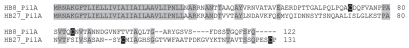Abstract
A lytic phage, designated as ϕTMA, was isolated from a Japanese hot spring using Thermus thermophilus HB27 as an indicator strain. Electron microscopic examination showed that ϕTMA had an icosahedral head and a contractile tail. The circular double-stranded DNA sequence of ϕTMA was 151,483 bp in length, and its organization was essentially same as that of ϕYS40 except that the ϕTMA genome contained genes for a pair of transposase and resolvase, and a gene for a serine to asparagine substituted ortholog of the protein involved in the initiation of the ϕYS40 genomic DNA synthesis. The different host specificities of ϕTMA and ϕYS40 could be explained by the sequence differences in the C-terminal regions of their distal tail fiber proteins. The ΔpilA knockout strains of T. thermophilus showed simultaneous loss of sensitivity to their cognate phages, pilus structure, twitching motility and competence for natural transformation, thus suggesting that the phage infection required the intact host pili. Pulsed-field gel electrophoresis analysis of the ϕTMA and ϕYS40 genomes revealed that the length of their DNA exceeded 200 kb, indicating that the terminal redundancy is more than 30% of the closed circular form. Proteomic analysis of the ϕTMA virion using a combination of N-terminal sequencing and mass spectrometric analysis of peptide fragments suggested that the maturation of several proteins involved in the phage assembly process was mediated by a trypsin-like protease. The gene order of the phage structural proteins was also discussed.
Key words: Thermus thermophilus, myovirus, genomics, antagonistic coevolution, proteomics
Introduction
Bacteria-infecting phages are ubiquitous in our world, especially in the ocean, and are probably the most abundant biological entity on earth.1 It is believed that they drive microbial evolution via lateral gene transfer and also influence biogeochemical cycles.2 Among these phages, tailed phages are the largest, most widespread and most diverse group of phages.3 Phages in one of the families, known as Myoviridae, have a contractile tails. They show the most complex structure, where the phage particle is formed in a complicated and sophisticated manner. T4, the archetype of the T-even phages, is one of the most intensively studied large myoviruses.4 Phages related to T4 are widespread in the biosphere, and probably they have diverged from a common ancestor by acquiring the ability to infect different host bacteria.5
Phages found in the hot water environment also show large diversity.6 For example, thermophilic archaeal viruses display an exceptional degree of diversity with regard to both morphotype and genome.7 In addition to evolutionary interest, the gene products of the thermophilic phages that are involved in nucleotide modification are also of great interest for various molecular biological applications.8 So far, more than one hundred Thermus infecting phages, the largest number for any thermophilic bacteria, have been isolated. These phages are morphologically diverse, as seen in mesophilic phages, including the Myoviridae, Siphoviridae, Tectiviridae and Inoviridae families, and more than half of the isolates are tail-less.9 The complete genome sequences of a myovirus, ϕYS40,10 two siphoviruses with the longest known tails, P23-45 and P74-26,11 two tail-less tectiviruses, P23-77,12 and T. aquaticus phage ϕIN93,13 have already been reported. Notably, of all the predicted genes, only approximately 25% have been assigned any putative function, indicating the novelty of their gene products. Purification and subsequent N-terminal amino acid sequencing of the lysozyme encoded in the genome of ϕIN93 demonstrated that its primary sequence was very different from the known lysozymes.14 The membrane-containing, icosahedral phage P23-77 of T. thermophilus is evolutionarily related to the viruses and plasmids of the halophilic archaea, all of which share two major capsid proteins and a putative packaging ATPase.12 Among the Thermus phages, ϕYS40 is the only one whose genome is larger than 100 kb, and has been extensively characterized at the molecular level that includes determining the transcriptional profile15 and host gene response after the infection.16
In general, the role of lytic phages in cellular evolution is not as clear as that of temperate phages, which are thought to be important vehicles for lateral gene transfer via transduction. Lytic phages of Thermus species, however, seem to contribute to the evolution of their host cells because DNA released from the lysed cells following infection could serve as good substrates for uptake by other Thermus cells. In this regard, it is noteworthy that because of their natural competence,17 T. thermophilus can take up DNA without any sequence specificity from all domains of life,18 regardless of the growth phase.19
To gain evolutionary insights into the Thermus large myoviruses through comparative genomic analysis, we isolated a T. thermophilus phage, ϕTMA, whose morphology and genome length were similar to those of the well-characterized phage ϕYS40, except that it showed broader host cell specificity. Subsequently, we determined and compared the complete genome sequences of ϕTMA and ϕYS40. Analysis of spontaneously occurring phage-resistant strains and gene-knockout strains revealed that the phage infection was dependent on the presence of type IV pili on the host cell surface. The linear size of the genome of the phage particle, estimated by pulsed-field gel electrophoresis, revealed an exceptionally large redundancy for the circular permutation. Proteomic analysis of the ϕTMA particle was performed to identify the putative gene products and determine the maturation process of the protein components involved in phage head formation.
Results
Isolation of a phage infectious to T. thermophilus HB27.
In order to isolate a novel myovirus, we screened hot water of Atagawa hot spring with HB27 as an indicator strain. Several plaques were found and one of them was cloned by two serial passages of phage dilutions. The newly isolated phage, designated as ϕTMA, formed clear plaques on the lawn of HB27 cells spread on double agar layer plates. Clear plaques were also formed with similar efficiency on the lawns of HB8 and T. flavus AT-62 cells, but not on T. aquaticus YT-1 cells.
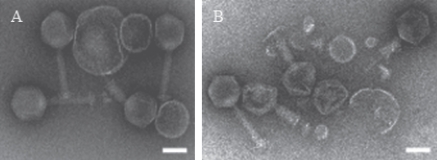
Electron micrograph of negatively stained ϕTMA showed that its morphology was very similar to that of ϕYS40 (Fig. 1). Accordingly, ϕTMA had an isometric, rather than prolate, head, a contractile tail and a thick baseplate with tail fibers. Like a typical myovirus, such as T4, the tail tube of ϕTMA protrudes from the bottom of the baseplate when the tail sheath contracts. There were several cases where lipid vesicles appeared to be bound to the bottom of the baseplate of the phage particles (Fig. 1).
Figure 1.
Electron micrographs of the two T. thermophilus phages: (A) ϕTMA and (B) ϕYS40. The bar indicates 100 nm.
Genome features of ϕTMA and ϕYS40.
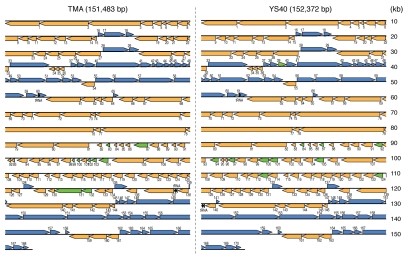
The genomes of ϕTMA and ϕYS40 consisted respectively of 151,483 and 152,372 base pairs (bp) long double-stranded DNAs (Fig. 2), and were predicted to contain 168 and 170 protein-coding genes, respectively (Table S1). The GC content (32.63%) of the ϕTMA genome was very similar to that (32.59%) of the ϕYS40 genome. One hundred and fifty-nine of the protein-coding genes were common to both genomes. Just as in ϕYS40, approximately 25% of the predicted proteins in ϕTMA showed sequence similarity to proteins of known function, including nucleotide metabolism (nine genes), DNA replication/recombination (nine genes), components of phage particles (nine genes) and others (13 genes). The amino acid sequence identity between the orthologs in the two genomes ranged from 43% to 100% and the average was 94%. Three tRNA genes were found in the ϕTMA genome, and the positions and nucleotide sequences of the three tRNA genes of ϕTMA were identical to those of ϕYS40. The gene order was also conserved between these two genomes (Fig. 2 and Table S1).
Figure 2.
Linear representations of ϕTMA and ϕYS40 genomes. Genes are shown using colored arrows indicating the transcription direction: blue, protein-coding genes on the positive strand; orange, protein-coding genes on the negative strand; green, protein-coding genes in one genome whose orthologs are missing from the other genome; and black, tRNA genes.
One of the noticeable differences between these two genomes is the presence of the genes for the putative transposase (TMA_131) and resolvase (TMA_132) in the ϕTMA genome (see Materials and Methods for the locus_tag prefixes). The gene product of TMA_131 showed 57% amino acid identity to the transposase (accession no. ACN98377) of the hyperthermophile Sulfurihydrogenibium azorense Az-Fu1, a member of the Aquificales family. The gene product of TMA_132 showed 59% amino acid identity to the resolvase-like protein (accession no. BAG02822) of the cyanobacteria Microcystis aeruginosa NIES-843.
It has been reported that the YS40_065 protein of ϕYS40 has low sequence similarity to a portion of the terminal protein for DNA replication of Bacillus subtilis phage ϕ29 and that the serine residue essential for the protein-primed replication of linear dsDNA genome of ϕ29 is conserved in the YS40_065 protein of ϕYS40.10 Thus, YS40_065 was annotated as a gene encoding the terminal protein for DNA replication. However, its ortholog (TMA_064) protein in ϕTMA showed only 65% amino acid identity to the YS40_065 protein, and contained an asparagine residue in place of the serine residue that was essential for the DNA priming (Fig. 3), suggesting that the TMA_064 protein is unlikely to function as a terminal protein for DNA replication in ϕTMA.
Figure 3.
Amino acid sequence alignment of the putative terminal protein of ϕYS40 (YSP_065) and its corresponding ortholog in ϕTMA (TMA_064). The serine residue whose hydroxyl group has been presumed to form a phosphoester bond with the 5′-terminal dAMP of the ϕYS40 genome for DNA replication is highlighted in black. Gray shading indicates identical amino acid residues.
ϕTMA formed clear plaques on both HB8 and HB27 efficiently, whereas ϕYS40 formed clear plaques only on HB8. The gene encoding the long tail fiber protein is highly likely to be responsible for discriminating the host-phage recognition specificity. Consistent with this idea, comparison of amino acid sequences between the TMA_001 and YSP_001 proteins showed a striking difference, especially in the C-terminal region, including a deletion of 30 amino acid residues in the TMA_001 protein and low amino acid identity of the sequence following the deletion (66%) (Fig. 4).
Figure 4.
Amino acid sequence alignment of the tail fiber proteins of ϕTMA (TMA_001) and ϕYS40 (YSP_001). Gray shading indicates identical amino acid residues.
Both ϕTMA and ϕYS40 are lytic phages. Although lysozyme and holin are generally involved in the lysis process in double-stranded lytic phages,20 we have not found the genes for lysozyme and holin in ϕTMA and ϕYS40.
Resequencing of the ϕYS40 genome.
ϕYS40 was originally isolated by our group,21 and we determined the complete genome sequence of ϕYS40. However, during the preparation of this manuscript, the genome sequence (152,372 bp, 170 protein-coding genes) of ϕYS40 has also been reported by another group (accession no. DQ997624).10 Genome sequence alignment analysis showed that eight nucleotides were different in the protein-coding regions of these two reported ϕYS40 sequences, resulting the indicated amino acid replacements in the following genes: YSP_004 (I88L, ATT→CTT), YSP_011 (I102V, ATA→GTA), YSP_031 (E202D, GAG→GAT), YSP_032 (S30L, TCG→TTG), YSP_068 (T153K, ACA→AAA), YSP_082 (H243Q, CAT→CAG), YSP_152 (I278S, AAT→AAG), and YSP_153 (E71K, GAA→AAA). Furthermore, a deletion of one nucleotide was found between the genes YSP_066 and YSP_067 and an insertion of one nucleotide was found between the genes YSP_124 and YSP_125. Another difference was that the gene YSP_096 (our annotation, this study), which is encoded in the same strand as the flanking genes (Fig. 2), was not found in the latter study reported by Naryshkina et al. who, instead, assigned YS40_096 (accession no. ABJ91490) to a gene encoded in the strand opposite to the strand encoding the flanking genes. Thus, the genes YSP_096 and YS40_096 are not the same genes.
Type IV pili-dependent infection of ϕTMA and ϕYS40.
Several spontaneous mutant strains of HB27 resistant to ϕTMA were selected in the presence of excess ϕTMA as described in Materials and Methods. These mutants arose at a frequency of approximately 10−5. A well-isolated colony was cloned by serial passage of diluted cells, and was designated as 27R1. Similarly, a mutant strain resistant to ϕYS40 was isolated from HB8, and was designated as 8R. The resistance of 27R1 to ϕTMA and that of 8R to ϕYS40 were further confirmed using the plaque-forming assay.
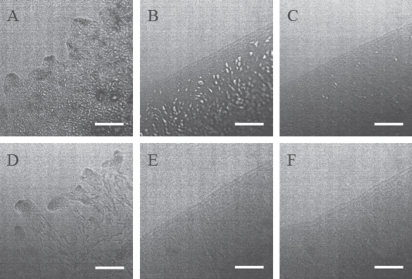
The wild-type HB8 and HB27 grown on agar plates showed spreading zones at the edges of their growing areas, which is a characteristic of the twitching motility (results not shown). Visual inspection showed that the extent of spreading of HB27 was smaller than that of HB8, whereas the phage-resistant mutant strains 8R and 27R1 showed dome-shaped colonies with sharp edges (results not shown). Micromorphology of the twitching or non-twitching zone edge was also examined by light microscopy. The wild-type strains, HB27 and HB8, showed the characteristic motile rafts of cells at the leading edge of the moving zone (Fig. 5A and D). The motility of HB27 was less intense than HB8, which is consistent with the results obtained by visual inspection. In the phage-resistant mutants 27R1 and 8R, the motility was completely abolished (Fig. 5B and E).
Figure 5.
Twitching motility of wild-type and mutant T. thermophilus. (A) HB27 (wild-type), (B) 27R1 (ϕTMA-resistant), (C) KA271 (pilA), (D) HB8 (wild-type), (E) 8R (ϕTMA- and ϕYS40-resistant), (F) AU81 (TTHA 1221-). The bar indicates 50 µm.
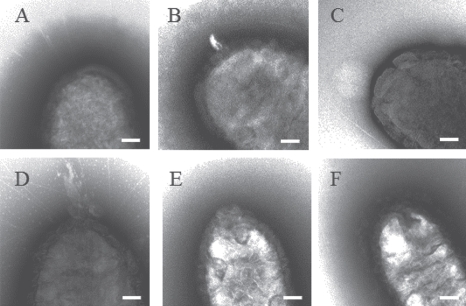
Electron micrographs of the wild-type cells are shown in Figure 6. As shown previously in reference 22, we confirmed the pilus filament at the poles of the HB27 cells (Fig. 6A). The pilus filaments with uniform width were also found on the surface of HB8, radiating especially from the poles (Fig. 6D). Both phage resistant strains 27R1 and 8R lacked such pilus (Fig. 6B and E).
Figure 6.
Electron micrograph of type IV pili of wild-type and mutant T. thermophilus. (A) HB27 (wild-type), (B) 27R1 (ϕTMA-resistant), (C) KA271 (pilA), (D) HB8 (wild-type), (E) 8R (ϕTMA- and ϕYS40-resistant), (F) AU81 (TTHA 1221-). The bar indicates 100 nm.
It was previously shown that the uptake of DNA by T. thermophilus HB27 is dependent on the presence of the pili.22 Because both 8R and 27R1 strains lacked their pili, we tested their competence for natural transformation using a gene knockout vector that conferred kanamycin resistance as described in Materials and Methods. We consequently found that neither of these two mutant strains produced any transformed colonies, whereas the wild-type strains HB8 and HB27 were transformed with efficiencies of 2.0 × 104 and 1.3 × 104 CFU/mg plasmid DNA, respectively.
The pilA gene of HB27 encoding the pilus structural protein pilin has been identified previously in reference 23. In order to confirm that the presence of the pili is required for competency, a pilA deletion mutant strain, KA271, was constructed. We subsequently confirmed that this pilA mutant strain lacked both the pilus fiber on the cell surface (Fig. 6C) and competence for natural transformation. The pilA mutant strain showed resistance to infection by ϕTMA and non-motility on agar plate (Fig. 5C). The HB8 genome also contained the pilin gene (TTHA1221, accession no. NC_006461) and the pilin gene in HB8 showed only 39% amino acid sequence identity to its counterpart in HB27 (Fig. 7), which is much lower than those of many other orthologs of HB8 and HB27. Notably, the location of the cysteine residues, supposed to be involved in the disulfide-bonded loop formation, and which are conserved in many type IV pili, was very different. To confirm that the pilin gene of HB8 is involved in the phage infection process, a mutant strain AU81, in which this gene was disrupted, was constructed. Electron microscopic analysis showed that AU81 lacked the pilus filament (Fig. 6F). The mutant strain AU81 was also resistant to both ϕYS40 and ϕTMA, non-motile (Fig. 5F) and noncompetent for DNA uptake as was seen with 8R. These results demonstrated that the pilin gene of HB8 is one of the requirements for infection with ϕYS40 and ϕTMA phages.
Figure 7.
Amino acid sequence alignment of the pre-pilins of HB8 and HB27. Gray shading indicates identical amino acid residues. The cysteine residues involved in the formation of a putative C-terminal disulfide-bonded loop are highlighted in black.
Pulsed-field gel electrophoresis.
To determine the topology and sizes of the packaged DNAs of ϕTMA and ϕYS40, the capsid DNA was digested separately with a number of different restriction enzymes, each enzyme recognizing its own unique restriction site, and subsequently the intact and restriction enzyme-digested capsid DNAs were subjected to pulsed-field gel electrophoresis (PFGE). As shown in Figure 8, the intact genomes of the thermophilic phages were larger than 200 kb (average DNA length, as determined from three independent experiments, was 220 ± 4 kb for ϕYS40 and 203 ± 2 kb for ϕTMA) and the digested product appeared as smeared bands (Fig. S1), which suggests the existence of variable DNA termini in phage particles. These results indicate that the genomic DNA is linear and the terminal redundancy is 68 ± 4 kb for ϕYS40 and 52 ± 2 kb for ϕTMA, which corresponded to 44% and 34%, respectively, of their closed circular forms.
Figure 8.
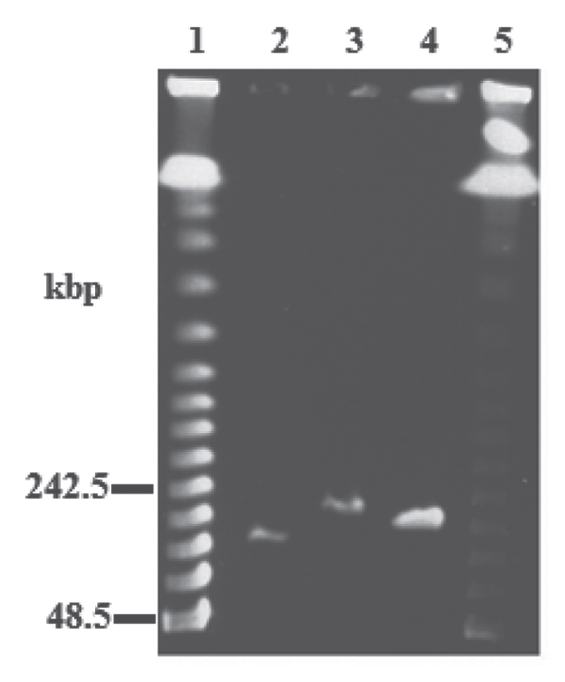
PFGE of the genomic DNAs of ϕYS40 and ϕTMA. lanes 1 and 5, lambda DNA ladders; lane 2, T4; lane 3, ϕYS40; lane 4, ϕTMA.
Protein composition of the ϕTMA virion.
To identify the structural proteins of ϕTMA, the ϕTMA virions were purified by cesium chloride density gradient ultracentrifugation. Based on the sequence analysis (by Edman degradation) of the bandforming proteins isolated from the SDS-PAGE (Fig. 9), we identified three N-terminal sequences: ALN AAG QVA E for the 47 kDa protein, MTS QGY SLK Y for the 45 kDa protein and SVV DVT VEG for the 30 kDa protein. As described below, peptides identical to each of these three N-terminal sequences were found respectively in three separate proteins encoded by the putative ORFs of the ϕTMA genome, and none of them showed any significant homology to proteins encoded by the other ORFs of ϕTMA. The peptide sequence ALN AAG QVA E was found in the protein encoded by the TMA_072 gene and started at position 16 (A16) of the encoded protein. This result indicated that the TMA_072 protein, the major head protein, is processed between residues K15 and A16. The peptide sequence MTS QGY SLK Y was found in the N-terminal region of the protein encoded by the TMA_019 gene. The peptide sequence SVV DVT VEG was found in the protein encoded by the TMA_166 gene and started at position 110 (S110) of the encoded protein. This result also suggested that, similar to the TMA_072 protein, the TMA_166 protein was processed between the residues K109 and S110. We, however, were unable to determine the N-terminal amino acid sequence of the 24 kDa protein by Edman degradation even though this protein band was prominent on the SDS-PAGE, suggesting that the N-terminus of the 24 kDa protein might be blocked.
Figure 9.
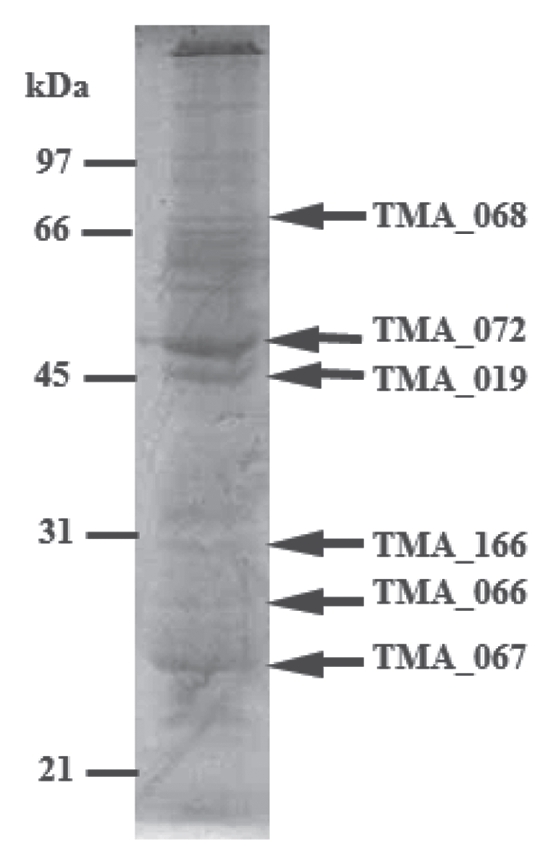
SDS-PAGE analysis of the ϕTMA virion proteins. The CBB-stained SDS/polyacrylamide gel showing protein bands of purified ϕTMA virions. Proteins (bands indicated using arrows) were identified by Edman degradation and/or mass spectrometry. Relative migrations of MW marker proteins are indicated on the left.
We also identified five phage subunits by mass spectrometric analysis of peptide fragments derived from in-gel trypsin digestion. The mass spectrometric results showed that the trypsin digested peptide fragments from the 71 kDa protein band were derived from the product of the TMA_068 gene. In the similar manner, the peptide fragments obtained from in-gel trypsin digested 47 kDa, 45 kDa, 26 kDa and 24 kDa proteins (N-terminal sequence of the last one was found to be blocked by Edman degradation analysis as described above) were also analyzed by mass spectrometry. Based on these analyses, we found that the 47 kDa, 45 kDa, 26 kDa and 24 kDa proteins were derived from the TMA_072, TMA_019, TMA_066 and TMA_067 genes, respectively. These results were consistent with the N-terminal sequencing results of the TMA_072 and TMA_019 proteins.
Discussion
After tectivirus, myovirus is the second most ubiquitous family of Thermus phages isolated so far. However, all of the collected Thermus myoviruses, except for ϕYS40 (which has a genome size of >100 kb), resemble coliphage Mu or P2 with a genome size of 28–34 kb.9 This report describes the isolation of another myovirus, ϕTMA, with large genome size and morphology similar to those of the ϕYS40, but with different host specificity.
The closed circular map of both ϕTMA and ϕYS40 genomes consisted of approximately 152 kb long DNA. This value is, however, inconsistent with the previous report in which the molecular weight of the ϕYS40 virion DNA, as determined by sucrose density gradient centrifugation, was estimated to be 1.36 × 108 (approximately 206 kb, assuming that the average molecular weight of a single DNA base pair is 660) using the T4 DNA as an internal marker (1.3 × 108, approximately 197 kb),21 suggesting that the ϕYS40 genome is longer than the T4 genome. In this study, the length of the genomic DNA purified from the phage particles was determined more precisely using PFGE, and was found to be more than 200 kb. In the T4-related phages, DNA packaging starts from any free end of a long concatemer duplicated in the host cell and terminates when the head is full, giving rise to a linear DNA with various terminally redundant ends within their capsids.24 The redundancy accounts for the circular permutation of the genome and is usually several percent of the circular map, which, for example, is approximately 3% in T4. In contrast, the length of the DNA within the phage particles of ϕTMA and ϕYS40 is more than 50 kb longer than the circular map, and thus, the redundancy is more than 30%. The low mobility of the ϕYS40 genomic DNA in the PFGE assay could not be explained on the basis of its covalent binding to a protein, such as the YSP_065 protein that was predicted as the protein primer in DNA replication, because the DNA sample used in the PFGE analysis was pre-treated with proteinase K. The large redundancy may facilitate homologous recombination, contributing to the repair of frequently occurring mutations at a high temperature. Consistent with this idea, several genes supposedly involved in the DNA repair, such as the putative recA and recB genes, were found in the ϕTMA and ϕYS40 genomes. It is, however, unclear whether, among all the Thermus myoviruses, the large redundancy is specific to ϕYS40 and ϕTMA, because we currently do not have appropriate information on the precise genome sizes and complete genome sequences of other Thermus myoviruses. Another possible explanation for such a long-terminal repeat is that when the thermophilic phages are propagated in environments other than their native environment (i.e., in the laboratory), many genes are no longer needed and are lost while the terminal repeat is increased in length to physically compensate for the lost genes in their capsids and to maintain the efficiency of DNA packaging and injection. Results of the PFGE assay also suggest a larger difference in length between the DNAs of ϕTMA and ϕYS40 genomes than their nucleotide sequence suggest. The difference might reflect the difference in capsid lengths, one of the determinants of DNA length in T4.25 It has been shown that the head length of the T4 capsids could be controlled by mutations in certain amino acids of the major structural protein, and the head length is determined by a vernier-type mechanism that involves interaction between the core and shell proteins.26 The putative major head proteins of ϕTMA and ΔYS40, encoded by TMA_072 and YSP_073 genes, respectively, could be responsible for the differences in space within the capsid. However, other capsid proteins could also be responsible for determining the length and shape of the capsid.27
T4 has approximately 290 probable protein-coding genes packed into its 169 kb long genome whereas ϕTMA and ϕYS40 have 168 and 170 protein-coding genes, respectively, in their 152 kb long genomes. The average length of ORFs in the ϕTMA and ϕYS40 genomes are 837 bp and 836 bp, respectively, which are longer than the average length of ORFs (588 bp) in the T4 genome, suggesting that the protein-coding genes in the thermophilic phages are longer than those in the T4 phage. The presence of smaller number of protein-coding genes in ϕTMA and ϕYS40 compared with those in the counterpart mesophilic large virulent phages suggests that the thermophilic phages might have a simpler life cycle than the mesophilic ones. Alternatively, it can be argued that the thermophilic phages might require longer proteins to attain stability at higher temperature; however, comparative analysis of complete genomes of mesophilic, thermophilic and hyperthermophilic organisms indicated a trend toward shortened thermophilic proteins relative to their mesophilic homologs,28 and this trend seems to hold true for proteins of thermophilic phages.29
One of the significant differences between the genomes of the two Thermus phages is the presence of the transposase and resolvase genes in the ϕTMA genome. Some transposons have a pair of transposase and resolvase, where the transposase helps to form a cointegrate between the donor and recipient replicons by carrying a directly repeated copy of the transposable unit, and then the intermediate is separated by the resolvase into donor and recipient replicons, each containing one copy of the transposon.30 The amino acid sequence of the ϕTMA transposase is most similar to that of the hyperthermophile S. azorense Az-Fu1 and very similar to that found in the T. scotoductus SA-01 genome (accession no. YP_004201456). In addition, the resolvase gene in the ϕTMA genome is considerably similar to that of T. scotoductus SA-01 (accession no. YP_004201455). The transposase and resolvase genes of T. scotoductus SA-01 reside next to each other and their order is same as that in ϕTMA. Thus, genetic exchange could occur between the (hyper)thermophiles and the phages. This speculation is in accordance with an earlier suggestion that diverse Thermus phages have access to a common gene pool,11 although as of now there is no report describing the presence of a pair of transposase and resolvase in other phage genomes. In sharp contrast to the host cell's GC content (69%), the GC contents of the transposase (32%) and resolvase (31%) genes of ϕTMA are very similar to the average GC content of the ϕTMA genomic DNA (32.6%), suggesting that the elements may have transposed from other genomes with low GC content and that the transposition may have occurred over a time long enough for each gene to ameliorate to a lower GC content. It however remains unclear whether the DNA region that contains the genes for the transposase and resolvase is transposable. Our sequence analysis did not reveal any inverted repeats flanking these genes to function as an insertion sequence.
In T-even related phages, host range specificity can be changed by amino acid substitution or duplication/mutational alteration of the His-boxes found in the C-terminal portion of gene 37 tail fibers that bind to receptors on the host bacterial surface.31 ϕTMA showed broader host specificity than ϕYS40. The amino acid sequence of the long tail fiber proteins of ϕTMA and ϕYS40 strikingly differed in their C-terminal portion, including a deletion of 30 amino acid residues in the ϕTMA protein. Thus, the C-terminal region of the long tail fiber protein of the thermophilic phages might also be critical for the host discrimination, as was found with the mesophilic phages. Because the tail fiber proteins of ϕTMA and ϕYS40 did not have any Hisboxes, their structures might be considerably different from that of the mesophilic phages. There are other possibilities for the host range specificity, including the T. thermophilus DNA modification/restriction system. However, we have observed that ϕTMA, which can infect both HB8 and HB27, could form plaques on both strains with virtually same efficiency irrespective of the host used for the previous passage (results not shown).
The phage resistant strains selected in the presence of excess ϕTMA and ϕYS40 lacked the pilus fiber on the host cell surface. Deletion of the pilA gene led to the phage resistance in HB27 and HB8 (this study). These results demonstrate that the myoviruses infect T. thermophilus via the pilus of the host strain. It has been shown that the phage PO4 binds to the type IV pili expressing on the surface of Pseudomonas aeruginosa32 and also to the pili heterologously expressed on the cell surface of Neisseria gonorrhoeae,33 suggesting that the P. aeruginosa pili are the primary receptors for the phage PO4. Because infection of the T. thermophilus strains by ϕTMA and ϕYS40 was dependent on the existence of the host pili, they could also be the primary receptors for the thermophilic phages. The amino acid sequence of the pilA gene products of HB8 and HB27 differed from each other, especially in the C-terminal region that included the putative disulfidebonded loop region, which has been shown to be critical for the pilus assembly and twitching motility in P. aeruginosa.34 It has been hypothesized that the sequence diversity of pilin found in pathogenic bacteria reflects an evolutionary compromise between the retention of the function as a retractile tether for twitching motility and antigenic variation against host immune system.34 On the other hand, the significant difference found in the pilus structural protein of the T. thermophilus likely reflects competition against the phages. The astonishing divergences in the primary sequences of pilins of the thermophiles and putative tail fiber proteins of the phages must have resulted partly from the competition between the thermophiles and the phages in the hot springs. In addition to the differences in the type IV pili, comparative genomics of HB8 and HB27 have shown striking differences in the cell surface determinants, including the S-layer proteins and cell envelope-modifying enzymes, such as the glycosyltransferases.35 These strain-specific surface structures other than the pili might also act as countermeasures against phages in the natural environment.
SDS-PAGE analysis of the ϕTMA structural proteins suggested that the proteins in the ϕTMA phage particles are very stable. We found that proper denaturation of proteins by boiling prior to the sample loading on the gel was very important for reproducibility. Short time boiling (<3 min) caused poor reproducibility, as a result of which few protein bands went missing in the SDS-PAGE analysis. In the previous report in reference 10, the protein composition of the ϕYS40 virions was analyzed only by mass spectrometry. We identified six ϕTMA virion structural subunits that corresponded to the products of TMA_019, TMA_066, TMA_067, TMA_068, TMA_072 and TMA_166 genes on the basis of their N-terminal sequence and mass spectrometric analyses. These gene products were also found in ϕYS40, and the relative abundances of these proteins in these two phages were similar (i.e., TMA_072 protein was most abundant, followed by the TMA_019 protein). We found that both the TMA_072 and TMA_166 proteins were processed at the C-terminal side of a lysine residue. This result suggests that a trypsin-type protease is involved in the phage assembly process. The major capsid proteins of T4 phage36 and 201ϕ2-1,37 one of ϕKZ-related phages, also exhibited posttranslational cleavage, although in those cases proteolysis occurred after a glutamate residue. Based on the abundance and posttranslational cleavage data we conclude that the TMA_072 gene encodes for the precursor of the major capsid subunit, even though the encoded TMA_072 protein did not show any sequence homology with the subunits of the registered virus particles available in the public databases, except for the ortholog found in ϕYS40.
We could also speculate on the function of some of the other capsid proteins. First, the primary sequence of the protein encoded by the TMA_068 gene revealed low but significant homology with the tail sheath proteins of other Myoviridae family of phages having a contractile tail and a linear double-stranded DNA. Although the Myoviridae virion generally contains high copies of the sheath protein, the TMA_68 gene product was not abundant in the SDS-PAGE analysis. The transmission electron microscopic analysis of ϕTMA showed frequent contracted phage particles, which could have caused by osmotic shock during the dialysis following the cesium chloride gradient ultracentrifugation step. Another reason for contraction might be the sensitivity of the phage particles to high concentration of CsCl used for the density gradient centrifugation. It has been shown previously that ϕYS40 is easily inactivated at high salt concentration.21 A similar sensitivity to high salt concentration could be responsible for the contraction of ϕTMA tail, leading to aggregation of the sheath protein. We speculate that the aggregate is resistant to denaturation by the standard SDS/PAGE sample buffer, and is too large to enter the separating gel (see Fig. 9), which could be a major reason for the observed low abundance of the TMA_068 gene product. Second, the product of the TMA_067 gene could be a tube protein because of its abundance and molecular weight (24 kDa), which is close to the tube proteins of other phages (e.g., 18 kDa and 15.9 kDa for the T4 phage and K phage, respectively). Finally, products of the TMA_073 gene and that of its counterpart YSP_074 gene of ϕYS40 showed partial similarity to the HK97 prohead protease. A strictly conserved pair of serine and histidine residues found in the prohead protease superfamily38 was also conserved in the proteins encoded by the TMA_073 and YSP_074 genes. This gene is located next to the one encoding the phage major head structural protein described above. We, therefore, hypothesize that the TMA_073 protein of ϕTMA (and also the YSP_074 protein of ϕYS40) is the protease involved in the head maturation. In this way, the order of the genes encoding the head protease, major capsid protein tail-related sheath and tube proteins are highly conserved among other Myoviridae (Table 1). It is hard to speculate on the function of the TMA_066 protein in ϕTMA virion morphogenesis, because its primary sequence did not share any homology with the sequences of proteins involved in virion morphogenesis that are available in the public database. However, it showed a weak homology (22% amino acid sequence identity) with the putative tube protein encoded by the TMA_067 gene, a probable tube protein-encoding gene (see above). The gp54 protein of T4, whose amino acid sequence shows partial similarity to the tail tube structural protein of T4, is believed to function as a tail tube initiator.39 It is possible that the TMA_066 protein could help in the formation of the tail tube of the thermophilic phage.
Table 1.
Gene orders for the capsid proteins of Thermus, Staphylococcus and Escherichia phages
| Thermus phage TMA | Staphylococcus phage K | Escherichia phage Mu | ||||||
| Gene no. | Length (aa) | Gene product | Gene no. | Length (aa) | Gene product | Gene no. | Length (aa) | Gene product |
| 73 | 544 | prohead protease | 43 | 316 | 33 | 182 | Scaffold | |
| 72 | 470 | capsid precursor | 44 | 463 | capsid precursor | 34 | 305 | capsid precursor |
| 71 | 171 | 45 | 305 | 35 | 136 | |||
| 70 | 220 | 46 | 292 | 36 | 141 | |||
| 69 | 236 | 47 | 206 | 37 | 182 | |||
| 48 | 278 | 38 | 67 | |||||
| 68 | 648 | sheath | 49 | 587 | sheath | 39 | 495 | Sheath |
| 67 | 216 | tube | 50 | 142 | 40 | 118 | Tube | |
| 66 | 246 | virion component | 51 | 152 | 41 | 144 | ||
In conclusion, we isolated a thermophilic myovirus, ϕTMA, evolutionarily related to ϕYS40. Their divergence seems to be partly driven by coevolution with the host thermophile that proceeds via interaction between the tail fiber of the phages and the pili of the host cells and involves a mobile element encoding a transposase. The presence of small number of the predicted ORFs suggests a unique simple life cycle for the thermophilic large myoviruses. The unexpectedly large terminal redundancy in their genomes suggests a role in the maintenance of genetic information through circular permutation and also implies novel significance in processes such as in DNA repair. The gene order and processing of the head proteins of these phages resemble those of the mesophilic phages.
Materials and Methods
Bacterial strains, media, phages and DNA.
The Thermus strains used in the present study are described in Table 2. The rich medium used for growing T. thermophilus consisted of 0.8% (w/v) polypepton, 0.4% (w/v) yeast extract, 0.2% (w/v) NaCl, 0.35 mM CaCl2 and 0.4 mM MgCl2, and the composition of the synthetic medium used for its growth has been described previously in reference 40. Escherichia coli JM109 was used for plasmid propagation. The T. thermophilus phages used in this study were ϕTMA and ϕYS40.21 ϕTMA was isolated from Atagawa hot spring, Japan, with HB27 as an indicator strain. For plaque formation experiment, the top and bottom agar medium contained 0.75% and 1.5% agar, respectively. Phage T4 was propagated and purified as described previously in reference 41, to prepare the genomic DNA that was used as a reference for the pulsed-field gel electrophoresis. Synthetic oligonucleotides used as PCR primers are summarized in Table 3.
Table 2.
T. thermophilus strains used in this study
| Parental | Source or | Phage sensitivity | |||
| Strain | Relevant genotype | ||||
| strain | reference | ϕTMA | ϕYS40 | ||
| T. thermophilus | |||||
| HB8 | Wild-type | (47) | + | + | |
| HB27 | Wild-type | (21) | + | − | |
| 8R | HB8 | Phage resistant | This study | − | − |
| 27R1 | HB27 | Phage resistant | This study | − | − |
| MT111 | HB27 | ΔpyrE | (48) | + | − |
| AM114 | HB8 | ΔpyrE | This study | + | + |
| KA271 | MT111 | pilA::pyrE | This study | − | − |
| AU891 | AM114 | pilA(TTHA1221):: | This study | − | − |
| pyrE | |||||
| T. flavus AT-62 | Wild-type | (49) | + | − | |
| T. aquaticus YT-1 | Wild-type | (50) | − | − | |
Table 3.
Oligonucleotides used in this study
| Primer name | Sequencea |
| 5′-GGGGTACCTTGAGCGTGCAGACCTTCCTC-3′ | |
| prm1-Kpm | |
| 5′-GGAAGCTTCACCTCCTCCCACACTCCCTC-3′ | |
| prm1-Hin | |
| 5′-GGGAATTCGTGGACATTGACCCCAATGGTC-3′ | |
| prm3-Eco | |
| 5′-GGTCTAGACTCTAAGCCGCCTGAGCTTC-3′ | |
| prm4-Xba | |
| 5′-GGTGAGCAATAACTAGGAGGACCATATGGACGTC-3′ | |
| 3TSDN | |
| P5 | 5′-CAGGAAACAGCTATGAC-3′ |
| TTHA1221Kpn | 5′-GCGGGTACCACCTTCCAGGTCAACGGGACTGT-3′ |
| TTHA1221Nde | 5′-GCGTTCCGCATATGTCTCTATCCTCC-3′ |
| TTHA1221Eco | 5′-GCGGAATTCAGCTTCCAGTAACGAGGCAGCCT-3′ |
| TTHA1221Bam | 5′-GATGGGATCCACCTCCTGGCGTAGGAGGTCCA-3′ |
| pilA27Kpm | 5′-GCGGGTACCGGAGGGTTCAGGTACACCTACAC-3′ |
| pilA27Nde | 5′-GCGTTCCGCATATGTCATTTCCTC-3′ |
| pilA27Eco | 5′-GCGGAATTCTAAGTTAAAGTATGGCCTCGGCGC-3′ |
| pilA27Bam | 5′-GATGGGATCCGTACGAAGGCCTCCGTACGGTTT-3′ |
Electron microscopy.
An aliquot of the sample was directly applied to a copper grid covered with a thin carbon film. After blotting off the excess liquid, the phages were immediately stained with 2% uranyl acetate and the bacterial cells were stained with 1% phosphotungstenic acid. The stained samples were observed using a JEM-1230 transmission electron microscope (JEOL) operated at 100 kV, and the images were recorded using a CCD camera, Fastscan F114T (TVIPS) or BioScan model 792 (Gatan).
Genome sequencing and informatics.
The genome sequences of ϕTMA and ϕYS40 were determined using a whole-genome shotgun strategy. For this purpose, we constructed small-insert (∼2 kb) genomic libraries, and generated nucleotide sequences of 4,224 genomic clones of ϕTMA (12-fold coverage) and 9,024 genomic clones of ϕYS40 (14-fold coverage) from both ends using the ABI 3700 Sequencer (Applied Biosystems). Sequence reads were assembled with the help of the Phred-Phrap-Consed program and gaps were closed by direct sequencing of clones that spanned the gaps or sequencing of PCR products amplified with oligonucleotide primers designed to anneal to each end of the neighboring contigs. The overall accuracy of the finished sequence was estimated to have an error rate of <1 per 10,000 bases (Phrap score of ≥40).
An initial set of predicted protein-coding genes was identified using Glimmer 3.0.42 All predicted proteins were searched against a non-redundant protein database (nr, NCBI) using BLASTP with a bit-score cutoff of 60. The start codon of each protein-coding gene was manually refined from the BLASTP alignments. The tRNA genes were predicted using tRNAscan-SE.43 Orthology across whole-genome was determined using BLASTP reciprocal best hits in all-against-all comparisons of amino acid sequences. The genome sequence data of ϕTMA and ϕYS40 have been deposited in DDBJ/GenBank/EMBL and the accession numbers are as follows: AP011617 (ϕTMA) and AP011616 (ϕYS40). The locus_tag prefixes of ϕTMA and ϕYS40 are TMA_ and YSP_, respectively, while the locus_tag prefix of ϕYS40 reported by another group10 (accession no. DQ997624) is YS40_.
Characterization of twitching motility.
The T. thermophilus colony to be tested was grown overnight on 1.2% agar (w/v) containing nutrient medium at 70°C, after which the edge morphology of the colonies were examined with a dissecting microscope (BX60, Olympus, Japan).
Transformation studies.
The natural transformation efficiency of T. thermophilus was obtained using a knockout vector, pRMAHTK7, which contains the T. thermophilus ribosomal L11 methyltransferase encoding prmA gene interrupted by the thermostabilized kanamycin resistant gene as the marker. To construct the plasmid, a DNA fragment containing the upstream region of the prmA gene was amplified by PCR using the primers prm1-Kpn and prm2-Hin. Another DNA fragment containing the downstream region of the prmA gene was amplified by PCR using the primers prm3-Eco and prm4-Xba. Two PCR amplified fragments were purified. The upstream fragment was digested with the restriction enzymes KpnI and HindIII and the downstream fragment was digested with the restriction enzymes EcoRI and XbaI, and the digested fragments were then sequentially cloned into the corresponding sites of the plasmid pBHTK1.44 The procedure for natural transformation of T. thermophilus was described previously in reference 17. Transformants were selected on 100 µg/mL kanamycin supplemented T. thermophilus nutrient medium agar (1.2%) plate.
Isolation of T. thermophilus mutant strains resistant to ϕTMA and ϕYS40.
An overnight culture of T. thermophilus HB27 or HB8 cells (about 1 × 107 cells) was mixed with 1 × 108 ϕTMA or ϕYS40. After overnight incubation at 70°C, a surviving colony on the plate was purified and isolated. The mutant strains resistant to ϕTMA and ϕYS40 derived from HB27 and HB8 were designated as 27R1 and 8R, respectively. Resistance to the respective cognate phage was confirmed by plaque forming experiments.
Construction of T. thermophilus pilA strains.
A T. thermophilus ΔpyrE strain, AM114, was constructed from HB8 as described previously in reference 45, with a slight modification. In brief, the wild-type HB8 was naturally transformed with pKN605, which contains the pyrimidine biosynthetic operon except the entire pyrE gene, by putting a few drops of the plasmid solution on the top of the cells grown on a non-selective plate. After overnight incubation at 70°C, the cells were streaked on a selective plate containing the minimum medium supplemented with 100 µg/ml uracil and 200 µg/ml 5-fluoroorotic acid, and transformants were selected and purified. The deletion of the pyrE gene in the selected cell was confirmed by DNA gel blot hybridization analysis (results not shown).
The pyrE gene, amplified by PCR with the primers 3TSDN and P5 and using pT8L2P46 as a template, was cloned in the EcoRV site of pBluescript II SK+ (Agilent Technologies, 21205) in the opposite direction of the lacZ gene by a blunt-ended ligation reaction. The resultant plasmid was designated as p3TSDN1. For replacement of the TTHA1221 gene with the pyrE gene in the HB8 genome, a knockout vector was constructed as follows. A DNA fragment encoding the upstream region of the TTHA1221 gene was amplified by PCR using the primers TTHA1221Kpn and TTHA1221Nde. Another DNA fragment of the downstream region of the TTHA1221 gene was amplified by PCR using the primers TTHA1221Eco and TTHA1221Bam. After purification, the upstream fragment was digested with the restriction enzymes KpnI and NdeI and the downstream fragment was digested with the restriction enzymes EcoRI and BamHI. The digested fragments were then sequentially cloned into the corresponding sites of the plasmid p3TSDN1. The resultant plasmid (ppilA1-pyrE) was used to transform AM114 (ΔpyrE) and one of the transformant was designated as AU81. Similarly, a knockout vector for replacing the pilA gene of HB27 with the pyrE gene was constructed using the two sets of PCR primers, pilA27Kpn/pilANde and pilA27Eco/pilA27Bam, and the plasmid p3TSDN1. The resultant plasmid (ppilA4-pyrE) was used to transform MT111 (ΔpyrE) and one of the transformant was designated as KA271. Replacement of the pilA gene with the pyrE gene in AU81 and KA271 was confirmed by DNA gel blot analysis (results not shown).
Pulsed-field gel electrophoresis.
ϕYS40 and ϕTMA particles were purified by sucrose gradient centrifugation as described in reference 21. The purified phage particles were dialyzed against a buffer (10 mM Tris-HCl, pH 7.5–10 mM MgCl2) for more than six hours, following which they were mixed with liquefied 1% low melting agarose (BioRad) in 1.2 M Sorbitol-0.1 M EDTA (pH 8.0), and then cooled to room temperature to solidify. The solidified agarose was then cut into adequate sized blocks. The blocks were treated with 0.5 M EDTA (pH 8.0)-1% N-lauroylsarcosin-1 mg/ml protease K at 50°C overnight and then washed with TE buffer. The phage DNA was then separated on 1% pulsed-field certified agarose gel in 0.5x TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH 8.0) at 4°C for 22 h at an angle of 120° using CHEF MAPPER CHEF DR-II (BioRad). Switch time and voltage used for running the gel were 60 sec and 4.5 V/cm, respectively. Lambda-DNA ladder (BioRad, 170-3635) was used as the size marker.
Protein sequence analysis.
Purification of ϕTMA for proteomic analysis was performed as follows. Phage particles from one-liter culture were purified by ultracentrifugation using a sixstep cesium chloride gradient. Tris-M buffer (82 ml of 1 M HCl, 12.1 g of Tris base, 5 g of NaCl, 1 g of NH4Cl) was used for the preparation of the cesium chloride stock solution (100 g of CsCl, 7.5 ml of Tris-M buffer, 66 ml of H2O), which was diluted with an appropriate ratio of water to prepare a gradient layer. Following six different cesium chloride solutions of various density (r) were prepared: r = 1.13 (stock solution:water = 2:8), r = 1.22 (3:7), r = 1.29 (4:6), r = 1.36 (5:5), r = 1.46 (6:4) and r = 1.55 (7:3). Approximately 8 ml of each solution was layered in a SPR28SA rotor tube (HITACHI) and phage suspension was placed on the top of the gradient. Ultracentrifugation (20,000 rpm) was performed in a HITACHI SCP70H for 20 min at 4°C. The liquid content inside the tube was fractionated into 12 tubes and each fraction was examined by SDS-PAGE. The fraction containing ϕTMA phage was dialyzed overnight against phosphate-buffered saline, pH 7.4.
For protein sequencing, the phage solution was mixed with four times its volume of cold (−20°C) acetone. After centrifugation at 10,000x g for 15 min, the phage pellet was dissolved in 150 µl of 1% SDS and boiled for 5 min. We added 200 µl of 6 M urea, 1 M thiourea, 2% Triton X-100 and 1% 2-mercaptoethanol. Proteins were precipitated by chloroform and then washed with methanol. After centrifugation at 12,000x g for 2 min, the precipitate was dissolved in SDS sample buffer and applied onto a 12.5% SDS-PAGE. For Edman degradation, all proteins were transferred to a PVDF membrane following the electrophoresis and then the membrane was stained with 0.1% CBB in 50% methanol. Pieces of the blotted membrane, each containing a protein band, were directly subjected to protein sequence analysis using a protein sequencer (Applied Biosystems, Procise). For mass spectrometric analysis, in-gel protease digestion was performed as follows. After SDS-PAGE, protein bands were excised from the gel and the pieces were washed with 2 µl of 25 mM NH4HCO3 in aqueous 50% acetonitrile. Tryptic digestion was initiated with the addition of 5 µl of 5 µg/ml modified trypsin (Promega, V5113) in 25 mM NH4HCO3. Samples were incubated overnight at 37°C with gentle shaking. After centrifugation, the resultant peptide-containing supernatants were subjected to ESIIT-MS analysis (Esquire 3000 plus, Bruker Daltonik GmbH, Bremen, Germany).
Acknowledgements
We thank M. Takahashi (Tokyo University of Pharmacy and Life Sciences), K. Oshima, K. Furuya, C. Yoshino, H. Inaba, K. Motomura and Y. Hattori (University of Tokyo), A. Tamura and N. Itoh (Kitasato University) for technical assistance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Suttle CA. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 2.Rohwer F, Prangishvili D, Lindell D. Roles of viruses in the environment. Environ Microbiol. 2009;11:2771–2774. doi: 10.1111/j.1462-2920.2009.02101.x. [DOI] [PubMed] [Google Scholar]
- 3.Ackermann HW. 5,500 Phages examined in the electron microscope. Arch Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 4.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tétart F, Desplats C, Kutateladze M, Monod C, Ackermann HW, Krisch HM. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J Bacteriol. 2001;183:358–366. doi: 10.1128/JB.183.1.358-66.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoenfeld T, Patterson M, Richardson PM, Wommack KE, Young M, Mead D. Assembly of viral metagenomes from Yellowstone hot springs. Appl Environ Microbiol. 2008;74:4164–4174. doi: 10.1128/AEM.02598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nat Rev Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 8.Blondal T, Thorisdottir A, Unnsteinsdottir U, Hjorleifsdottir S, Ævarsson A, Ernstsson S, et al. Isolation and characterization of a thermostable RNA ligase 1 from a Thermus scotoductus bacteriophage TS2126 with good single-stranded DNA ligation properties. Nucleic Acids Res. 2005;33:135–142. doi: 10.1093/nar/gki149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu MX, Slater MR, Ackermann HW. Isolation and characterization of Thermus bacteriophages. Arch Virol. 2006;151:663–679. doi: 10.1007/s00705-005-0667-x. [DOI] [PubMed] [Google Scholar]
- 10.Naryshkina T, Liu J, Florens L, Swanson SK, Pavlov AR, Pavlova NV, et al. Thermus thermophilus bacteriophage ϕYS40 genome and proteomic characterization of virions. J Mol Biol. 2006;364:667–677. doi: 10.1016/j.jmb.2006.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minakhin L, Goel M, Berdygulova Z, Ramanculov E, Florens L, Glazko G, et al. Genome comparison and proteomic characterization of Thermus thermophilus bacteriophages P23-45 and P74-26: siphoviruses with triplex-forming sequences and the longest known tails. J Mol Biol. 2008;378:468–480. doi: 10.1016/j.jmb.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jalasvuori M, Jaatinen ST, Laurinavicius S, Ahola-Iivarinen E, Kalkkinen N, Bamford DH, et al. The closest relatives of icosahedral viruses of thermophilic bacteria are among viruses and plasmids of the halophilic archaea. J Virol. 2009;83:9388–9397. doi: 10.1128/JVI.00869-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsushita I, Yanase H. The genomic structure of Thermus bacteriophage ϕIN93. J Biochem. 2009;146:775–785. doi: 10.1093/jb/mvp125. [DOI] [PubMed] [Google Scholar]
- 14.Matsushita I, Yanase H. A novel thermophilic lysozyme from bacteriophage ϕIN93. Biochem Biophys Res Commun. 2008;377:89–92. doi: 10.1016/j.bbrc.2008.09.101. [DOI] [PubMed] [Google Scholar]
- 15.Sevostyanova A, Djordjevic M, Kuznedelov K, Naryshkina T, Gelfand MS, Severinov K, et al. Temporal regulation of viral transcription during development of Thermus thermophilus bacteriophage ϕYS40. J Mol Biol. 2007;366:420–435. doi: 10.1016/j.jmb.2006.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J Mol Biol. 2010;395:270–281. doi: 10.1016/j.jmb.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 17.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol. 1986;166:338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarzenlander C, Averhoff B. Characterization of DNA transport in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 2006;273:4210–4218. doi: 10.1111/j.1742-4658.2006.05416.x. [DOI] [PubMed] [Google Scholar]
- 19.Hidaka Y, Hasegawa M, Nakahara T, Hoshino T. The entire population of Thermus thermophilus cells is always competent at any growth phase. Biosci Biotechnol Biochem. 1994;58:1338–1339. doi: 10.1271/bbb.58.1338. [DOI] [PubMed] [Google Scholar]
- 20.Young I, Wang I, Roof WD. Phages will out: strategies of host cell lysis. Trends Microbiol. 2000;8:120–128. doi: 10.1016/S0966-842X(00)01705-4. [DOI] [PubMed] [Google Scholar]
- 21.Sakaki Y, Oshima T. Isolation and characterization of a bacteriophage infectious to an extreme thermophile, Thermus thermophilus HB8. J Virol. 1975;15:1449–1453. doi: 10.1128/jvi.15.6.1449-1453.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedrich A, Prust C, Hartsch T, Henne A, Averhoff B. Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl Environ Microbiol. 2002;68:745–755. doi: 10.1128/AEM.68.2.745-55.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedrich A, Rumszauer J, Henne A, Averhoff B. Pilin-like proteins in the extremely thermophilic bacterium Thermus thermophilus HB27: implication in competence for natural transformation and links to type IV pilus biogenesis. Appl Environ Microbiol. 2003;69:3695–3700. doi: 10.1128/AEM.69.7.3695-700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Streisinger G, Emrich J, Stahl MM. Chromosome structure in phage T4, III. Terminal redundancy and length determination. Proc Natl Acad Sci USA. 1967;57:292–295. doi: 10.1073/pnas.57.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doermann AH, Eiserling FA, Boehner L. Genetic control of capsid length. I. Isolation and preliminary description of four new mutants. J Virol. 1973;12:374–385. doi: 10.1128/jvi.12.2.374-385.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lane T, Serwer P, Hayes SJ, Eiserling F. Quantized viral DNA packaging revealed by rotating gel electrophoresis. Virology. 1990;174:472–478. doi: 10.1016/0042-6822(90)90101-V. [DOI] [PubMed] [Google Scholar]
- 27.Keller B, Dubochet J, Adrian M, Maeder M, Wurtz M, Kellenberger E. Length and shape variants of the bacteriophage T4 head: mutations in the scaffolding core genes 68 and 22. J Virol. 1988;62:2960–2969. doi: 10.1128/jvi.62.8.2960-2969.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson MJ, Eisenberg D. Transproteomic evidence of a loop-deletion mechanism for enhancing protein thermostability. J Mol Biol. 1999;290:595–604. doi: 10.1006/jmbi.1999.2889. [DOI] [PubMed] [Google Scholar]
- 29.Goulet A, Spinelli S, Blangy S, van Tilbeurgh H, Leulliot N, Basta T, et al. The thermo- and acido-stable ORF-99 from the archaeal virus AFV1. Protein Sci. 2009;18:1316–1320. doi: 10.1002/pro.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed RR. Transposon-mediated site-specific recombination: a defined in vitro system. Cell. 1981;25:713–719. doi: 10.1016/0092-8674(81)90178-1. [DOI] [PubMed] [Google Scholar]
- 31.Tétart F, Repoila F, Monod C, Krisch HM. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J Mol Biol. 1996;258:726–731. doi: 10.1006/jmbi.1996.0281. [DOI] [PubMed] [Google Scholar]
- 32.Bradley DE. A pilus-dependent Pseudomonas aeruginosa bacteriophage with a long noncontractile tail. Virology. 1973;51:489–492. doi: 10.1016/0042-6822(73)90448-0. [DOI] [PubMed] [Google Scholar]
- 33.Winther-Larsen HC, Wolfgang MC, van Putten JPM, Roos N, Aas FE, Egge-Jacobsen WM, et al. Pseudomonas aeruginosa Type IV pilus expression in Neisseria gonorrhoeae: effects of pilin subunit composition on function and organelle dynamics. J Bacteriol. 2007;189:6676–6685. doi: 10.1128/JB.00407-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey H, Habash M, Aidoo F, Burrows LL. Single-residue changes in the C-terminal disulfide-bonded loop of the Pseudomonas aeruginosa type IV pilin influence pilus assembly and twitching motility. J Bacteriol. 2009;191:6513–6524. doi: 10.1128/JB.00943-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brüggemann H, Chen C. Comparative genomics of Thermus thermophilus: Plasticity of the megaplasmid and its contribution to a thermophilic lifestyle. J Biotechnol. 2006;124:654–661. doi: 10.1016/j.jbiotec.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 36.Isobe T, Black LW, Tsugita A. Protein cleavage during virus assembly: A novel specificity of assembly dependent cleavage in bacteriophage T4. Proc Natl Acad Sci USA. 1976;73:4205–4209. doi: 10.1073/pnas.73.11.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas JA, Weintraub ST, Hakala K, Serwer P, Hardies SC. Proteome of the large Pseudomonas myovirus 201ϕ2-1: delineation of proteolytically processed virion proteins. Mol Cell Proteomics. 2010;9:940–951. doi: 10.1074/mcp.M900488-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng H, Shen N, Jimin P, Grishin NV. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 2004;13:2260–2269. doi: 10.1110/ps.04726004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishimoto LK, Ishimoto KS, Cascino A, Cipollaro M, Eiserling FA. The structure of three bacteriophage T4 genes required for tail-tube assembly. Virology. 1988;164:81–90. doi: 10.1016/0042-6822(88)90622-8. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka T, Kawano N, Oshima T. Cloning of 3-isopropylmalate dehydrogenase gene of an extreme thermophile and partial purification of the gene product. J Biochem. 1981;89:677–682. doi: 10.1093/oxfordjournals.jbchem.a133245. [DOI] [PubMed] [Google Scholar]
- 41.Carlson K, Miller ES. Molecular biology of bacteriophage T4 General Procedures. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. Washington DC: ASM Press; 1994. pp. 427–437. [Google Scholar]
- 42.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nureki O, Shirouzu M, Hashimoto K, Ishitani R, Terada T, Tamakoshi M, et al. An enzyme with a deep trefoil knot for the active-site architecture. Acta Crystallogr D Biol Crystallogr. 2002;58:1129–1137. doi: 10.1107/S0907444902006601. [DOI] [PubMed] [Google Scholar]
- 45.Tamakoshi M, Uchida M, Tanabe K, Fukuyama S, Yamagishi A, Oshima T. A new Thermus-Escherichia coli shuttle integration vector system. J Bacteriol. 1997;179:4811–4814. doi: 10.1128/jb.179.15.4811-4814.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamakoshi M, Nakano Y, Kakizawa S, Yamagishi A, Oshima T. Selection of stabilized 3-isopropylmalate dehydrogenase of Saccharomyces cerevisiae using the host-vector system of an extreme thermophile. Extremophiles. 2001;5:17–22. doi: 10.1007/s007920000168. [DOI] [PubMed] [Google Scholar]
- 47.Oshima T, Imahori K. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a non-sporulating thermophilic bacterium from a Japanese thermal spa. Int J Syst Bacteriol. 1974;24:102–112. doi: 10.1099/00207713-24-1-102. [DOI] [Google Scholar]
- 48.Tamakoshi M, Yaoi T, Oshima T, Yamagishi A. An efficient gene replacement and deletion system for an extreme thermophile, Thermus thermophilus. FEMS Microbiol Lett. 1999;173:431–437. doi: 10.1111/j.1574-6968.1999.tb13535.x. [DOI] [PubMed] [Google Scholar]
- 49.Saiki T, Kimura R, Arima K. Isolation and characterization of extremely thermophilic bacteria from hot springs. Agr Biol Chem. 1972:362357–362366. [Google Scholar]
- 50.Brock TD, Freeze H. Thermus aquaticus gen. n. and sp. n., a non-sporulating extreme thermophile. J Bacteriol. 1969;98:289–297. doi: 10.1128/jb.98.1.289-297.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.