Abstract
The chemical diversity of nature has tremendous potential for discovery of new molecular probes and medicinal agents. However, sensitivity of HTS assays to interfering components of crude extracts derived from plants, macro- and microorganisms has curtailed their use in lead discovery efforts. Here we describe a process for leveraging the concentration-response curves (CRCs) obtained from quantitative HTS to improve the initial selection of “actives” from a library of partially fractionated natural product extracts derived from marine actinomycetes and fungi. By using pharmacological activity, the first-pass CRC paradigm aims to improve the probability that labor-intensive subsequent steps of re-culturing, extraction and bioassay-guided isolation of active component(s) target the most promising strains and growth conditions. We illustrate how this process identified a family of fungal metabolites as potent inhibitors of firefly luciferase, subsequently resolved in molecular detail by x-ray crystallography.
INTRODUCTION
Natural products (NPs) are unparalleled in representation among both drugs and chemical probes, and as many as 63% of the new chemical entities for drugs discovered between 1981–2006 have been traced to or based-on these structurally diverse secondary metabolites (Carlson, 2010; Newman, 2008; Newman and Cragg, 2009). Recent studies have revealed that 34 NPs-based drugs have been approved between 1998–2007 (Butler, 2008). However, natural-derived small molecules have received less attention in recent years, due in part to the laborious process involved in identification, isolation, and follow-up compared to leads derived from synthetic libraries (Li and Vederas, 2009). Screening of NP libraries often includes a time-demanding workflow including deconvolution, and validation effort that became unsustainable when compared to the timelines achievable from HTS of synthetic chemical libraries. Paradoxically, despite the prevalence of NPs among successful drugs and powerful chemical probes, this burden resulted in the curtailing or termination of NP extract (NPE)-based discovery efforts at many pharmaceutical companies over the last decade (Harvey, 2007; Koehn and Carter, 2005; Lam, 2007; Newman, 2008).
To sample maximum chemical diversity in NPE-based screening, the library should remain sufficiently rich in chemical matter but devoid of undesirable characteristics that make it difficult to test with highly sensitive HTS assays (Harvey, 2007; Inglese et al., 2007; Koehn and Carter, 2005). For this reason we focused on NPE libraries where partial or pre-fractionation of the crude extracts was accomplished and accompanied by elution profile data. Fractionated extracts help reduce or eliminate artifacts associated with crude material. For example, crude organic solvent extracts are relatively straightforward to prepare, but the high concentrations of salts, pigments and polymeric constituents can interfere significantly with sensitive detection outputs of modern HTS. Compound-mediated phenomena complicating the interpretation of HTS data are becoming increasingly well-characterized (Baell and Holloway, 2010; Inglese et al., 2007; Thorne et al., 2010a).
To improve efficiency of NP identification, we explored a new strategy that integrates recent advances in HTS and NPE library production to enable a more efficient means of isolating active components from NP sources. Specifically we chose “pre-fractionated” NPEs from culturable microorganisms that are prepared by differential solvent extraction of initially XAD-resin bound NPEs into three “pre-fractions” that are transferred to a 384-well plate (Figure S1a). We also employed a titration-based screening paradigm, quantitative HTS (qHTS) (Inglese et al., 2006), where samples in large chemical libraries can be rapidly tested at different concentrations and concentration-response curves (CRCs) are fitted to the data. Titration of the different solvent extractions provides a further dimension of resolution where pharmacological activity of the CRCs is correlated with the NPE elution profile data. Further, in such an approach samples can be formatted into a 1536 well-based titration archive suitable for rapid screening (Figure S1b, Yasgar et al., 2008). The use of a CRC classification scheme (Inglese et al., 2006; Shukla et al., 2009), derived from qHTS to select active wells provides a unique means to track active components in association with the sample elution profile, and provides higher confidence data to ensure that typical HTS artifacts are largely avoided. A successful major effort using our screening paradigm could re-initiate and improve the application of this fertile area of natural product chemical probe and drug discovery. Here we report our methodology and initial validation involving qHTS in the testing of NPE libraries derived from Costa Rica marine microbial resources.
RESULTS
Evaluation of an NPE library by qHTS
In an effort to improve the process of identifying biological activity from NPE libraries we tested extracts in qHTS format across 35 diverse assays that were optimized for 1536-well format (Table S1). To reduce the ionic, optical and aggregation-based interference from high salt concentration, pigments, polymeric organics and other resinous materials, we prepared the extracts using XAD-resin to enrich the NPs from these genetically diverse and pure culture actinomycete and fungal strains while reducing the content of biomass at the extremes of polarity (e.g. non-organic or highly lipophilic).
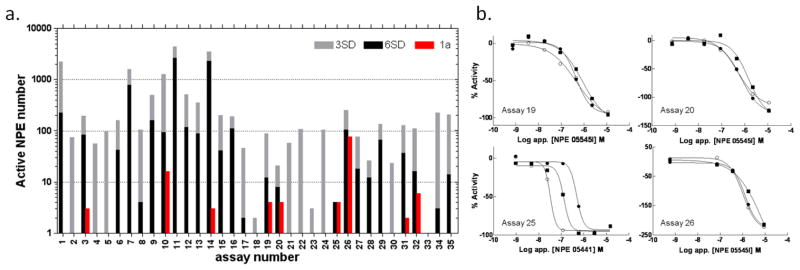
The NPE library we subsequently prepared for qHTS contained 15,704 samples that comprised 13 separate 7-concentration inter-plate titration series, such that overall 91 individual 1536-well plates were maintained as a titration archive for screening (Figure S1b). As a dynamic library that increases in size over time, a subset of the archived library (~5,300 samples) was tested in the diverse assay panel to estimate the relative activity of the extracts. By examining data from a single tested concentration, qHTS data can also be examined as a traditional single-point high throughput screen. A retrospective analysis of the activity from the preliminary testing of the NPE library across the assays in this manner resulted in a broad range of activity from none to several thousand actives (Figure 1a) at either 3 or 6 standard deviations (SD) as a cut-off for biological activity. From this subset, ‘hit’ rates spanning 0–80% for 3SD or an average of 11%, and 0–50% for 6SD or average of 4.5% were observed. The highest hit rates, those above 5% (3SD) were all limited to biochemical assays except for one that measured cellular viability (assay 26, Figure 1a). Using qHTS analysis among the assays tested, 11 gave high quality class 1a CRCs (see Figure 1a, b). These assays were comprised of two cell-based assays and nine biochemical assays that employed detection formats of fluorescence, absorbance and bioluminescence, with five of the 11 based on firefly luciferase (FLuc) outputs. Initially we decided to follow-up actives from screens employing purified molecular targets, based primarily on a high quality pharmacological response in a single assay. A particular extract (Papua New Guinea 05441) with high apparent potency 1a curves in all three solvent extracts was identified for Calmodulin Kinase IIa (assay #25, Figure 1a, b), however a re-test with a new extract from the same strain did not yield the original activity (Figure S2a), illustrating the challenges of working with uncharacterized mixtures and the importance of initial selection criteria limiting the number of follow-ups. However, several assays (#19, #20, #26) based on a common FLuc output showed CRCs from a number of NPEs suggesting to us that these NPEs potentially contained FLuc inhibitors (Figure 1 and S2b). More restricted in prevalence, however, were class 1a CRCs (e.g., Costa Rica strain 05545 and 06085). Using this observation to strengthen the commitment to proceed with de-replication, we re-cultured strain 05545 and initiated the isolation of the active constituents of this NPE (Figure 2).
Figure 1. Activity analysis of NPEs in various assays.
a. Bar chart summarizing active NPEs in assays ordered by format (1–15 are fluorogenic; 16–18 are fluorescent polarization; 19–30 are bioluminescent; 31–32 ALPHA based chemluminescent; 33–34 use fluorescent protein expression and 35 is an absorbance output). The number of active NPEs per assay are indicated on the y-axis, where red represents a class 1a CRC (as previously defined [11]), while 3SD and 6SD cutoffs for activity at a single apparent concentration of 10 μM are given by the grey and black bars. b. Plots showing qHTS class 1a CRC results from assays selected for further analysis (see also Figure S2b). Assays 19, 20 and 26 displayed activity from a common strain, NPE 05545. Additional assay details can be found Table S1
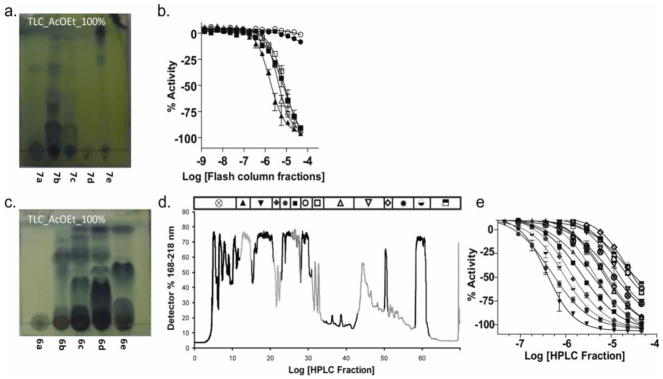
Figure 2. Bioactivity guided de-replication of NPE 05545.
a. Thin layer chromatography analysis of flash column fractions from an acetone extract of XAD-16-bound culture extract from strain 05545. b. Activity of flash column fractions shown in a in FLuc enzyme assay (open square, X+A; solid triangle, 7a; open triangle, 7b, closed square, 7c; solid circle, 7d; open circle, 7e). c. Thin layer chromatography analysis of flash column fractions from an independent culture of 05545. d. Reverse HPLC of the components of flash column fraction 6c in panel C. e. Activity from the HPLC separation shown in panel C.
Structural characterization of a series of aspulvinones
The strain 05545, a marine fungus from the genus Aspergillus (determined by 18S rRNA gene sequence analysis, data not shown) was isolated from Costa Rica marine sediments, collected in Isla Despensa, Guanacaste Conservation Area in December 2005. The terrestrial Aspergillus sp. is known by the production of pulvinone derivatives, unsaturated tetronic acids with an aryl substituent at C2, an arylmethylene substituent at C4 and isoprene residues in the aryl rings (Nobutoshi Ojima, 1975).
The marine fungus was scaled-up to 5L, and the active extracts were subjected to chromatographic separation using a combination of MPLC and HPLC (Figure 2). The fractionation of active extracts was followed by secondary bioassay to guide isolation of six new derivatives, aspulvinones I-CR to M-CR (4–9), in addition to known aspulvinones E, F and H (1–3), as well as (Hiroshi Sugiyama, 1979; Nobutoshi Ojima, 1975) butyrolactone I and III (10–11) and (K. V. Rao, 2000; Rajesh R. Parvatkar, 2009; Xuemei Niu, 2008) benzofuran (12)(Dervilla M. X. Donnelly, 1988; Hung-Yi Huang, 2008) (See Figure 3). Aspulvinone F (2) was reported in 1975 with an incorrect structure (Nobutoshi Ojima, 1975), and in 1979 it was reconsidered by Begley et al. (1979), who suggested that aspulvinone F likely bears a dihydrofuran ring instead of an epoxide. Our NMR data and X-ray crystallographic analysis reveals that the proposed revised structure is formally confirmed, and the absolute configuration of the chiral center was established as R (See Figure 4a-b).
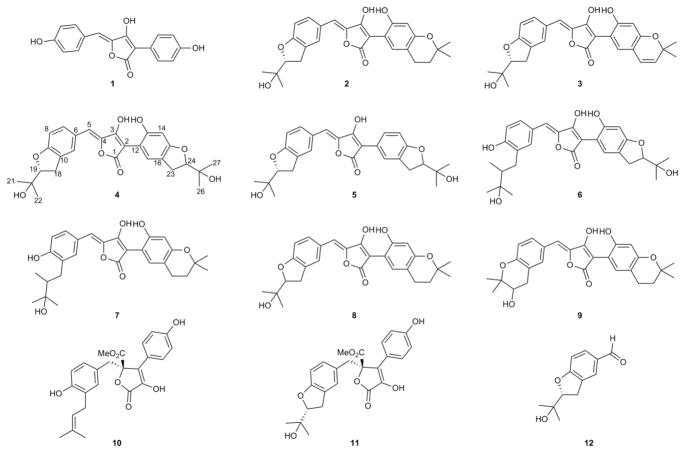
Figure 3. Structures of isolated metabolites.
Aspulvinone E (1), aspulvinone F (2), aspulvinone H (3), aspulvinone I-CR (4), aspulvinone J-CR (5), aspulvinone K-CR (6), aspulvinone L-CR (7), aspulvinone M-CR (8), aspulvinone N-CR (9), butyrolactone I (10), butyrolactone III (11) and benzofuran (12). 1H, 13C NMR, COSY and HSQC spectra for compounds 2, 4–9 are provided in Figures S4-S31 and Tables S3 and S4.
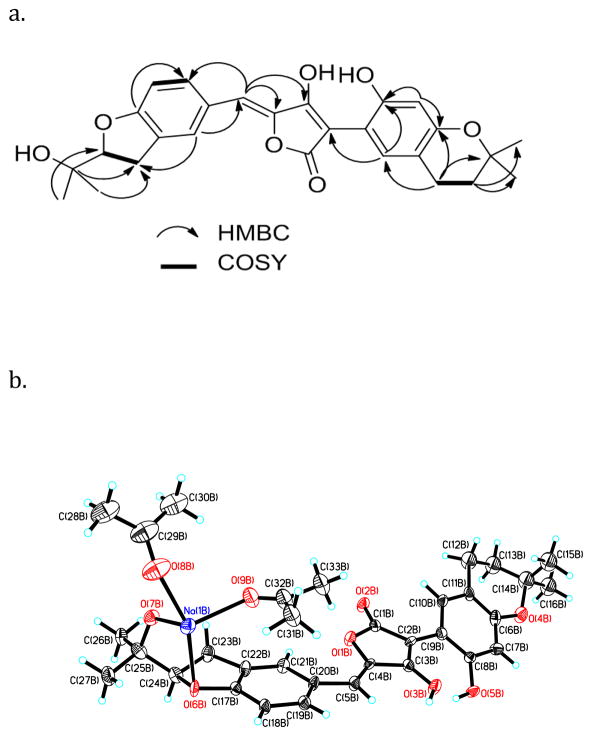
Figure 4. NMR and X-ray analysis of aspulvinone F (2).
a. Key 1H-1H COSY and HMBC correlations observed in 2, b. X-ray structure of 2. Perspective views showing 50% probability displacement ellipsoids of an independent aspulvinone F molecule. Na+ is chelated with four oxygens, two from acetone molecules and two from aspulvinone F (2). Carbon atoms are shown in black, oxygen atoms in red, protons in white, and Na+ in blue.
Aspulvinone I-CR (4) was isolated as a pale yellow solid. The HREIMS gave an [M]− ion at m/z 479.1659, consistent with the molecular formula C27H28O8, requiring 14 sites of unsaturation and 16 amu more than compound 2 (C27H28O7). The 1H NMR and 13C NMR data indicated that the structure of 4 is very similar to compound 2. The most significant differences in the NMR data reside in the high-field shift effect of the signal at C-23 (δH 3.19, δC 31.8) and the presence of a sp3 methine instead of a sp3 methylene at C-24, (δH 4.61, δC 91.4). These data indicated that compound 4 bears a dihydrofuran ring fused to the benzene ring, as opposed to the dihydropyran ring present in compound 2. This structure was further supported by COSY, HSQC and HMBC spectra. COSY correlations between H2-23 and H2-24, as well as between H2-18 and H2-19 supported the presence of two dihydrofuran ring systems. HMBC correlation from H2-18 to C-9 and C-10; H-8 to C-10; H-7 and H-11 to C-9; H2-23 to C-15 and C-17; and H-17 to C-23 confirmed the presence of a dihydrofuran fused to each of the benzene rings.
Aspulvinone J-CR (5) was obtained as a pale yellow solid. Its molecular formula C27H28O7 was established by HREIMS [M+Na]+ ion at 487.1733, indicating 14 degrees of unsaturation. The NMR features of 5 were similar to those of 4 except that the right hand benzene ring is trisubstituted, showing a sp2 methine at C-13 in 5 (δH 7.81, δC 127.8). A COSY cross-peak between H-13 and H-14 as well as HMBC correlations between H-13 and C-2, C-11 and C-15 confirmed these assignments. These assignments were re-confirmed by co-crystal structure of 5 with the FLuc (see below).
Aspulvinone K-CR (6) was isolated as a brown-yellow optically active oil. The [M+Na]+ ion at m/z 521.1788 in the HREIMS suggested C27H30O9 as the molecular formula, which indicated 18 additional amu, with one less unsaturation site than compound 4. A significant change in the 1H NMR and 13C NMR was also observed in the methine C-19, which shifted from δH 4.63 and δC 90.7 in 4 to δH 3.64 and δC 79.8 in 6. These modifications together with the shift of C-9, from δC 161.0 in 4 to δC 157.2 in 6, suggested that there are a hydroxyl and 2,3-dihydroxy-3-methylbutyl group upon C-9 and C-10, respectively, instead of the dihydrofuran ring.
Aspulvinone L-CR (7) isolated as a yellow oil, gave a [M+Na]+ ion at m/z 505.1838 in the positive ion HREIMS, consistent with the molecular formula of C27H30O8, and requiring one less unsaturation site than compound 2. NMR data of compound 7 were similar to those of compound 2 except for C-18 and C-19. The shifts of C-18 from δH 3.18 and δC 30.0 in 2 to δH 2.96, 2.66 and δC 34.8 in 7, as well as the change in C-19 from δH 4.60 and δC 89.9 in 2 to δH 3.63 and δC 81.1 in 7, suggests that in this compound the formation of the dihydrofuran ring did not occur. Instead, there is the open form where C-10 bears a 2,3-dihydroxy-3-methylbutyl group, as in compound 6.
Aspulvinone M-CR (8) was isolated as a yellow solid. The HREIMS gave an [M+Na]+ ion at m/z 503.1682 consistent with the molecular formula C27H28O8 and the same degrees of unsaturation as compound 2. The only difference compared with compound 2 was the shift of C-24 from δH 1.77 and δC 33.3 in 2 to δH 3.72 and δC 71.0 in 8 due to a secondary hydroxyl group at C-24.
Aspulvinone N-CR (9) was obtained as a yellow crystalline oil with the molecular formula C27H28O7 on the basis of HREIMS data, [M+H]+ ion at m/z 465.1913. The level of unsaturation is the same as in compound 2, but the difference in the shifts of methylene C-18 and methine C-19 indicated the presence of a dihydropyran with a hydroxyl group at C-19 instead of the dihydrofuran ring present in 2. Compared with 2, compound 9 showed shifts in the signal H2-18 from δH 3.18 to 3.03, 2.72, and H-19 signal shifted from δH 4.60 to 3.75. Also, the 13C signals of C-18 and C-19 shifted from δC 89.9 to 68.7 and from δC 71.1 to 77.2, respectively. COSY correlations between H2-18 and H-19 and HMBC correlations between H2-18 and C-9, C-10, C-11, C-19 and C-20, as well as between H-19 with C-10, C-18, C-21 and C-22 were consistent with a hydroxyl group at C-19.
Pulvinones 11–12 (Hiroshi Sugiyama, 1979; K. V. Rao, 2000; Nobutoshi Ojima, 1975; Rajesh R. Parvatkar, 2009) have been isolated from terrestrial and marine Aspergillus species, while 1–3 (Hiroshi Sugiyama, 1979; Xuemei Niu, 2008) only from the terrestrial fungus. Here we report the isolation of aspulvinones 1–9 from a marine Aspergillus sp., indicating that although derived from a completely different environment, this fungus maintains the capacity for synthesizing this class of compounds.
The aspulvinone family of natural products has a more complex carbon framework than the related pulvinones due to the incorporation of two isoprene units. The ortho position to the hydroxyl group serves as the site for alkylation by a dimethylallyl diphosphate mediated by a presumed prenyltransferase. Based on isolation of aspulvinones K-CR and L-CR (compounds 6 and 7), we surmise that prenylation is followed by epoxidation or di-hydroxylation of the double bond with subsequent cyclization, resulting in the five or six membered ring heterocycles. Whether this represents a common biosynthetic theme that includes pyran ring formation in the notoamide/stephacidin/paraherquamide class of fungal derived alkaloid natural products remains to be explored in detail (Ding et al., 2010).
Specificity of aspulvinone activity
The aspulvinones demonstrated a wide range of activity across five luciferase species and variants (Table 1). Two of these luciferases (LmLuc and UltraGlo) were related to the commonly utilized luciferase derived from Photinus pyralis (FLuc) and two were unrelated luciferases that accommodate different substrates and enzymatic mechanisms to produce bioluminescence (RLuc and VLuc). Analogs of the natural product displayed binding affinity for the related luciferases FLuc, LmLuc and the thermostable (UltraGlo, Promega) firefly luciferases in the submicromolar range while potencies decreased to undetectable for the two species of luciferases that were genetically unrelated to FLuc.
Table 1.
Aspulvinone analogs tested against five lucifease variants.
| Cmp | FLuc | LmLuc | Ultra | RLuc | VLuc |
|---|---|---|---|---|---|
| 1 | 1.4±1.6 | 3.6±1.1 | 0.76±0.12 | Inactive | >57 |
| 2 | 3.6±1.5 | 2.5±0.5 | >57 | >57 | inactive |
| 3 | 1.8±0.9 | 1.9±0.8 | 19.0±4.6 | >57 | >57 |
| 4 | 0.72±0.40 | 0.50±0.20 | 4.1±1.8 | Inactive | 12.0±2.3 |
| 5 | 0.1±0.02 | 0.1±0.03 | 4.5±3.8 | >57 | 6.5±3.3 |
| 8 | 1.7±1.6 | 5.3±5.9 | 33±2.1 | Inactive | 18±8 |
| 10 | inactive | inactive | inactive | >57 | >57 |
| 11 | inactive | inactive | inactive | Inactive | >57 |
| 12 | 18.6±8.8 | 14.8±2.8 | 31.7±3.0 | Inactive | inactive |
All values are in μM; >57, %inhibition between 20–50% at the highest tested concentration of 57 μM; inactive, %inhibition <20% observed at the highest tested concentration. Data is mean ±SD values, from at least 4 determinations. FLuc = Photinus pyralis luciferase. Related luciferases: LmLuc = Luciola mingrelica luciferase (82% identical); Ultra = Ultra-Glo luciferase (derived from Photirus pennsylvanica (68% identical); unrelated luciferases: RLuc = Renilla reniformas luciferase; VLuc = Vibrio fischeri luciferase (Ye et al., 1997)
X-ray co-crystal of aspulvinone J-CR bound to FLuc
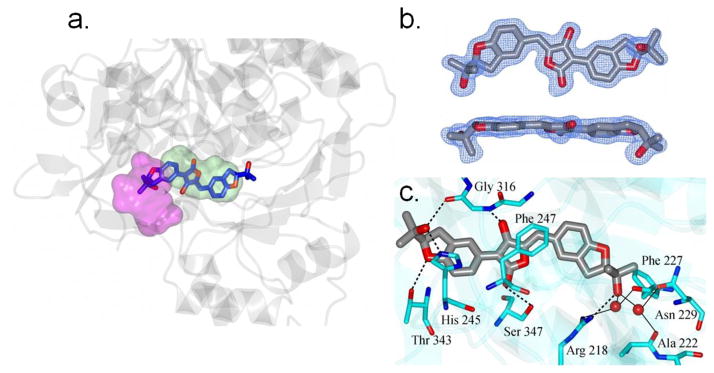
To further characterize the physical interaction of the aspulvinone series with Fluc, apo crystals of the Photinus pyralis luciferase protein were soaked in the presence of aspulvinone J-CR (5) one of the more potent aspulvinone congeners (Table 1). Following structural determination by molecular replacement and subsequent refinement to a resolution of 1.7 Å, the resulting electron density maps were examined for ligand binding. Aspulvinone J-CR (5) was clearly bound as the observed difference electron density map (Fo-Fc), which displayed prominent peaks greater than 3σ that were consistent with this compound (Figure 5b).
Figure 5. Structure of FLuc containing bound aspulvinone J-CR (5).
a. View of 5 (blue and red cylinders) in the active site of luciferase (grey ribbons). The ATP and luciferin binding regions are colored magenta and green respectively. b. Two views of the Fo-Fc electron density map for 5 contoured at 3sσ. c. Hydrogen bonding between 5 (grey/red) and luciferase (cyan). Water molecules are drawn as red spheres. Direct contacts between luciferase and 5 are shown as dashed lines and water mediated interactions are indicated by the solid lines. Crystallographic data for 5 can be found in Table S2. Coordinates and structure factors have been deposited to the Protein Databank with the accession code 3RIX.
Aspulvinone J-CR (5) fully occupies the D-luciferin binding site with the isopropyl alcohol at one terminus of the structure positioning deeply into the D-luciferin binding pocket and the other isopropyl alcohol extending into the ATP binding site (Figure 5a). The aspulvinone molecule adopts a mostly planar arrangement across the three-ring system. However, the plane of the benzofuran ring that is positioned in the D-luciferin binding region is angled 18.9° relative to the mean plane defined by the hydroxyl-butyrolactone core (Figure 5b). The benzofuran ring in the ATP binding region is only 5.8° relative to the hydroxyl-butyrolactone mean plane. The isopropyl termini are oriented on the same side of the three-ring system. In general, the driving force of aspulvinone binding to luciferase is highly enriched by hydrogen bond interactions (Figure 5c). Two oxygen atoms of the hydroxyl-butyrolactone core are able to form hydrogen bonds to the backbone –NH of Gly316 and the hydroxyl group of Ser347, with the optimal H-bonding distance of 2.73Å and 2.85Å, respectively. Notably, the isopropyl alcohol on both ends of the aspulvinone J-CR (5) structure could form tight binding with the surrounding residues through a hydrogen bond matrix. The one fitting deeply into the D-luciferin binding pocket forms a direct hydrogen bond to Arg218 and also a few water-mediated hydrogen bonds to Arg218, Ala222, Phe227 and Asn229 through two structural water molecules. In contrast, the other isopropyl alcohol extending into the AMP binding site is involved in hydrogen bonding with His245 and the backbone carbonyl oxygen of Gly316. Additionally, the oxygen of the dihydrobenzofuran ring could further help the molecule lock on the correct binding mode through the hydrogen bond interaction to Thr343.
Based on the co-crystal structure of Fluc with aspulvinone J-CR (5), we further examined aspulvinone F (2) which is ~30-fold less potent than 5 as an FLuc inhibitor (Table 1). The docking model of 2 indicated that it could adopt the same binding orientation and maintain nearly all key interactions as 5. However, the isopropyl alcohol extending into the AMP binding site was missing in 2. As this isopropyl alcohol forms hydrogen bonds with His245 and the backbone carbonyl oxygen of Gly316 in 5, the absence of such a moiety in 2 could explain this significant potency shift (Figure S32).
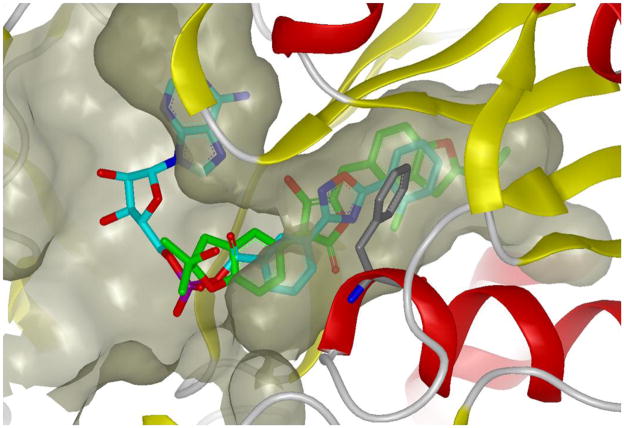
In a previous study, we elucidated inhibitor-based protein stabilization as the mechanism by which the small molecule PTC124 increases the activity of a FLuc reporter protein used in an assay designed to discover nonsense codon suppressor compounds (Auld et al., 2009a; Auld et al., 2008b). We found that PTC124 stabilized the half-life of the enzyme by reacting at the FLuc active site with ATP to form a multi-substrate adduct inhibitor (MAI), which we determined from crystallographic studies (Auld et al., 2010; Thorne et al., 2010b). This potent inhibitor can be displaced from the enzyme through the use of typical luciferase detection reagents that contain high concentrations of substrates including CoASH which presumably thiolytically cleaves the MAI (Auld et al., 2010), thus allowing detection of FLuc enzyme activity in the assay. The PTC124-AMP adduct effectively fills the active site of FLuc. Alignment of the aspulvinone co-crystal structure and the structure of PTC124-AMP adduct in the luciferase active site revealed a similar binding orientation in the D-luciferin pocket (Figure 6). The hydroxyl-butyrolactone core of aspulvinone occupies a similar position to the oxadiazole of PTC124. However it was found that the central ring system of aspulvinone J-CR forms an angle of 50.8 degrees between the mean planes defined by the butyrolactone ring (5-membered) and the aryl ring of Phe247. This is too large for a π-π (face-to-face, quadrupole-quadrupole) interaction or the aromatic stacking observed in the PTC124-AMP-FLuc structure, and more closely approximates a herringbone (edge-to-face, dipole-quadrupole) interaction (Burley and Petsko, 1985, 1988). The planar nature of both ligands makes the molecules well accommodated in the unusually long and linear binding pocket of D-luciferin.
Figure 6. Superposition of aspulvinone J-CR (5) and PTC124-AMP adduct within FLuc binding pocket.
Protein is shown in ribbon representation and the binding pocket is depicted by molecular surface, 5 is shown in green and PTC124-AMP is shown in cyan. Phe247 is shown in grey. This figure was prepared with the program VIDA (OpenEye Scientific Software).
DISCUSSION
Selection of initial NPEs for bioassay-guided fractionation were considered carefully given the considerable effort in dereplication required for each follow-up. Our primary consideration was an apparent pharmacological response as indicated by high quality CRCs from one or more of the three solvent extracts. Data of this character was significantly more discriminating than activity based on a threshold inhibition (e.g., compare analysis of % activity vs high quality curve class activity in Figure 1a) where even a low false positive rate can lead to unacceptable confirmation rates. In addition, given the rare and unique nature of the NPE samples, we employed the qHTS approach to measure the activity in as comprehensive a manner as possible. Comparison of assays using a common detection technology allowed us to flag extracts that interfered with the detection output and also enabled the identification of the specific FLuc inhibitors shown in this study. However, in general the diversity of detection modalities, assay formats and conditions employed across the sample of MLPCN assays surveyed here limited our ability to make optimal use of qHTS data to ascertain whether a CRC was the result of a pharmacologically-mediated event or resulted from compound-mediated interference in the assay format or detection modality (Inglese et al., 2006; Thorne et al., 2010b).
Based on these findings, it is evident that another criterion for follow-up should be whether an NPE is either independently observed in several cross-validating or orthogonal assay formats having a common target, or is differentially active within a series of targets tested in a common assay platform. Coupled with a qHTS approach assay panels that provide such a reinforcing or contrasting result should further improve the efficiency and probability of identifying isolable active compounds. In the present study this approach lead us to three assays in which high quality CRCs were observed where the common target was firefly luciferase. Dereplication of the active extract proceeded in a straightforward manner leading to the isolation of several new aspulvinone congeners. Follow-up studies against a series of bioluminescent enzymes demonstrated that these molecules are specific for bioluminescent enzymes related to the wild-type variety of firefly luciferases. The lower overall activity against UltraGlo luciferase is consistent with a previous study that suggested a more restricted inhibitor profile with this thermostable optimized luciferase (Auld et al., 2009b).
The molecular basis for the high potency and selectivity of aspulvinone J-CR (5) toward FLuc is revealed in the X-ray co-crystal structure. Here a flat planar structure that binds largely to the D-luciferin pocket but also can span into the AMP pocket is observed. The general structural architecture of the aspulvinone core bears some resemblance to the diaryl oxidazole found in molecules like PTC124, which bind to the luciferin pocket of FLuc, both using a heteroaromatic five membered ring to bridge two substituted phenyl moieties. Unlike PTC124 however, 5 does not form an MAI through reaction with ATP. In fact ATP is not bound to FLuc in the presence of 5 presumably due to protrusion of the isopropyl alcohol of 5 into space normally occupied by the terminal phosphoryl groups of ATP. Further, the deep pocket present within FLuc, which is not fully occupied by either luciferin or PTC124 is exploited by 5 and evident by the extension of the second isopropyl alcohol on the opposite end of 5 reaching into this deep pocket (see Figure 6). These findings illustrate the chemical diversity capable of interacting with this pocket and help to further explain the frequent occurrence of small molecule modulators of FLuc identified in chemical screening libraries.
In future uses of NPE libraries we are exploring incorporation of a titration-based screening paradigm across common assay platforms, for example, in biochemical target-based screens across gene families (Veith et al., 2009) or in cell-based assays using the same detection output (Yuan et al., 2009). Such data would be augmented when properly designed assay panels are configured that differentiate between NPEs bearing a technically-based inhibition pattern vs. NPEs that are genuinely active on a biologically relevant mechanism (e.g. contain orthogonal assay pairs; Inglese et al., 2007). The combination of qHTS with panels containing both parallel and orthogonally designed assays should facilitate the identification of natural products selective for the target or cellular pathway being explored from NPE libraries, in the same way this strategy is used for synthetic chemical libraries (Johnson et al., 2009; Xia et al., 2009).
We view this study as a work in progress aimed at defining an improved process for exploring of the vast potential of natural products held within complex mixtures. Focusing here on the use of microbial derived NPs pre-fractionated to remove the majority of HTS-interfering components, we have employed the pharmacological resolving power of qHTS to reveal apparent IC50s from these mixtures. By using a pattern of high quality CRCs (curve class 1a) we demonstrated that a reproducible activity could be targeted from among many “active” NPEs and reduced to a candidate NPE for follow-up dereplication. Incorporating the wealth of metadata associated with NPEs can further enhance the CRC information. Metadata information includes strain, growth conditions, broth or cellular fraction, and solvent extraction method, which can all be used to guide the choice of fractions for follow-up. For this purpose, we developed a browser to help visualize and track the metadata information (see Methods for full description). In this study we were able to identify a series of aspulvinones that selectively inhibited the P. pyralis FLuc, showing that the widespread modulation of FLuc activity observed in synthetic chemical libraries (Auld et al., 2008a) extends to NPEs as well. By applying the strategy we have outlined here we are now in a position to improve the efficiency of NPE screening in drug discovery applications across a wide range of human and animal disease targets.
SIGNIFICANCE
The natural world remains a source of great chemical diversity producing compounds demonstrated to have tremendous medicinal value. However, their use in discovery efforts has been decreasing because HTS approaches that may be sufficient for testing of pure synthetic compounds remain suboptimal for complex mixtures such as NPEs. For example, typical extracts may harbor optically active pigments with relatively low concentrations of bioactive substances. This interference can prevent effective testing of crude NPEs at high concentrations in sensitive fluorescent or luminescent HTS-based assays. Therefore, both the preparation of the NPE and enhancements to the screening paradigm is needed. Increasing efficiency for assessing the biological activity of secondary metabolites derived from plants, macro- and microorganisms will likely re-energize their use in lead discovery efforts. Here, a library of pre-fractionated NPEs lead to rapid identification of active natural product molecules. Quantitative HTS (qHTS) enabled effective investigation of the extracts as the technique integrates testing of a concentration series that can accommodate relatively high apparent uM concentrations for the detection of low abundance compounds. NPEs containing relevant bioactive molecules were revealed by sigmoidal CRCs and readily discriminated from inactive samples. Inclusion of cross-validation assays for a specific pathway or target, or a common assay platform for testing multiple cell lines or targets improved identification of target-directed or target-selective NPEs, respectively. Given the effort required to process active fractions derived from NPEs, the general approach described here should facilitate the discovery of novel natural products employing on-going advances in assay development and functional genomics.
METHODS
Preparation of NPE Libraries and Control Plates
Isolated strains from Costa Rican marine sediments were cultured (100) ml in two different media (see below). After cell growth, broth was separated by centrifugation and agitated by shaking overnight with amberlite XAD-16, which was further extracted with CH3OH, acetone and EtOAc sequentially. The NPEs were transferred to the NIH Chemical Genomics Center (NCGC) on dry ice in 384-well plates containing 60μL of a 15mg/mL solution in DMSO. NCGC reformatted the extracts in the HTS compound management core using the qHTS dilution paradigm (Yasgar et al., 2008) resulting in an inter-plate or ‘through-plate’ titration series in 1536-well format in DMSO (Figure S1b). Briefly, 50 384-well NPE library plates were titrated using a 1:5 dilution factor, to generate a 7-point titration series. The resulting 350 384-well plates were compressed to 91 1536-well plates for screening, with the first four columns of each plate reserved for assay specific controls. Unique identifiers were auto-generated by registering the sample IDs and strain origins from an accompanying data file into ActivityBase (ID Business Solutions Ltd., Guildford, UK). ActivityBase was also used to track the sample locations in the barcoded 384-well plates.
Controls were added from columns 1–4 of a separate 1536-well compound plate. Depending on the assay titrations, max/min and/or neutral controls are used (e.g., col 1–2, 16-point titrations in duplicate of test compounds starting at 20 mM in DMSO); column 3, neutral control (DMSO); column 4, control inhibitor (20 mM resveratrol).
qHTS and Follow-up Data Analysis
The NPE 1536-well titration archive was tested against assays of the MLPCN using an automated qHTS process described previously (Inglese et al., 2006; Michael et al., 2008) and described further in the Supplemental Information.
Curve Classification Analysis
The curve classification used is the same as the one described elsewhere (Inglese et al., 2006). Briefly, CRCs are categorized into four classes. Class 1 contains complete CRC having both upper and lower asymptotes and r2 values >0.9. Class 2 contains incomplete concentration-response curves lacking the high concentration asymptote and shows r2 values >0.9. Being supported by a single concentration having activity, Class 3 curves are considered low confidence. For Class 3 the minimal acceptable activity is set at 3 SD of the mean activity calculated as described above. Curves are classified as negative or positive, depending on whether they exhibit a signal decrease (apparent inhibition) or increase (apparent activation). Finally, class 4 contains compounds that do not show any activity that define a CRC and are therefore classified as inactive. Active compounds were identified as a range of curve classes from 1 through 3 to select for compounds showing signal decreases.
Luciferase assays
Vibrio fischeri luciferase (VLuc) was purchased from Sigma (cat# L8507) which is a partially purified preparation of VLuc that contains the necessary NADH/NADPH-dependent FMN reductases. Luciferase from Luciola mingrelica (LmLuc) was purchased from Sigma (cat# L4899). Purified wild-type luciferase from Photinus pyralis (FLuc) was obtained from Sigma (cat. no. L9506) and purified Ultra-Glo luciferase was obtained from Promega (Auld et al., 2009b). Renilla reniformis luciferase (RLuc) was purchased from Nanolight.
The 1536-well RLuc and FLuc assays were performed as previously described (Auld et al., 2009a; Auld et al., 2008b) using either 5μM coelenterazine or 10 μM D-luciferin and ATP, respectively. The assay for LmLuc used the same protocol as FLuc with a final concentration of 5 nM LmLuc enzyme. The assay for Ultra-Glo also used 10 μM D-luciferin and ATP and was performed as described (Auld et al., 2009b). The assay for the FMN-dependent luciferase VLuc followed the procedure described in Lavi et al. (Lavi et al., 1981) but used a miniaturized assay volume of 4 μL in 1536-well plates. For the assay, 2 μL of substrate buffer (0.1 M sodium phosphate pH 7.0, 0.005% decanal, 0.1% BSA, 0.1 mM NADH, and 2 μM FMN) was dispensed into Kalypsys white medium binding 1536-well plates. Compounds were then added using a Kalypsys pintool. The assay was started by adding enzyme buffer (0.1 M sodium phosphate pH 7.0, 0.2 μM VLuc) and the luminescent signal was read immediately on a Perkin Elmer Viewlux (180 seconds exposure, 6x binning).
Fungal Isolation, Identification and Cultivation
The fungus Aspergillus sp. (strain 05545) was isolated from a marine sponge collected in 2005 in La Cruz, Costa Rica, by J. Cortez (authorization accession permit R-CM-08-2006-OT for samples donated by CIMAR to INBio, collection permit 106-2005-SINAC). The isolated fungus was preserved with 20% glycerol at −80 °C. It was cultured in ISP2 media (10 g of malt extract, 4g of yeast extract, 4g of dextrose and 30g of NaCl per liter of distilled water) at 28 °C on a rotary shaker at 200 rpm in five replicates of 2.5 L Fernbach flasks, each containing 1 L of culture media
Extraction and Isolation
After eight days of culture, Aspergillus sp. (strain 05545) fungus cells were harvested by centrifugation at 3000 × g, sonicated and extracted with MeOH. The culture broth was extracted by adding 20 g/L amberlite XAD-16. After shaking overnight, the resin was collected and washed with 1L of DI water before it was sequentially extracted with methanol, acetone and ethyl acetate. The resulting extracts were filtered and concentrated to obtain 1.82 g, 0.87 g and 0.26 g of crude extract, respectively. The MeOH extract was subjected to successive chromatography procedures. Firstly a medium-pressure reversed-phase LOBAR B (Ø=25 mm × 310 mm) LiChroprep column was used, eluted with MeOH-H2O (7:3) to yield three active fractions, b, c and d. Fractions b (539 mg), c (510 mg) and d (364 mg) were further subjected to HPLC purification on a XBridge Prep C18 column (Ø=10 mm × 250 mm) with CH3CN-H2O gradient. The subfractions were further purified on the same column using phase MeOH-H2O gradient as the mobile phase to give: aspulvinone E (1, 3.2 mg), aspulvinone F (2, 40 mg), aspulvinone H (3, 6 mg), aspulvinone I-CR (4, 13 mg), aspulvinone J-CR (5, 4.2 mg), aspulvinone K-CR (6, 7.4 mg), aspulvinone L-CR (7, 1.8 mg), aspulvinone M-CR (8, 2 mg), aspulvinone N-CR (9, 5 mg), butyrolactone I (10, 1.6 mg), butyrolactone III (11, 6.6 mg) and benzofuran (12, 1.8 mg).
General Experimental Procedures for Natural Product Structure Elucidation
Optical rotations were measured on an Autopol III polarimeter. IR spectra were recorded with a Perkin Elmer BX FT-IR infrared spectrometer using NaCl plates. Mass spectra were carried out with a Micromass AutoSpec Ultima Magnetica mass spectrometer. 1H, 13C and 2D NMR spectra were recorded in CD3OD on a Varian INOVA 600 MHz NMR spectrometer at 600 MHz for 1H NMR and 150 MHz for 13C, using TMS as an internal standard. Medium pressure chromatography was performed using pre-packed columns, including LOBAR® GRÖBE B (Ø 25 mm × 310 mm) LiChroprep RP-18 from MERCK®. HPLC separations were performed on a Beckman Coulter system (Fullerton, CA) equipped with a diode-array detector, using a XBridge Prep C18 column (Ø=10 mm × 250 mm). TLC was performed using TLC silica gel 60 F254 from EMD, and plates were visualized by spraying with PMA and heating. Specific characterization and spectral data for Aspulvinone F (2), Aspulvinone I-CR (4), Aspulvinone J-CR (5), Aspulvinone K-CR (6), Aspulvinone L-CR (7), Aspulvinone M-CR (8), and Aspulvinone N-CR (9), are provided in Supplemental Information.
Crystallographic Data of Aspulvinone F-CR (2)
Yellow, C132H156Na4O36, monoclinic, space group P21, a = 21.470(6) Å, b = 10.861(3) Å, c = 27.520(7) Å, β = 93.705(4)°, V = 6404(3) Å3, Z = 2, crystal size 0.32 × 0.28 × 0.08 mm3. The intensities of the reflections were collected by means of a Bruker APEX II CCD diffractometer (MoKα radiation, λ=0.71073 Å), and equipped with an Oxford Cryosystems nitrogen flow apparatus. The collection method involved 0.5° scans in ω at 26° in 2θ. Data integration down to 0.82 Å resolution was carried out using a Bruker SAINT V7.46 A diffractometer (Madison, WI) with reflection spot size optimization. Absorption corrections were made with the Bruker program TWINABS (Madison, WI). The structure was solved by the direct methods procedure and refined by least-squares methods again F2 using SHELXS-97 and SHELXL-97 (Sheldrick, 2008). Non-hydrogen atoms were refined anisotropically, and hydrogen atoms were allowed to ride on the respective atoms. The Ortep plots were produced with SHELXL-97 program, and the other drawings were produced with Accelrys DS Visualizer 2.0 (San Diego, CA).
Co-crystallization and Data Collection for FLuc-Aspulvinone J-CR
Luciferase from Photinus pyralis was purchased from Sigma (Cat. # L9506, Lot # 017K74201) concentrated to 10.2 mg/mL in (200mM (NH4)2SO4, 1mM EDTA, 1mM DTT, 10% glycerol, 25% ethylene glycol, 25mM Tris pH 7.8) as previously described (Conti et al., 1996). Apo crystals were obtained at 4 °C in Clover Jr. (Emerald BioSystems) sitting drop plates using equal volumes of protein and crystallization solution (25% (v/v) PEG 400, 20% (v/v) PEG 3350, 0.1 M MgCl2, 0.1 M Tris pH 8.5, Precipitant Synergy #41, Emerald BioSystems) equilibrated against 100 μL of the latter. Apo crystals were soaked for 2.5 hours in the presence of 10 mM NCGC183923 (Aspulvinone J-CR (5)) and 10 mM NCGC183923 (Aspulvinone J-CR (5))/2.5 mM ATP dissolved in crystallization solution. Diffraction data were collected at 100K at the Advanced Photon Source (APS) IMCA-CAT beamline 17ID using a Dectris Pilatus 6M pixel array detector.
Structure Solution and Refinement
Diffraction data were integrated and scaled with XDS (Kabsch, 1988) and Scala (Evans, 2006) respectively. The protein only coordinates from a previously determined luciferase structure (using PDB: 3IES) was used for initial refinement against the processed diffraction data. Refinement and model building were carried out with Phenix (Adams, 2010) and Coot (Emsley, 2004) respectively. Disordered side chain residues were truncated to the point to which electron density could be observed. Structure validation was conducted with Molprobity (Lovell, 2003) and figures were prepared with CCP4mg (Potterton, 2004). Following initial refinement, prominent electron density (Fo-Fc) consistent with NCGC183923 was observed in the active site for the inhibitor and inhibitor/ATP soaked crystals. However, no electron density consistent with ATP was observed for the latter sample. Final structure refinement was conducted using the diffraction data collected for the NCGC183923/ATP soaked crystal as it diffracted to the highest resolution. Crystallographic data are provided in Table S2.
NPE Informatics Browser
Our current primary screening data visualization platform is a two-stage solution implemented in LabVIEW (Austin, TX): a parser that collates the qHTS data by microbial strain variant, and a browser that displays the collated data as a global overview of assays tested against the NPE collection, see
Supplementary Material
Highlights.
Quantitative high-throughput screening of natural product extracts
Retrospective single concentration HTS analysis across 35 assays
Strategy for identification, isolation and structural elucidation of new secondary metabolites
Co-crystal structure of potent natural product inhibitor with firefly luciferase
Acknowledgments
The authors gratefully acknowledge Shao-Liang Zheng for the aspulvinone F X-ray diffraction studies and Christopher Rath for assistance with mass spectra interpretation (University of Michigan). At the NCGC, we acknowledge Sam Michael and Paul Shinn for automation and sample management support, and Adam Yasgar, Anton Simeonov, Ron Johnson and Wei Zheng for examining NPEs in several MLPCN assays. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the University of Kansas COBRE Protein Structure Laboratory was supported by NIH Grant Number P20 RR-17708 from the National Center for Research Resources. This work was supported by the NIH Roadmap for Medical Research (J.I. & D.S.A.), NIH grant U01 TW007404 as part of the International Cooperative Biodiversity Group initiative at the Fogarty International Center, and the Hans W. Vahlteich Professorship (D.H.S.). P.G.C. gratefully acknowledges the Spanish Foundation of Science and Technology (FECYT) for a postdoctoral fellowship.
Footnotes
Supporting Information Available. 1H, 13C NMR, COSY and HSQC spectra for compounds 1, 4–9 are provided in Figures S4-S31. Coordinates and structure factors of the 1.7 Å resolution structure of a firefly luciferase-aspulvinone J-CR (5) inhibitor complex are available at PDB ID code 3RIX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld DS, Lovell S, Thorne N, Lea WA, Maloney DJ, Shen M, Rai G, Battaile KP, Thomas CJ, Simeonov A, et al. Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124. Proc Natl Acad Sci U S A. 2010;107:4878–4883. doi: 10.1073/pnas.0909141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld DS, Southall NT, Jadhav A, Johnson RL, Diller DJ, Simeonov A, Austin CP, Inglese J. Characterization of chemical libraries for luciferase inhibitory activity. J Med Chem. 2008a;51:2372–2386. doi: 10.1021/jm701302v. [DOI] [PubMed] [Google Scholar]
- Auld DS, Thorne N, Maguire WF, Inglese J. Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression. Proc Natl Acad Sci U S A. 2009a;106:3585–3590. doi: 10.1073/pnas.0813345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld DS, Thorne N, Nguyen DT, Inglese J. A specific mechanism for nonspecific activation in reporter-gene assays. ACS Chem Biol. 2008b;3:463–470. doi: 10.1021/cb8000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld DS, Zhang YQ, Southall NT, Rai G, Landsman M, Maclure J, Langevin D, Thomas CJ, Austin CP, Inglese J. A Basis for Reduced Chemical Library Inhibition of Firefly Luciferase Obtained from Directed Evolution. J Med Chem. 2009b;52:1450–1458. doi: 10.1021/jm8014525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Begley MJ, Gedge DR, Knight DW, Pattenden G. Re-investigation of Structures by X-Ray Crystallographic and Spectroscopic Analysis. J C S Perkin I; 1979. Aspulvinones, a New Class of Natural Products from Aspergillus terreus; pp. 77–83. [Google Scholar]
- Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Burley SK, Petsko GA. Weakly polar interactions in proteins. Adv Protein Chem. 1988;39:125–189. doi: 10.1016/s0065-3233(08)60376-9. [DOI] [PubMed] [Google Scholar]
- Butler MS. Natural products to drugs: natural product-derived compounds in clinical trials. Nat Prod Rep. 2008;25:475–516. doi: 10.1039/b514294f. [DOI] [PubMed] [Google Scholar]
- Carlson EE. Natural products as chemical probes. ACS Chem Biol. 2010;5:639–653. doi: 10.1021/cb100105c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Lloyd LF, Akins J, Franks NP, Brick P. Crystallization and preliminary diffraction studies of firefly luciferase from Photinus pyralis. Acta crystallographica. 1996;52:876–878. doi: 10.1107/S0907444996002405. [DOI] [PubMed] [Google Scholar]
- Donnelly Dervilla MX, NF, Kouno Isao, Martin Maria, O’Reilly Joseph. Dihydrobenzofurans from Heterobasidion Annosum. Phytochemistry. 1988;27:2709–2713. [Google Scholar]
- Ding Y, de Wet JR, Cavalcoli J, Li S, Greshock TJ, Miller KA, Finefield JM, Sunderhaus JD, McAfoos TJ, Tsukamoto S, et al. Genome-based characterization of two prenylation steps in the assembly of the stephacidin and notoamide anticancer agents in a marine-derived Aspergillus sp. J Am Chem Soc. 2010;132:12733–12740. doi: 10.1021/ja1049302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley PaKC. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta crystallographica. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Harvey AL. Natural products as a screening resource. Curr Opin Chem Biol. 2007;11:480–484. doi: 10.1016/j.cbpa.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Sugiyama Hiroshi, NO, Kobayashi Masaki, Senda Yasuhisa, Ishiyama Junichi, Seto Shuichi. C-13 NMR Spectra of Aspulvinones. Agric Biol Chem. 1979;43:403–404. [Google Scholar]
- Huang Hung-Yi, TI, Peng Chien-Fang, Tsai Ian-Lih, Chen Ih-Sheng. Constituents of the Root Wood of Zanthoxylum wutaiense with Antitubercular Activity. J Nat Prod. 2008;71:1146–1151. doi: 10.1021/np700719e. [DOI] [PubMed] [Google Scholar]
- Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Huang R, Jadhav A, Southall N, Wichterman J, MacArthur R, Xia M, Bi K, Printen J, Austin CP, et al. A quantitative high-throughput screen for modulators of IL-6 signaling: a model for interrogating biological networks using chemical libraries. Mol Biosyst. 2009;5:1039–1050. doi: 10.1039/b902021g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao KV, AKS, Veerender M, Ravikumar V, Mohan EVS, Dhanvantri SD, Sitaramkumar M, Moses Babu J, Vyas K, OM Reddy G. Butyrolactones from Aspergillus terreus. Chem Pharm Bull. 2000;48:559–562. doi: 10.1248/cpb.48.559. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic indexing of rotation diffraction patterns. Journal of Applied Crystallography. 1988;21:67–72. [Google Scholar]
- Koehn FE, Carter GT. Rediscovering natural products as a source of new drugs. Discov Med. 2005;5:159–164. [PubMed] [Google Scholar]
- Lam KS. New aspects of natural products in drug discovery. Trends Microbiol. 2007;15:279–289. doi: 10.1016/j.tim.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Lavi JT, Lövgren TNE, Raunio RP. Comparison of luminous bacteria and their bioluminescence-linked enzyme activities. FEMS Microbiology Letters. 1981;11:197–199. [Google Scholar]
- Li JW, Vederas JC. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- Lovell SC, et al. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Michael S, Auld D, Klumpp C, Jadhav A, Zheng W, Thorne N, Austin CP, Inglese J, Simeonov A. A robotic platform for quantitative high-throughput screening. Assay Drug Dev Technol. 2008;6:637–657. doi: 10.1089/adt.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ. Natural products as leads to potential drugs: an old process or the new hope for drug discovery? J Med Chem. 2008;51:2589–2599. doi: 10.1021/jm0704090. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural product scaffolds as leads to drugs. Future Med Chem. 2009;1:1415–1427. doi: 10.4155/fmc.09.113. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981–2002. J Nat Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- Ojima Nobutoshi, STaSS Structures of pulvinone derivatives from Aspergillus Terreus. Phytochemistry. 1975;14:573–576. [Google Scholar]
- Potterton L, et al. Developments in the CCP4 molecular-graphics project. Acta crystallographica. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- Parvatkar Rajesh R, CDS, Tripathi Ashootosh, Naik Chandrakant G. Aspernolides A and B, butenolides from a marine-derived fungus Aspergillus terreus. Phytochemistry. 2009;70:128–132. doi: 10.1016/j.phytochem.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Shukla SJ, Nguyen DT, Macarthur R, Simeonov A, Frazee WJ, Hallis TM, Marks BD, Singh U, Eliason HC, Printen J, et al. Identification of pregnane X receptor ligands using time-resolved fluorescence resonance energy transfer and quantitative high-throughput screening. Assay Drug Dev Technol. 2009;7:143–169. doi: 10.1089/adt.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne N, Auld DS, Inglese J. Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr Opin Chem Biol. 2010a;14:315–324. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne N, Inglese J, Auld DS. Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chem Biol. 2010b;17:646–657. doi: 10.1016/j.chembiol.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith H, Southall N, Huang R, James T, Fayne D, Artemenko N, Shen M, Inglese J, Austin CP, Lloyd DG, et al. Comprehensive characterization of cytochrome P450 isozyme selectivity across chemical libraries. Nat Biotechnol. 2009;27:1050–1055. doi: 10.1038/nbt.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Huang R, Guo V, Southall N, Cho MH, Inglese J, Austin CP, Nirenberg M. Identification of compounds that potentiate CREB signaling as possible enhancers of long-term memory. Proc Natl Acad Sci U S A. 2009;106:2412–2417. doi: 10.1073/pnas.0813020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Xuemei, HMD, Menzel Klaus-Dieter, Lozach Olivier, Walther Grit, Meijer Laurent, Grabley Susanne, Sattler Isabel. Butyrolactone I Derivatives from Aspergillus terreus Carrying an Unusual Sulfate Moiety. J Nat Prod. 2008;71:689–692. doi: 10.1021/np070341r. [DOI] [PubMed] [Google Scholar]
- Yasgar A, Shinn P, Jadhav A, Auld D, Michael S, Zheng W, Austin CP, Inglese J, Simeonov A. Compound Management for Quantitative High-Throughput Screening. JALA (Charlottesville, Va) 2008;13:79–89. doi: 10.1016/j.jala.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Buck LM, Schaeffer HJ, Leach FR. Cloning and sequencing of a cDNA for firefly luciferase from Photuris pennsylvanica. Biochim Biophys Acta. 1997;1339:39–52. doi: 10.1016/s0167-4838(96)00211-7. [DOI] [PubMed] [Google Scholar]
- Yuan J, Johnson RL, Huang R, Wichterman J, Jiang H, Hayton K, Fidock DA, Wellems TE, Inglese J, Austin CP, et al. Genetic mapping of targets mediating differential chemical phenotypes in Plasmodium falciparum. Nat Chem Biol. 2009;5:765–771. doi: 10.1038/nchembio.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.