Abstract
Phages are among the simplest biological entities known and simultaneously the most numerous and ubiquitous members of the biosphere. Among the three families of tailed dsDNA phages, the Myoviridae have the most structurally sophisticated tails which are capable of contraction, unlike the simpler tails of the Podoviridae and Siphoviridae. Such “nanomachines” tails are involved in both efficient phage adsorption and genome injection. Their structural complexity probably necessitates multistep morphogenetic pathways, involving non-structural components, to correctly assemble the structural constituents. For reasons probably related, at least in part, to such morphological intricacy, myoviruses tend to have larger genomes than simpler phages. As a consequence, there are no well-characterized myoviruses with a size of less than 40 kb. Here we report on the characterization and sequencing of the 23,931 bp genome of the dwarf myovirus ϕ1402 of Bdellovibrio bacteriovorus. Our genomic analysis shows that ϕ1402 differs substantially from all other known phages and appears to be the smallest known autonomous myovirus.
Key words: Bdellovibrio phage, dwarf myovirus, complete genome, terminase, capsomers
Introduction
Bdellovibrio bacteriovorus is a diminutive, motile, curved, rod-shaped, Gram-negative bacterium of the δ-Proteobacteria class. Bdellovibrios live in freshwater, seawater and soil, but are best known for their predatory activity in which they can penetrate into the periplasmic space of Gram-negative bacteria and utilize their constituents as carbon and energy sources.1 The first Bdellovibrio phage described in B. bacteriovorus was a tailless icosahedral virion with a ssDNA genome and a double capsid,2 an otherwise unheard of phage capsid structure that was probably an artifact of the uranyl acetate staining. Additional Bdellovibrio phages were isolated and described in reference 3–5 in 1972, including another ssDNA phage and two small phages of the Myoviridae family. Unfortunately, none of these viruses are currently available.
Thirty years later a new isolate, ϕMH2K, became the first Bdellovibrio phage to have its genome sequenced.6 This bacteriophage is morphologically similar to the classical coliphage ?X174 of the Microviridae family, but phylogenetically it is quite distant from it, being more closely related to the Microviridae phage Ch-1 of Chlamydia. In 2000, B. Fane of the University of Arizona at Tucson isolated a small Bdellovibrio myovirus, ϕ1402, from sewage (Fane B, personal communication). Soon afterwards this phage's morphology was examined by electron microscopy by H.W. Ackermann and its DNA was preserved in his laboratory collection at Laval University. Given the paucity of information available about the phages infecting this unusual and interesting group of predatory bacteria, we have now sequenced the genome of ϕ1402. Here we report on both phage ϕ1402's morphology and genome sequence. This phage represents the smallest and simplest autonomous myovirus currently known. Of the 42 putative ORFs encoded by its genome, only two are clear homologs of any known phage genes.
Results and Discussion
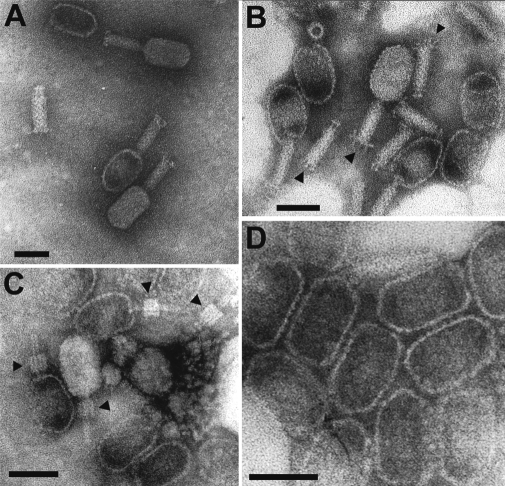
Phage ϕ1402 has a contractile tail and thus is classified as a myovirus (Fig. 1). As measured on 20 particles, it has an elongated 68 × 40 nm head and a tail that is, in its extended state, 62 × 17 nm. The tail has a neck that lacks a collar and, hence, collar-associated appendages. In the contracted state, the tail measures 20 × 20 nm and shows a base plate of 17 × 2 nm that separates from the sheath. The base plate has a set of 5–7 nm long fibers. Electron microscopy of empty heads reveals capsomers of 5 nm diameter that are unusually easy to distinguish, presumably because of the absence of any supplementary head decoration proteins. The precise number and exact pattern of organization of these capsomers in the capsid has not yet been determined.
Figure 1.
Electron micrographs of ϕ1402. (A and B) Complete phages with extended tails and some empty heads. Arrows in (B) indicate tail fibers. The ring structure in the upper left of (B) is a contracted tail sheath standing on its end. (C) Particles with contracted tails (arrows). (D) Enlargement of empty heads clearly showing capsomers. All photos: phosphotungstate; scale bars indicate 50 nm.
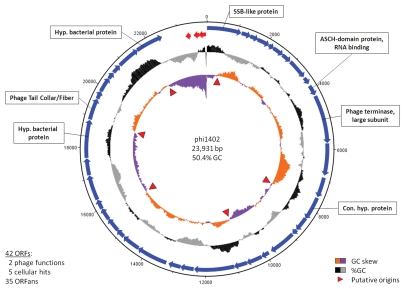
The phage genome is a 23,931 bp, circularly permuted dsDNA molecule with a GC content of 50.4% (Fig. 2). Restriction digest profiles (data not shown) identified a fragment containing a pac site which is diagnostic of phages using a terminally redundant, head-full packaging strategy.11 The DNA sequence encodes 42 ORFs, which are nearly all located on the positive strand and have no significant overlaps. The genome has no obvious phylogenetic relationship to any other phage in public databases. Remarkably, only two of its ORFs can clearly be considered as phage-related: ORF11, the large subunit of the phage terminase and ORF33 which is related to various fibrous phage proteins (Table 1). Both these sequences are among the most highly conserved and ubiquitous sequences in phage genomes. Five other ϕ1402 proteins had significant E-values (≤10−4) when compared to cellular proteins: ORF01 is related to ssDNA-binding proteins; ORF10 belongs to an RNA-binding Conserved Domain Database (CDD) protein family; and three ORFs have homologies to hypothetical cellular proteins. The remaining 35 proteins (83%) are ORFans—database entries without any known homologs. Iterative PSI-BLAST, applying less stringent selection criteria, found weak hits to three additional ORFs (ORF21/24/38). These are putatively identified as dehydrogenases/reductases, potassium/proton antiporters and peptidoglycan-binding domain proteins, respectively, but the significance of such weak homologies is dubious.
Figure 2.
Genome sequence of ϕ1402. The circles, from outermost to innermost, represent: the scale in kilobases; the rightward-transcribed ORF s (blue); the leftward-transcribed ORF s (red); the %GC content (black for above-average and gray for below-average); the GC skew (orange for positive and purple for negative). Putative origins are indicated with red triangles and the 7 ORF s with assignable homologs are indicated.
Table 1.
ϕ1402 ORF s with identifiable homologs/gene functions
| ORF | Strand | Start | End | ORF length | aa length | E-value | Homologs/functiona |
| 01 | + | 1 | 1098 | 1098 | 365 | 6e-07 | Erf family protein [Enterococcus faecalis]; SSB-like pfam04404 |
| 10 | + | 3886 | 4224 | 339 | 112 | 7e-08 | ASCH domain protein [Listeria monocytogenes]; RNA-binding cd186302 |
| 11 | + | 4253 | 5797 | 1545 | 514 | 2e-11 | Phage terminase, large subunit [Enterococcus phage phiFL2A] |
| 14 | + | 7582 | 8316 | 735 | 244 | 2e-07 | Hypothetical protein [Populus trichocarpa]; many other conserved hyp. proteins |
| 32 | + | 17666 | 18874 | 1209 | 402 | 8e-04 | BNR domain protein [Azotobacter vinelandii] |
| 33 | + | 18884 | 19576 | 693 | 230 | 2e-16 | Phage tail collar domain protein [Ralstonia phage RSL1]; many phage fibers |
| 39 | + | 21591 | 22475 | 885 | 294 | 3e-29 | Hypothetical protein [Microcystis aeruginosa] |
| Weak hits | |||||||
| 21 | + | 11093 | 12373 | 1281 | 426 | >1e-04 | Many dehydrogenases/reductases |
| 24 | + | 13284 | 14273 | 990 | 329 | >1e-04 | Potassium/proton antiporter [Bradyrhizobium] |
| 38 | + | 20863 | 21594 | 732 | 243 | >1e-04 | Peptidoglycan binding domain protein [Roseiflexus] |
Generally listed are the closest phage (if any) homologs of identifiable function, followed by identifiable cellular homologs, and then hypothetical proteins (if no function could be deduced) from blastp searches against the nr (or phage restricted) database using an E-value cutoff of <10−4. Also included are additional hits/homologs of interest and hits to the Conserved Domain Database (CDD) with their cd/pfam identifiers.
The highly compact ϕ1402 genome (95% coding) contains no tRNAs and dot-plot analysis reveals no significant direct- or inverted-repeat sequences. A comparison of the ϕ1402 genome and that of its host revealed that both had similar GC contents and codon usage patterns (data not shown). The intergenic regions of the phage genome contained no obvious consensus promoter sequences; however, the host promoter sequences of Bdellovibrio spp. have not yet been defined either. The ϕ1402 genome contains a single large (∼1 kb) untranslated region (UTR) located between the end of the forward-transcribed genes and the small cluster of genes transcribed leftward. Although this UTR has a substantially reduced GC ratio, there are no additional indications that it corresponds to an attP site of a temperate phage. ϕ1402 always behaved as a standard lytic phage, with no particularities regarding plaque morphology, stock production, nor latent period (Fane B, personal communication). However, BLASTn did find an identical 16 bp sequence (GTA ACT CCT CAA GAA T) in the phage UTR and in a Bdellovibrio bacteriovorus intergenic sequence (coordinates 3,528,980–3,528,995 on NC_005363) that is located between an asparaginase gene and a gene for a hypothetical protein, thus quite unlike the site of many temperate phage attP sites that are often in close proximity to tRNA sequences.12 The function of this large phage UTR region, therefore, remains unknown. Although an investigation of the DNA sequence with Ori-Finder was inconclusive, there are 6 putative shifts in GC skew indicating possible origins of replication, with the strongest candidate being near the 0 coordinate.
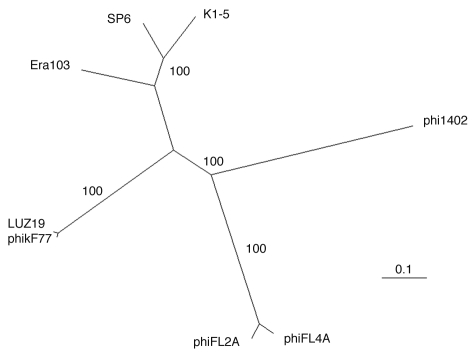
In conclusion, phage ϕ1402 differs from other myoviruses by its extremely small size, the presence of visible capsomers, and its very small dsDNA genome. Tailed-phage capsids are generally smooth; the visualization of capsomers after negative staining is exceptional in tailed phages and many indicate structural differences separating ϕ1402 from other phages. Among the myoviruses, only the satellite coliphages P4 and ϕR73 have smaller DNAs, of 11.6 and 12.7 kb, respectively. Both of these have small isometric heads (∼45 nm diameter) and tails of approximately 140 nm in length.13–15 A phylogenetic analysis of the ϕ1402 terminase protein (Fig. 3) further reinforces the notion that this unusual Bdellovibrio phage has diverged considerably from all other currently known phage groups. In the absence of any evidence that ϕ1402 is a satellite phage, it appears to represent the smallest known autonomous myovirus and thus an interesting example of reductionism carried to the extreme. It will be interesting to identify all the functions that ϕ1402 has been able to jettison on its evolutionarily reductionist pathway and, equally important, what few genes it was constrained to retain. It seems likely that even this small “simplistic” myovirus would require at least 25 genes to encode all the essential virion components and the associated genes for correct assembly of the virion. Such a conservative estimate leaves only 17 functions unaccounted for in the ϕ1402 genome, but many of these must be involved in the phage's takeover of host macromolecular synthesis and phage transcription/replication. It will be interesting to see what the few remaining unaccounted for phage functions do and how they do it with such modest genetic resources.
Figure 3.
Large subunit terminase phylogeny. Neighbor-joining tree of the ϕ1402 terminase with its 7 closest phage homologs. Values at the nodes indicate the results of 100 bootstrap replicates and the scale bar indicates 0.1 substitutions per site.
Materials and Methods
Electron microscopy.
The phage was pelleted at 25,000x g for 1 hour, using a Beckman high-speed centrifuge and a JA-18.1 fixed-angle rotor (Beckman, 347824). The phage pellet was washed twice in neutral 0.1 M ammonium acetate and re-suspended. The phages were then deposited on copper grids with carboncoated Formvar films and stained with 2% uranyl acetate (pH 4) and 2% phosphotungstate (pH 7.2). They were then examined in an electron microscope (Philips, EM300) whose magnification was calibrated using T4 phage tails as size standards.
DNA extraction and sequencing.
Phage DNA was extracted 3 times with 1:1 v/v phenol: chloroform, amended with 0.25 M (final concentration) sodium acetate, and then precipitated immediately with 100% room-temperature ethanol. After a spin at 15,000 g for 3 min, the pellet was washed with 70% ice-cold ethanol and re-spun. The pellet was dried at 37°C for 1 h and re-suspended in sterile Milli-Q water and left to rehydrate at 4°C. The resulting pure DNA was used for restriction digests, bar-coded library construction and 454 pyrosequencing that was performed according to the manufacturer's instructions on a quarter picotiter plate of a GS-FLX sequencer (Roche) at the IBIS/Université Laval Plate-forme d'Analyses Génomiques.
Bioinformatics.
Raw reads were assembled using the GS De Novo Assembler (Roche), resulting in one final contig with a 330-fold coverage. Analysis of the genome was done with the following programs: (1) GLIMMER7 (www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi; >100 nt; bacterial genetic code) and GeneMark8 (exon.gatech.edu/GeneMark; heuristic approach for prokaryotes and viruses; >90 nt) for ORF determinations; (2) tRNA search using tRNAscan-SE9 (lowelab.ucsc.edu/tRNAscan-SE); (3) Java Word Frequencies and Java Dot Plot Alignments (athena.bioc.uvic.ca) for the exploration of DNA “words”/patterns; (4) Graphical Codon Usage Analyser (gcua.schoedl.de) and the Codon Usage Database (www.kazusa.or.jp/codon) for the exploration of codon usage patterns; (5) the BLAST tools at NCBI (blast.ncbi.nlm.nih.gov) for the characterization of genes/proteins and untranslated regions (UTRs) of the DNA; (6) various phylogenetic tools of the Mobyle Project at the Institut Pasteur (mobyle.pasteur.fr/cgi-bin/portal.py); (7) DNAPlotter10 for generating the circular genome visualizations (www.sanger.ac.uk/resources/software/dnaplotter); (8) Ori-Finder (tubic.tju.edu.cn/Ori-Finder/) to explore replication origins; and (9) HHpred (toolkit.tuebingen.mpg.de/hhpred) for the homology detection and structure prediction of ORFs with weak BLAST homologs. The annotated genome sequence of ϕ1402 has been deposited in GenBank with the accession number JF344709.
Acknowledgements
The authors thank Dr. Bentley M. Fane (BIO5 Institute, University of Arizona-Tucson) for his kind gift of phage ϕ1402 and Dr. Brian Boyle at the IBIS/Université Laval Plate-forme d'Analyses Génomiques (Québec, QC) for sequencing and assembly advice. H.M.K.'s research was primarily supported by intramural funding from the INSB of the CNRS and by supplementary funding from the Kribu Foundation.
Abbreviations
- ORF
open-reading frame
- UTR
untranslated region
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Sockett RE. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu Rev Microbiol. 2009;63:523–539. doi: 10.1146/annurev.micro.091208.073346. [DOI] [PubMed] [Google Scholar]
- 2.Hashimoto T, Diedrich DL, Conti SF. Isolation of a bacteriophage for Bdellovibrio bacteriovorus. J Virol. 1970;5:97–98. doi: 10.1128/jvi.5.1.97-98.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Althauser M, Samsonoff WA, Anderson C, Conti SF. Isolation and preliminary characterization of bacteriophages for Bdellovibrio bacteriovorus. J Virol. 1972;10:516–523. doi: 10.1128/jvi.10.3.516-523.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schindler J, Ludvík J. A new bacteriophage specific for a saprophytic mutant of Bdellovibrio bacteriovorus. Acta Virol. 1972;16:501–502. [PubMed] [Google Scholar]
- 5.Varon M, Levisohn R. Three-membered parasitic system: a bacteriophage, Bdellovibrio bacteriovorus and Escherichia coli. J Virol. 1972;9:519–525. doi: 10.1128/jvi.9.3.519-525.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brentlinger KL, Hafenstein S, Novak CR, Fane BA, Borgon R, McKenna R, et al. Microviridae, a family divided: isolation, characterization and genome sequence of ϕMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J Bacteriol. 2002;184:1089–1094. doi: 10.1128/jb.184.4.1089-94.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucl Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucl Acids Res. 2005;33:451–454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casjens SR, Gilcrease EB. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. In: Clokie MRJ, Kropinski AM, editors. Bacteriophages: Methods and Protocols. New York, NY: Humana Press; 2009. pp. 91–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canchaya C, Fournous G, Brüssow H. The impact of prophages on bacterial chromosomes. Mol Microbiol. 2004;53:9–18. doi: 10.1111/j.1365-2958.2004.04113.x. [DOI] [PubMed] [Google Scholar]
- 13.Inouye S, Sunshine MG, Six EW, Inouye M. Retronphage phiR73: an E. coli phage that contains a retroelement and integrates into a tRNA gene. Science. 1990;252:969–971. doi: 10.1126/science.1709758. [DOI] [PubMed] [Google Scholar]
- 14.Lindqvist B, Dehó G, Calendar R. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol Rev. 1993;57:683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dehò BG, Ghisotti FD. The plasmid status of satellite bacteriophage P4. Plasmid. 2001;45:1–17. doi: 10.1006/plas.2000.1497. [DOI] [PubMed] [Google Scholar]