktail and clindamycin were also dominated by Proteobacteria. Animals that were challenged with C. difficile after antibiotic treatment harbored gut communities that were distinguished by the clinical disease that developed. Proteobacteria dominated sick antibiotic treated animals exposed to C. difficile while the communities of animals that remained well appeared to resemble controls with a predominance of Firmicutes. Dendrograms were constructed using Morisita-Horn community similarities based on >97% sequence similarity while taxonomic assignments were made using the Ribosomal Database Project Classifier. abx-antibiotic cocktail, clinda-clindamycin, C.diff-C. difficile.
Interestingly, subsequent changes in the gut microbial community structure following C. difficile challenge followed two distinct courses. Animals that developed severe clinical disease harbored a gut microbial community at the time of necropsy that remained dominated by Proteobacteria (Fig. 5B). Animals that remained clinically well and had significantly less severe histologic colitis at the time of necropsy possessed a gut micro-biota that appeared to be returning towards the baseline state. Members of the Firmicutes again became significant members of the community and Proteobacteria were no longer dominant. 16S rRNA gene sequences corresponding to C. difficile were detected in the gut communities of both clinically well and sick animals but in agreement with the quantitative PCR results, C. difficile sequences composed 8.5% (±7.7) of those recovered in clinically ill mice, but only 1.1% (±1.5) of the sequences in well mice. Thus, mice that were clinically well harbored an indigenous microbial community that was more similar to the baseline state seen in untreated controls than those with severe clinical CDI.
Cefoperazone treatment renders mice susceptible to colonization and colitis following C. difficile challenge.
We previously demonstrated that administration of the beta-lactam antibiotic cefoperazone had significant and long-lasting effects on the indigenous gut microbiota.26 Even after a six-week recovery period following a 10-day course of cefoperazone, the gut microbiota exhibited altered community structure and diminished diversity. The 5 antibiotic cocktail did not have such long-lasting effects as gut microbial composition returned to baseline within four weeks of discontinuing the drug (data not shown).
To determine if the greater disturbance of the gut microbiota associated with cefoperazone administration differentially altered the course of experimental C. difficile infection, six to eight week old C57BL/6 mice were treated with cefoperazone prior to oral challenge with C. difficile. One group of mice received cefoperazone in drinking water for 10 days followed by a 2-day period on plain water (Fig. 1C). Another group received the same cefoperazone treatment followed by a single dose of clindamycin after 2 days (Fig. 1D). Two final groups received 10 days of cefoperazone followed by a 6-week period without the drug with or without a single dose of clindamycin (Fig. 1E and F). All four groups of animals then received a challenge of 1 × 105 CFU of C. difficile via oral gavage. All animals in the 2 groups of mice that were challenged 2–3 days after cefoperazone treatment (with or without clindamycin) were moribund by 2 days post challenge while one animal died between 1–2 days post challenge. Similarly, all animals treated with cefoperazone followed by a 6-week recovery period that received a single dose of clindamycin and C. difficile challenge also exhibited signs of CDI and were moribund by 4 days post challenge. However, animals that were challenged with C. difficile after the 6-week recovery period without a dose of clindamycin were not colonized (data not shown).
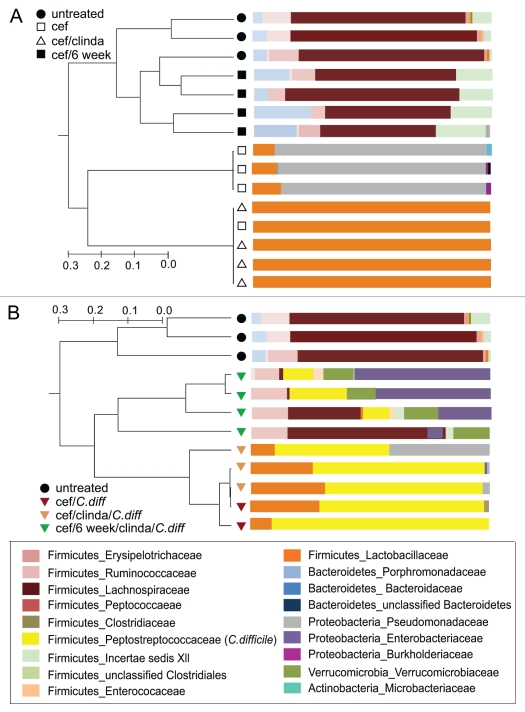
16S rRNA-encoding gene analysis on tissue collected from cefoperazone-treated animals indicated that a 10-day treatment with this antibiotic resulted in a bacterial gut community dominated by bacteria in the family Pseudomonadaceae (Fig. 6A). Following a 2-day period without cefoperazone and a single dose of clindamycin, the gut microbiota became dominated by members of the Lactobacillaceae (Fig. 6A). After infection with C. difficile the communities of all animals pretreated with cefoperazone with or without clindamycin became dominated by C. difficile (Fig. 6B) with between 48–92% (69 ± 15%) of the 16S rRNA-encoding gene sequences retrieved corresponding to this organism. C. difficile 16S rRNA-encoding gene sequences were detected in 3 of the 4 animals infected 6 weeks after cefoperazone treatment followed by a dose of clindamycin and C. difficile challenge (12.8 ± 10%).
Figure 6.
Shifts in microbial community structure and composition associated with cefoperazone administration and C. difficile infection. The composition of the gut microbiota was determined by 16s rRNA clone library construction. (A) The microbial communities in animals that were treated with cefoperazone with or without clindamycin were altered from that seen in untreated controls. Six weeks after cefoperazone treatment, the communities returned to a community structure that resembled untreated controls. (B) Animals that were challenged with C. difficile within three days after stopping cefoperazone treatment (with or without clindamycin treatment) had very high relative levels of C. difficile in their microbial communities. Animals that were challenged with C. difficile 6 weeks after antibiotic treatment was stopped followed by a single dose of clindamycin had less C. difficile present. The relative abundance of C. difficile appeared to directly correlate with the amount of Proteobacteria and inversely with the abundance of Lachnospiraceae. Dendrograms were constructed using Morisita-Horn similarities based on >97% sequence similarity while taxonomic assignments were made using the Ribosomal Database Project Classifier. cef-cefoperazone, clinda-clindamycin, C.diff-C. difficile.
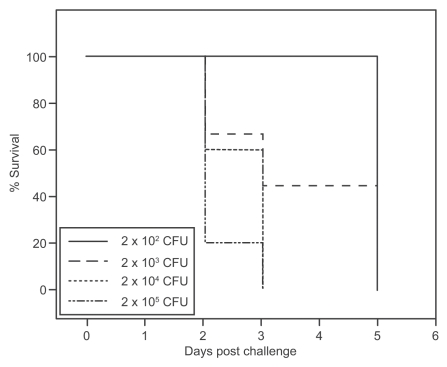
Given the apparent increased susceptibility to C. difficile infection after cefoperazone treatment, we determined if decreasing the challenge dose would alter the severity of the resultant CDI. Groups of 9 to 10 animals were treated with a 10-day course of cefoperazone followed by a 2-day recovery period without the drug, and were then challenged with varying doses of C. difficile ranging from 2 × 102 CFU to 2 × 105 CFU. The animals were monitored for weight loss and clinical signs of severe disease, and euthanized when the appropriate clinical endpoints were reached. There was a strict dose dependence on the rate at which clinical endpoints were reached (Fig. 7). The majority of animals receiving the highest dose of C. difficile became moribund within 2 days post challenge while all of the animals receiving the lowest dose of 102 CFU remained clinically well until 5 days post challenge at which time all became ill and required euthanasia.
Figure 7.
Dose dependence of disease in cefoperazone-treated mice infected with C. difficile. Kaplan-Meier survival plot for mice infected with different doses of C. difficile VPI 10463 (2 × 102, 2 × 103, 2 × 104, 2 × 105 CFU) after 10 days of cefoperazone pretreatment and 2-day recovery. (n = 9 for 102, 103 groups and n = 10 for 104, 105 groups). Survival curves are significantly different (p = 0.0123 by the Log-rank (Mantel-Cox) Test) and there is a significant trend (p = 0.001).
Discussion
The term “colonization resistance” was coined to refer to the ability of an established gut microbial community to resist invasion by additional microbes.27–29 Although this initially applied to pathogenic microbes, the concept was derived from concepts of community robustness derived from studies of classical ecologic systems (for example grasslands and lakes) and thus could be applied to any invading microbe.30 Current hypotheses suggest that the normal indigenous microbiota is not permissive for the establishment of colonization by C. difficile.15 In rare cases where normal individuals are colonized by C. difficile without overt clinical disease, it is further hypothesized that the normal indigenous microbiota can at least limit the production of toxin, perhaps by directly interfering with toxin production or limiting the population size of C. difficile and preventing significant amounts of toxin from accumulating in the gut.15,31 Accordingly, disruption of the indigenous microbiota by antibiotics leads to a loss of colonization resistance, making the gut vulnerable to colonization by exogenous C. difficile spores or, in previously colonized patients, expansion and toxin production. In support of this concept, Wilson and colleagues provided evidence for the ability of the normal gut microbiota to inhibit C. difficile by demonstrating that administration of normal cecal homogenates would decrease the number of viable C. difficile and prevent colitis in antibiotic-challenged hamsters.15,32
Using a murine model of C. difficile infection involving pretreatment of mice with antibiotics to overcome colonization resistance, we found that administration of 105 CFU of C. difficile to animals treated with a cocktail of five antibiotics and clindamycin results in uniform colonization and a mortality rate of about 60%. The initial description of this model noted that by increasing the challenge dose of C. difficile, mortality would increase in direct relationship to the dose of organism.23 In the current study, we also found that a more significant disruption of the microbiota, using the antibiotic cefoperazone could also result in uniform mortality in animals that were challenged with a dose of C. difficile that was lethal to only about half the animals that were treated with the five antibiotic cocktail and clindamycin.
Although the combination of the antibiotic cocktail and clindamycin was able to overcome colonization resistance, it is important that neither alone had the same effect. Therefore, loss of colonization resistance is not simply associated with creating an overall “depauperate” community but is dependent on the specific changes to the community structure as well. Administration of the antibiotic cocktail alone significantly decreased the overall biomass of the community, but this decrease in bacterial community size alone did not lead to a loss of colonization resistance. Furthermore, after administration of the antibiotic cocktail and clindamycin, at the time of successful challenge with C. difficile, the overall bacterial population had recovered, but the community structure was markedly altered from baseline. This further supports the idea that the specific changes brought on by antibiotic administration determine susceptibility to C. difficile colonization. This is consistent with the clinical observation that the risk of subsequent CDI differs with different antibiotics33 and in vitro and animal studies that also differentiate antibiotics on the basis of their ability to overcome colonization resistance against C. difficile.31,34,35
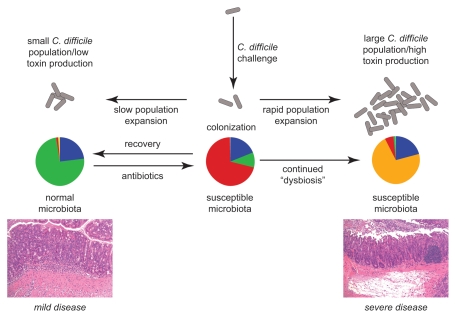
Taken together these results suggest that this murine infection model accurately represents competition between two opposing processes that are thought to be at the center of the pathogenesis of CDI (Fig. 8). On one hand there is the expansion of the population of C. difficile once it has colonized an altered/susceptible microbial community and the subsequent production of toxin. On the other hand, there is the tendency of stable microbial communities to return towards their baseline state following a perturbation, in this case, antibiotic administration.26,30,36,37 According to this model, the observation of 50% mortality in animals treated with the antibiotic cocktail and clindamycin and then challenged with 105 CFU of C. difficile reflects a point at which the two processes are in close balance. In this case, if the pathogen can grow and produce toxin more rapidly than the recovery of the indigenous microbiota clinically severe disease would result. Alternately, if the microbiota recovers prior to sufficient expansion of C. difficile there could be control of the infection. This balance can be shifted in favor of C. difficile colonization and severe disease either by administering a larger challenge dose of C. difficile or causing a greater perturbation in the microbial community structure by administering cefoperazone. Alternately, administration of a smaller inoculum of C. difficile results in less disease. In terms of the antibiotic cocktail with clindamycin, decreasing the inoculum prevented the development of clinical disease, but in the setting of cefoperazone, this merely delayed the onset of disease. This further supports the idea that cefoperazone administration results in a greater disturbance of the indigenous gut microbiota. However, if the gut microbial community is allowed to recover sufficiently from cefoperazone treatment (6 weeks), colonization resistance is restored and C. difficile did not colonize without further perturbation of the indigenous microbiota.
Figure 8.
Model of the interaction between dynamics of the gut microbiota and C. difficile in antibiotic-treated mice. Antibiotic administration alters the community structure of the indigenous gut microbiota to a state that is susceptible to colonization with C. difficile. Subsequent clinical outcome is determined by the balance between the recovery of the gut microbiota following withdrawal of antibiotics and the expansion of the population of C. difficile and toxin production.
The mechanisms by which the indigenous microbiota can resist colonization and limit disease are not clear. Although direct competition between organisms within the gastrointestinal tract is possible,38 it has been recently demonstrated that changes in the gut microbial community can indirectly affect colonization resistance via differential host responses. For example, decreasing the overall bacterial community through the administration of antibiotics can result in decreased host production of the antimicrobial peptide RegIIIγ.39 RegIIIγ binds the surface-exposed peptidoglycan layer of Gram-positive organisms with high affinity in a calcium-independent manner.40 Expression of RegIIIγ appears to be driven by the indigenous gut microbiota through host sensing of microbial-associated molecular patterns, primarily from Gram-negative organisms. RegIIIγ has been shown to be important in mediating colonization resistance to vancomycin resistant Enterococcus.41 However, in our animals we did not find a direct relationship between changes in RegIIIγ expression following antibiotic treatment and colonization resistance to C. difficile (data not shown).
The development of specific host immune responses against C. difficile appears to have an important role in determining the severity of CDI, including the development of recurrent disease.42 This observation underscores the exploration of C. difficile vaccines as a novel treatment/prevention modality.43–46 Our current model is characterized by the acute development of disease and employs naïve mice, and thus C. difficile-specific adaptive responses are not thought to play a role. However, since this model can be manipulated such that the disease is not uniformly fatal (unlike the hamster model of disease), it remains to be determined if this model will be useful for studying adaptive immunity in CDI. There are recent reports that this is a useful model for studying the role of innate immune responses in CDI.47,24 It is important to note that the role of immune responses in CDI is likely not independent of that of the indigenous microbes. It is clear that the gut microbiota has a key role in modulating the development of mucosal immune responses.6,7 Therefore, changes in the gut microbiota, can influence the response to pathogens by altering the nature of host immunity.48,49
Another mechanism by which altered gut microbial communities could affect a pathogen is by changing the overall chemical environment of the gut. Changes in the community structure of the gut microbiota can dramatically alter the concentrations of microbial metabolites.3,50 When comparing the gut microbial communities found in our animals that were clinically well versus those that were succumbing to CDI, the most obvious differences were the dominance of Proteobacteria in the ill animals and the return of Firmicutes, specifically members of the family Lachnospiraceae in well animals. These latter organisms are notable in that many are able to ferment complex carbohydrates to short-chain fatty acids (SCFA), which have an important role in maintaining intestinal homeostasis.51–53 With regards to C. difficile, SCFA are able to inhibit the growth of the organism and decrease toxin production in vitro.54
In summary, our results demonstrate that the community structure (not the absolute level) of the indigenous gut micro-biota plays a crucial role in shaping the outcome of C. difficile infection. The use of tractable murine models of disease should provide insight into the role that the indigenous gut microbiota plays in defense against pathogenic microbes. It remains to be seen which of the possible interactions between the host, indigenous microbiota and pathogen are important in determining the clinical outcome of infection. However, further study could lead to novel methods for the treatment and prevention of this increasing clinically important infection.
Experimental Procedures
Ethics statement.
All animal protocols used during the conduct of these experiments were reviewed and approved by the University Committee on Use and Care of Animals of the University of Michigan, Ann Arbor (protocol number 10212). The protocol was reviewed following guidelines for the care and use of laboratory animals set by the Office of Laboratory Animal Welfare, United States Department of Health and Human Services.
Animals and housing.
The infection studies were performed with wild type C57BL/6 mice from a breeding colony established using animals purchased from Jackson Laboratories. Mice were housed with autoclaved food, bedding and water. Cage changes were performed in a laminar flow hood. Animals experienced a cycle of 12 h of light and 12 h of darkness.
C. difficile growth conditions.
The reference strain of C. difficile, strain VP1 10463 (ATCC 43255) was obtained and cultured on brain heart infusion agar containing 5% cysteine. An anaerobic environment was maintained at all times using an anaerobic chamber (Coy Industries). An incubation temperature of 37°C was used for growth. C. difficile suspensions for animal challenge were prepared by inoculating a single colony of C. difficile from a culture plate into brain heart infusion (BHI) broth, containing 5% cysteine, and allowing for overnight growth. Cells were harvested by centrifugation (5,000 rpm for 15 min) and washed three times with pre-reduced PBS, pH 7.4. Bacterial enumeration was done to ensure that the correct dose of C. difficile vegetative cells was reached at the time of challenge.
Antibiotic administration and infection with C. difficile.
Mice were divided into treatment groups consisting of 5 to 8 animals that were six to eight weeks old. An antibiotic mixture of kanamycin (0.4 mg/mL), gentamicin (0.035 mg/mL), colistin (850 U/mL), metronidazole (0.215 mg/mL) and vancomycin (0.045 mg/mL) was prepared in sterile drinking water.23 Antibiotics were purchased from Sigma-Aldrich (cat# K1377, G1914, C4461, M1547, V2002, C5269 and C4292). The antibiotic cocktail was administered for 3 days then the animals were switched to regular autoclaved drinking water for 2 days. All mice in each experiment were housed under the same conditions and were fed standard autoclaved chow. A single dose of clindamycin (10 mg/kg) was administered intraperitoneally one day before C. difficile challenge. Cefoperazone (0.5 mg/ml) was prepared in sterile drinking water and administered for 10 days. The cefoperazone drinking water was replaced with a fresh supply every 48 hours for the duration of cefoperazone administration. Animals were then switched to regular autoclaved drinking water for 2 days. A single dose of clindamycin (10 mg/kg) was administered intraperitoneally one day before C. difficile challenge. Some animals were allowed to recover for 6 weeks after cefoperazone treatment then a single dose of clindamycin was administered prior to C. difficile infection. Animals were infected by oral gavage with 1 × 105 CFU of C. difficile strain VPI 10463. Animals were monitored daily for signs of disease such as diarrhea, hunched posture and weight loss.
Necropsy and histological procedures.
Mice were euthanized by CO2 asphyxiation. The tip of the cecum of each mouse was removed, halved and rinsed in phosphate-buffered saline to remove luminal contents. Approximately 5 mm of proximal colon and terminal ileal tissue and luminal contents were collected from each animal. All samples were snap frozen in liquid nitrogen and stored at −80°C. The remaining cecum, colon and ileal tissue were placed intact into histology tissue cassettes and stored in 10% buffered formalin for 24 hours then transferred to 70% ethyl alcohol.56 Tissue cassettes were then processed, paraffin embedded and then sectioned. Haematoxylin and eosin stained slides were prepared for histologic examination (McClinchey Histology Lab Inc.).
Histopathologic examination.
Histological slides were coded, randomized and scored in a blinded manner by one of the authors (ILB) who is a board-certified veterinary pathologist. A scoring system was adapted from a previously published method.23,57 Edema, cellular infiltration and epithelial damage in each tissue (colon, cecum, ileum) were scored from 0–4 according to the following defined criteria: Edema scores-(0): no edema; (1) mild edema with minimal (<2x) multifocal submucosal expansion; (2) moderate edema with moderate (2–3x) multifocal sub-mucosal expansion; (3) severe edema with severe (>3x) multifocal sub-mucosal expansion; and (4) same as score 3 with diffuse sub-mucosal expansion. Cellular infiltration scores were graded as follows: (0) no inflammation; (1) minimal multifocal neutrophilic inflammation; (2) moderate multifocal neutrophilic inflammation (greater submucosal involvement); (3) severe multifocal to coalescing neutrophilic inflammation (greater submucosal ± mural involvment); and (4) same as score 3 with abscesses or extensive mural involvement. Epithelial damage was scored as follows: (0) no epithelial changes; (1) minimal multifocal superficial epithelial damage (vacuolation, apoptotic figures, villus tip attenuation/necrosis); (2) moderate multifocal superficial epithelial damage (vacuolation, apoptotic figures, villus tip attenuation/necrosis); (3) severe multifocal epithelial damage (same as above) ± pseudomembrane (intraluminal neutrophils, sloughed epithelium in a fibrinous matrix); and (4) same as score 3 with significant pseudomembrane or epithelial ulceration (focal complete loss of epithelium).
DNA extraction.
Total DNA from fecal and tissue samples was extracted using the MagNA Pure DNA isolation protocol (Roche, cat# 03730964001). Samples were placed in an Ultra Clean fecal bead tube (MoBio) to which 500 µl of MagNA Pure bacterial lysis buffer (Roche) was added. Samples were bead beaten for 1 min with a mini bead beater (Biospec), digested with proteinase K, incubated at 65°C, bead beaten for 1 min and then was heat inactivated at 95°C. Samples were placed in the MagNA Pure (Roche) and the MagNA Pure nucleic isolation kit protocol for bacterial DNA was followed as recommended by the manufacturer.
16s ribosomal rRNA-encoding gene clone libraries.
The community structure of infected and uninfected mice was analyzed by the construction of 16S rRNA clone libraries.58,59 PCR targeting bacterial 16S rRNA genes using primers 8F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′) 60 was performed on each extracted DNA sample. PCR was performed using Illustra Pure Taq Ready-To-Go PCR beads (GE Healthcare, cat# 27955901). Reaction mixtures were set up with 100 ng of template DNA, 20 pmol of each primer, and water to a total volume of 25 µl. The reaction mixtures were subjected to amplification in a DNA thermal cycler (Eppendorf Mastercycler gradient) with the following cycling conditions: initial denaturation at 94°C for 2 min followed by 20 cycles of denaturation at 94°C for 30 sec, annealing at 58°C for 45 sec and extension at 72°C for 1.5 min. A final extension at 72°C for 10 min was performed. Control amplifications with sterile water were included in each amplification reaction and never gave visible amplicons. Amplicons were purified using a kit Illustra MicroSpin Column (GE Healthcare, cat# 27514001) according to the recommendations of the manufacturer. The purified PCR products were ligated into a plasmid vector (pCR 2.1; Invitrogen).
DNA sequencing and analysis.
Plasmid purification and DNA sequence determination of 96 randomly selected clones from each library were performed by the DNA Sequencing Core facilities at the University of Michigan. Each clone was sequenced with a single primer (8F) that typically yielded 750 bases of readable sequence. Sequences with numerous ambiguous base calls or with fewer than 350 total bases were excluded from further analysis.
Sequences were analyzed for the formation of chimeras using the Chimera Check program from the Ribosomal Database Project.60 Potential chimeric sequences were excluded from additional analysis. Sequences were also aligned to a phylogenetically diverse collection of 16S rRNA gene sequences using the RDP Classifier.60
Partial 16S rRNA sequences were initially analyzed using mothur61 to calculate pair wise Morisita-Horn distances which was exported to Mega4 62 software package and then UPGMA analysis was used to create dendrograms.
Quantitative PCR.
Quantitative PCRs were used to assay the quantity of rRNA operons in the DNA samples relative to a single-copy host gene (mouse tumor necrosis factor alpha [TNFα]) as detailed in Antonopoulos et al.27 Assays used the LightCycler 480 Probes Master reaction mixture (Roche, cat# 04707494001) at 1x concentration and appropriate primer-probe sets to increase the specificity of the signals detected from the sample DNA (100 ng). For detection of the bacterial signal, 100 nmol of each of the forward and reverse primers and the flourescent probe were included in the reaction mixtures. Sequences for the forward primer (5′-TCC TAC GGG AGG CAG CAG T-3′), the reverse primer (5′-GGA CTA CCA GGG TAT CTA ATC CTG TT-3′) and the probe (5′-[6-carboxyfluorescein]-CGT ATT ACC GCG GCT GCT GGC AC-[6-carboxytetramethylrhoda mine]-3′) were based on the work of Nadkarni et al. Signals were detected with a LightCycler 480 instrument (Roche). Detection of the host signal used 200 nmol of the forward (TNFα_mu_se; 5′-GGC TTT CCG AAT TCA CTG GAG-3′) and reverse (TNFα_mu_as; 5′-CCC CGG CCT TCC AAA TAA A-3′) primers and 100 nmol of the probe (TNFα_mu_probe; 5′-Cy5-ATG TCC ATT CCT GAG TTC TGC AAA GGG A-Iowa Black RQ-3′) adapted from Nitsche et al. Relative bacterial loads were compared via the CT method by normalizing the 16S rRNA gene signal to the host signal.65
Monitoring C. difficile colonization.
The colonization status of C. difficile infected animals was monitored using a C. difficile toxin multiplex qPCR assay of fecal pellets collected at various time points pre and post challenge from mice in each group. For the C. difficile Toxin Multiplex qPCR (LightCycler 480) 8 and 10 pmol/µl for tcdA and tcdB primers, respectively, were prepared from 200 pmol/µl original stocks. Primer and probe sets are as follows: tcdA_F: 5′-GGT AAT AAT TCA AAA GCG GCT, tcd_R: 5′-AGC ATC CGT ATT AGC AGG TG, tcdA_probe_FAM: 5′-6FAM-AGC CTA ATA CAG CTA TGG GTG CGA A-BHQ1, tcdB_F: 5′-GAA AGT CCA AGT TTA CGC TCA AT, tcdB_R: 5′-GCT GCA CCT AAA CTT ACA CCA, tcdB_probe_Hex: 5′-Hex-ACA GAT GCA GCC AAA GTT GTT GAA TT-BHQ1 (James Versalovic, personal communication, manuscript in preparation). For each 20 µl reaction, 4 µl template, 10 pmol tcdA primers, 12.5 pmol tcdB primer, 1.6 pmol tcdA probe and 2 pmol tcdB probe were used. The following cycling conditions were used for the qPCR run: Activation-95°C for 15 min, 95°C for 15 sec, Cycling (X45)-60°C for 20 sec, 72°C for 10 sec, Hold-37°C for 30 sec. Values were normalized to mouse TNFalpha gene content and the mean fold change of tcdA tcdB gene content were calculated using the 2−ΔCt method.66
C. difficile cytotoxin assay.
The assay was performed in 96-well flat-bottom microtiter plates (Corning) and was adapted from Corthier et al. Green African monkey kidney epithelial cells (Vero) (provided by M. Imperiale, University of Michigan) were grown to confluency in DMEM (GIBCO Laboratories, cat# 11965) containing 10% heat inactivated fetal bovine serum (GIBCO Laboratories, cat# 16140) and 1% penicillin streptomycin solution (GIBCO Laboratories, cat# 15140). The cells were trypsinized using 0.25% trypsin (GIBCO Laboratories, cat# 25200) and washed with 1 volume of DMEM medium. Cells were diluted in DMEM medium and approximately 1 × 105 cells were distributed per well and incubated at 37°C with 5% CO2 for 18–24 hours. Samples of luminal contents or intestinal tissue were weighed and 500 µl of 1x PBS was added. Intestinal tissue was homogenized using a Medimachine (Becton Dickenson). Samples were vortexed then spun at 13,000 rpm for 5 minutes and then the supernatant was filtered through a 0.2 µm membrane. Each sample was titrated in two-fold dilutions within the wells to a maximum dilution of 2−12 and each well had a corresponding control to which both antitoxin (TechLabs, cat# T5000) and sample were added. After an overnight incubation at 37°C, plates were fixed with 10% buffered formalin for 2 hours then stained with geimsa (50 µl per well) for 15 minutes followed by a wash with 1x PBS. Wells with approximately 100% round cells were easily recognized under 200x magnification. The cytotoxic titer was defined as the reciprocal of the highest dilution that rounds 100% of Vero cells per gram of sample. Vero cells with purified C. difficile toxin and antitoxin (TechLabs, cat# T5000) were used as controls.
Statistical analysis.
Statistical analyses were performed using Prism 5 for Mac OS X GraphPad Software. t tests were used for treatment group comparisons, except for categorical histology scores, where the nonparametric Krustal Wallis test was used. Statistical significance was set at a p value of < 0.05.
Acknowledgments
We thank Judy Opp for expert technical assistance. This work was funded by NIH grants DK070875 (V.B.Y.), AI090871 (V.B.Y./G.B.H.), AI083473 (G.B.H.) and HG005975 (P.D.S.). C.M.T. was supported by training grant AI07528.
Abbreviations
- CDI
C. difficile infection
- GI
gastrointestinal
- CFU
colony forming units
- PCR
polymerase chain reaction
- SCFA
short chain fatty acid
References
- 1.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yap IK, Li JV, Saric J, Martin FP, Davies H, Wang Y, et al. Metabolomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- 4.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Waaij D, Berghuis JM, Lekkerkerk-van der Wees JEC. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg. 1971;69:405–411. doi: 10.1017/s0022172400021653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekirov I, Finlay BB. The role of the intestinal microbiota in enteric infection. J Physiol. 2009;587:4159–4167. doi: 10.1113/jphysiol.2009.172742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall I, O'Toole E. Intestinal flora in newborn infants with the description of a new anaerobic pathogen, Bacillus difficilus. Am J Dis Child. 1935;49:390–402. [Google Scholar]
- 11.McDonald LC, Killgore GE, Thompson A, Owens RC, Jr, Kazakova SV, Sambol SP, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 12.Pepin J, Valiquette L, Alary ME, Villemure P, Pelletier A, Forget K, et al. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ. 2004;171:466–472. doi: 10.1503/cmaj.1041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly CP, Lamont JT. Clostridium difficile—more difficult than ever. N Engl J Med. 2008;359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 14.Paterson DL. “Collateral damage” from cephalosporin or quinolone antibiotic therapy. Clin Infect Dis. 2004;38:341–345. doi: 10.1086/382690. [DOI] [PubMed] [Google Scholar]
- 15.Wilson KH. The microecology of Clostridium difficile. Clin Infect Dis. 1993;16:214–218. doi: 10.1093/clinids/16.supplement_4.s214. [DOI] [PubMed] [Google Scholar]
- 16.Maroo S, Lamont JT. Recurrent Clostridium difficile. Gastroenterology. 2006;130:1311–1316. doi: 10.1053/j.gastro.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 17.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 18.Bartlett JG, Onderdonk AB, Cisneros RL, Kasper DL. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J Infectious Disease. 1977;136:701–705. doi: 10.1093/infdis/136.5.701. [DOI] [PubMed] [Google Scholar]
- 19.Onderdonk A, Cisneros RL, Bartlett JG. Clostridium difficile in gnotobiotic mice. Infect Immun. 1980;28:227–282. doi: 10.1128/iai.28.1.277-282.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson KH, Freter R. Interaction of Clostridium difficile and Escherichia coli with microfloras in continuousflow cultures and gnotobiotic mice. Infect Immun. 1986;54:354–358. doi: 10.1128/iai.54.2.354-358.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pawlowski SW, Calabrese G, Kolling GL, Freire R, AlcantaraWarren C, Liu B, et al. Murine model of Clostridium difficile infection with aged gnotobiotic C57BL/6 mice and a BI/NAP1 strain. J Infect Dis. 2010;202:1708–1712. doi: 10.1086/657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission and severe disease in immunocompromised hosts. Infect Immun. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008;135:1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Jarchum I, Liu M, Lipuma L, Pamer EG. Toll-Like receptor 5 stimulation protects mice from acute Clostridium difficile colitis. Infect Immun. 2011;79:1498–1503. doi: 10.1128/IAI.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voth D, Ballard J. Clostridium difficile toxins, mechanisms of action and role in disease. Clin Microbiol Rev. 2005;18:247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Waaij D, Berghuis-de Vries JM, Lekkerkerk Lv. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg (Lond) 1971;69:405–411. doi: 10.1017/s0022172400021653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freter R. In vivo and in vitro antagonism of intestinal bacteria against Shigella flexneri. II. The inhibitory mechanism. J Infect Dis. 1962;110:38–46. doi: 10.1093/infdis/110.1.38. [DOI] [PubMed] [Google Scholar]
- 29.Hentges DJ, Freter R. In vivo and in vitro antagonism of intestinal bacteria against Shigella flexneri. I. Correlation between various tests. J Infect Dis. 1962;110:30–37. doi: 10.1093/infdis/110.1.30. [DOI] [PubMed] [Google Scholar]
- 30.Robinson CJ, Bohannan BJ, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453–476. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pultz NJ, Donskey CJ. Effect of antibiotic treatment on growth of and toxin production by Clostridium difficile in the cecal contents of mice. Antimicrob Agents Chemother. 2005;49:3529–3532. doi: 10.1128/AAC.49.8.3529-3532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson KH, Silva J, Fekety FR. Suppression of Clostridium difficile by normal hamster cecal flora and prevention of antibiotic-associated cecitis. Infect Immun. 1981;34:626–628. doi: 10.1128/iai.34.2.626-628.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blondeau JM. What have we learned about antimicrobial use and the risks for Clostridium difficile-associated diarrhoea? J Antimicrob Chemother. 2009;63:238–242. doi: 10.1093/jac/dkn477. [DOI] [PubMed] [Google Scholar]
- 34.Jump RL, Li Y, Pultz MJ, Kypriotakis G, Donskey CJ. Tigecycline exhibits inhibitory activity against Clostridium difficile in the colon of mice and does not promote growth or toxin production. Antimicrob Agents Chemother. 2011;55:546–549. doi: 10.1128/AAC.00839-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nerandzic MM, Donskey CJ. Effect of ceftobiprole treatment on growth of and toxin production by Clostridium difficile in cecal contents of mice. Antimicrob Agents Chemother. 2011;55:2174–2177. doi: 10.1128/AAC.01612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stecher B, Chaffron S, Kappeli R, Hapfelmeier S, Freedrich S, Weber TC, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehotzky RE, Partch CL, Mukherjee S, Cash HL, Goldman WE, Gardner KH, et al. Molecular basis for peptidoglycan recognition by a bactericidal lectin. Proc Natl Acad Sci USA. 2010;107:7722–7727. doi: 10.1073/pnas.0909449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kyne L, Warny M, Qamar A, Kelly CP. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhoea. Lancet. 2001;357:189–193. doi: 10.1016/S0140-6736(00)03592-3. [DOI] [PubMed] [Google Scholar]
- 43.Permpoonpattana P, Hong HA, Phetcharaburanin J, Huang JM, Cook J, Fairweather NF, et al. Immunisation with Bacillus spores expressing toxin A peptide repeats protects against infection with toxin A+ B+ strains of Clostridium difficile. Infect Immun. 2011 doi: 10.1128/IAI.00130-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leav BA, Blair B, Leney M, Knauber M, Reilly C, Lowy I, et al. Serum anti-toxin B antibody correlates with protection from recurrent Clostridium difficile infection (CDI) Vaccine. 2010;28:965–969. doi: 10.1016/j.vaccine.2009.10.144. [DOI] [PubMed] [Google Scholar]
- 45.Pechine S, Janoir C, Boureau H, Gleizes A, Tsapis N, Hoys S, et al. Diminished intestinal colonization by Clostridium difficile and immune response in mice after mucosal immunization with surface proteins of Clostridium difficile. Vaccine. 2007;25:3946–3954. doi: 10.1016/j.vaccine.2007.02.055. [DOI] [PubMed] [Google Scholar]
- 46.Ghose C, Kalsy A, Sheikh A, Rollenhagen J, John M, Young J, et al. Transcutaneous immunization with Clostridium difficile toxoid A induces systemic and mucosal immune responses and toxin A-neutralizing antibodies in mice. Infect Immun. 2007;75:2826–2832. doi: 10.1128/IAI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasegawa M, Yamazaki T, Kamada N, Tawaratsumida K, Kim YG, Nunez G, et al. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J Immunol. 2011;186:4872–4880. doi: 10.4049/jimmunol.1003761. [DOI] [PubMed] [Google Scholar]
- 48.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghosh S, Dai C, Brown K, Rajendiran E, Makarenko S, Baker J, et al. Colonic microbiota alters host susceptibility to infectious colitis by modulating inflammation, redox status and ion transporter gene expression. Am J Physiol Gastrointest Liver Physiol. 2011;301:G39–G49. doi: 10.1152/ajpgi.00509.2010. [DOI] [PubMed] [Google Scholar]
- 50.Romick-Rosendale LE, Goodpaster AM, Hanwright PJ, Patel NB, Wheeler ET, Chona DL, et al. NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril) Magn Reson Chem. 2009;47:36–46. doi: 10.1002/mrc.2511. [DOI] [PubMed] [Google Scholar]
- 51.Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 53.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 54.May T, Mackie RI, Fahey GC, Jr, Cremin JC, Garleb KA. Effect of fiber source on short-chain fatty acid production and on the growth and toxin production by Clostridium difficile. Scand J Gastroenterol. 1994;29:916–922. doi: 10.3109/00365529409094863. [DOI] [PubMed] [Google Scholar]
- 55.Young VB, Knox KA, Pratt JS, Cortez JS, Mansfield LS, Rogers AB, et al. In vitro and in vivo characterization of Helicobacter hepaticus cytolethal distending toxin mutants. Infect Immun. 2004;72:2521–2527. doi: 10.1128/IAI.72.5.2521-2527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelly CP, Becker S, Linevsky JK, Joshi MA, O'Keane JC, Dickey BF, et al. Neutrophil recruitment in Clostridium difficile toxin A enteritis in the rabbit. J Clin Invest. 1994;93:1257–1265. doi: 10.1172/JCI117080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pace N, Stahl D, Lane D, Olsen G. Analyzing natural microbial populations by rRNA sequences. ASM News. 1985;51:4–12. [Google Scholar]
- 58.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt TM, Relman DA. Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences. Methods Enzymol. 1994;235:205–222. doi: 10.1016/0076-6879(94)35142-2. [DOI] [PubMed] [Google Scholar]
- 60.Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, et al. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 64.Nitsche A, Becker M, Junghahn I, Aumann J, Landt O, Fichtner I, et al. Quantification of human cells in NOD/SCID mice by duplex real-time polymerasechain reaction. Haematologica. 2001;86:693–699. [PubMed] [Google Scholar]
- 65.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 66.Corthier G, Dubos F, Raibaud P. Modulation of cytotoxin production by Clostridium difficile in the intestinal tracts of gnotobiotic mice inoculated with various human intestinal bacteria. Appl Environ Microbiol. 1985;49:250–252. doi: 10.1128/aem.49.1.250-252.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]