Abstract
Selective breeding of sheep for arginine (R) at prion gene (PRNP) codon 171 confers resistance to classical scrapie. However, other effects of 171R selection are uncertain. Ovine progressive pneumonia/Maedi-Visna virus (OPPV) may infect up to 66% of a flock thus any affect of 171R selection on OPPV susceptibility or disease progression could have major impact on the sheep industry. Hypotheses that the PRNP 171R allele is 1) associated with the presence of OPPV provirus and 2) associated with higher provirus levels were tested in an Idaho ewe flock. OPPV provirus was found in 226 of 358 ewes by quantitative PCR. The frequency of ewes with detectable provirus did not differ significantly among the 171QQ, 171QR, and 171RR genotypes (p > 0.05). Also, OPPV provirus levels in infected ewes were not significantly different among codon 171 genotypes (p > 0.05). These results show that, in the flock examined, the presence of OPPV provirus and provirus levels are not related to the PRNP 171R allele. Therefore, a genetic approach to scrapie control is not expected to increase or decrease the number of OPPV infected sheep or the progression of disease. This study provides further support to the adoption of PRNP 171R selection as a scrapie control measure.
Introduction
Scrapie is the prototypical prion disease and one of several described in animals and humans. Accumulation of disease associated prion protein (PrPSc), an abnormally folded form of normal host prion protein (PrPC), is central to disease and expression of the host prion gene (PRNP) is necessary in pathogenesis [1]. PRNP open reading frame (ORF) variants associate with disease incubation time [2] and relative disease susceptibility in sheep [3-7], goats [8-10], elk [11-13], deer [12,14] and humans [15-18].
Polymorphisms in sheep at PRNP codons 136 (Alanine/Valine), 154 (Arginine/Histidine), and 171 (Glutamine/Arginine) are involved in scrapie susceptibility (for review see [19]). Codon 171 is an important element of susceptibility in the United States (US) sheep population [6,7]. Sheep homozygous for glutamine at codon 171 (171QQ) are highly susceptible to Scrapie, whereas sheep heterozygous (171QR) or homozygous (171RR) for arginine are highly resistant to classical strains of US Scrapie.
The PRNP 171Q allele predominates in US sheep whereas the 171R allele and 171RR genotype are less common (the latter two occur at a frequency of about 37% and 16%, respectively [20]). Selective breeding for the 171R minor allele to produce animals with the 171QR or 171RR genotypes is sometimes used as a Scrapie control measure, however the functional consequences of 171R selection on other traits is uncertain. Genetic selection may have unexpected positive or negative effects as individual genes may have multiple biological roles (pleiotropy) or may be linked to other genes that impact overall biological functions. Uncertainty regarding PRNP selection effects (beyond Scrapie resistance) has led to investigation of multiple ovine traits related to reproduction, milk, meat, fiber and genetic diversity. However, PRNP selection effects on disease susceptibility (besides Scrapie) has only been studied for Salmonella resistance [21].
Ovine progressive pneumonia/Maedi-Visna virus (OPPV) is a monocyte/macrophage tropic lentivirus (a subclass of retrovirus) endemic in many US sheep flocks and causes pneumonia, mastitis, arthritis and encephalitis. One in five sheep are infected based on detection of anti-OPPV serum antibodies and seroprevalence can be as high as 66% in open rangeland environments [22,23]. As many as 76% of OPPV seropositive sheep may develop OPPV related diseases [24]. OPPV quantitative PCR (qPCR) is an alternative method to detect lentivirus and provides both diagnostic and prognostic information [25-27]. The qPCR assay measures the presence and amount of virus that has been reverse-transcribed and integrated into the host genome (provirus). The technique is a useful indicator of disease progression in the study of OPPV because OPPV provirus levels correlate with the severity of pulmonary lesions [28,29].
Scrapie is diagnosed in about one of every 500 culled sheep [20] thus OPPV has much greater prevalence. Uncertainty regarding whether PRNP selection would effect OPPV provirus levels can create producer reluctance to the implementation of 171R selection when OPPV is a more severe flock-health problem. A prion-retrovirus pathogenic relationship of undetermined mechanisms has been observed between PrPSc and Murine Leukemia Virus (MuLV) [30], PrPSc and Caprine Arthritis Encephalitis Virus (CAEV) [J Stanton, personal communication], PrPSc and mastitis presumptively caused by OPPV [31], and influence of PrPc expression on HIV infection [32]. In this study, the following two hypotheses were tested in an Idaho ewe flock: 1) the PRNP codon 171R allele is associated with the presence of OPPV provirus and 2) the PRNP 171R allele is associated with higher OPPV provirus levels. This study will help guide producer decisions and it provides information for future prion-retrovirus co-infection studies and advances knowledge of whether PRNP selection affects other infectious diseases.
Methods
Animals
Three hundred fifty eight ewes were sampled from a flock in southeastern Idaho in which OPPV is endemic and there are no reported cases of scrapie. Animals were cared for under guidelines of the United States Sheep Experimental Station Institutional Care and Use Committee. Breeding was performed without prior selection of prion genotype. The sample set was composed of 117 Columbia, 116 Polypay, and 125 Rambouillet sheep. Ages were three, four, five and six years with 39, 30, 31, and 17 Columbia; 27, 31, 33, and 25 Polypay; and 32, 32, 36, and 25 Rambouillet, respectively.
Nucleic acid extraction
Peripheral blood leukocytes (PBL) were isolated from whole blood as described [23]. Genomic DNA was extracted from PBL using a commercial kit (Gentra, Minneapolis, Minnesota).
PRNP Genotyping
DNA amplification and sequencing of the ovine PRNP ORF was performed similarly to previous experiments using forward primer 5'-GGCATTTGATGCTGACACC-3' and reverse primer 5'-TACAGGGCTGCAGGTAGAC-3' [33]. Reverse primer 5'-GGTGGTGACTGTGTGTTGCTGA-3' was used for standard dideoxynucleotide sequencing. All sequencing was performed at the Laboratory for Biotechnology and Bioanalysis (Washington State University, Pullman, WA). PRNP genotypes were analyzed using commercial software (Vector NTI, Invitrogen; Carlsbad, CA or Lasergene Seqman Pro v7.1, DNAstar, Inc, Madison, WI) and codon variants reported by single letter code (e.g. glutamine Q, arginine R, valine V, histidine, H, leucine L, phenylalanine F).
OPPV quantitative PCR
PPV provirus level was determined using a previously described quantitative real-time PCR (qPCR) assay [23]. The OPPV qPCR used primers TMENVCONf 5'-TCA TAG TGC TTG CTATCA TGG CTA-3' and TMENVCONr 5'-CCG TCC TTG TGT AGG ATT GCT-3' (Invitrogen Corporation, Carlsbad, CA) and a Taqman 5'-5'-hexachlorofluorescein-AGC AAC ACC GAG ACC AGC TCC TGC-3' Black Hole Quencher-1 probe (Integrated DNA Technologies, Coralville, IA) targeting the highly conserved transmembrane region within the envelope gene of the North American OPPV strains [34].
Statistical analyses
Two types of genotypic comparison were made using provirus data and PRNP genotype, with a minimum PRNP allele frequency of 10% required for analysis. Association between PRNP genotype and presence or absence of OPPV provirus was tested using logistic regression models from the logistic procedure of SAS v9.1 (SAS Institute, Cary, NC). Association between PRNP genotype and the level of logarithm (base 10)-transformed provirus in OPPV positive animals was tested using the glm procedure in SAS v9.1. In each case the association model included breed as a categorical predictor, age as a linear covariate, the interaction between breed and age, and the PRNP genotype of interest. Adjusted odds ratios and 95% confidence interval were calculated for the pair-wise comparison of the frequency of OPPV positive ewes in each PRNP genotype. Adjusted mean log-transformed provirus levels were reported with 95% confidence intervals. Stepdown Bonferroni p-value correction [35] was applied separately to each set of analyses.
Results
Distribution of PRNP genotypes
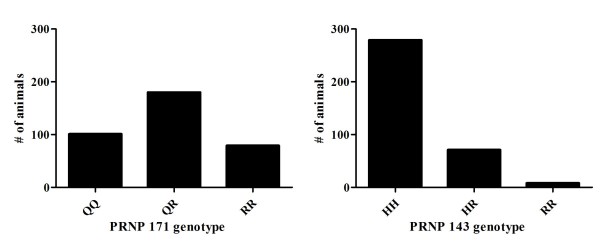
The PRNP genotypes were determined as the first step in testing association with the presence of OPPV provirus and OPPV provirus levels. PRNP ORF coding variants were identified at codons 101(Q/R), 136(A/V), 141(L/F), 143 (H/R), 154 (R/H), and 171 (Q/R) (Table 1). Of the 358 sheep sampled, 100 (28%) were 171QQ, 179 (50%) were 171QR and 79 (22%) were 171RR, providing a representation of all three genotypes (Fig. 1, left). Examination of the 171R allele relative to the overall PRNP ORF showed that in all animals with the 171RR genotype there were no other PRNP codon variants present. Codon changes at other positions only occurred in animals that had at least one wild type 171Q allele. Of the 358 sheep, 279 (78%) were 143HH, 71 (20%) were 143HR and 8 (2%) were 143RR (Fig. 1, right). Since codons 143 and 171 had amino acid substitutions with a minor allele frequency of at least 10% they were further analyzed, except for the rare 143RR genotype. Codons 101, 136, 141, and 154 had a minor allele frequency of less than 10% and therefore these four codons were excluded from further association analysis.
Table 1.
Distribution of PRNP ORF codon variants among individual breeds and in cumulative sample set
| PRNP genotype | Columbia | Polypay | Rambouillet | Total |
|---|---|---|---|---|
| 101QQ | 96 | 115 | 112 | 323 |
| 101QR | 21 | 1 | 12 | 34 |
| 101RR | 0 | 0 | 1 | 1 |
| 136AA | 97 | 116 | 123 | 336 |
| 136AV | 20 | 0 | 2 | 22 |
| 136VV | 0 | 0 | 0 | 0 |
| 141LL | 94 | 110 | 112 | 316 |
| 141LF | 23 | 5 | 13 | 41 |
| 141FF | 0 | 1 | 0 | 1 |
| 143 HH | 63 | 110 | 106 | 279 |
| 143 HR | 46 | 6 | 19 | 71 |
| 143 RR | 8 | 0 | 0 | 8 |
| 154RR | 106 | 114 | 98 | 318 |
| 154RH | 11 | 2 | 26 | 39 |
| 154HH | 0 | 0 | 1 | 1 |
| 171 QQ | 55 | 13 | 32 | 100 |
| 171 QR | 56 | 51 | 72 | 179 |
| 171 RR | 6 | 52 | 21 | 79 |
Figure 1.
Number of sheep distributed among PRNP genotypes. Left = codon 171, Right = codon 143, y-axis = number of animals.
Frequency of OPP provirus among PRNP genotype
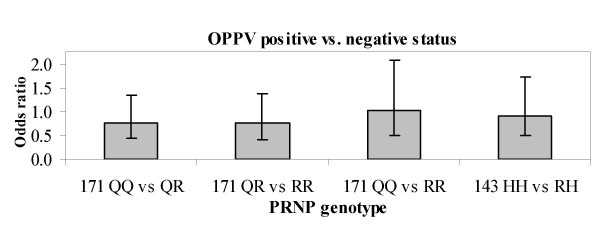
The presence or absence of OPPV provirus was compared among the PRNP 171 and PRNP 143 genotypes, using a statistical model accounting for age and breed, to determine if minor alleles within those genotypes affected the number of sheep that had detectable OPPV provirus. In the flock, 226 of 358 (63.1%) sheep had detectable OPPV provirus. Over half of the ewes were positive for OPPV provirus within each PRNP 171 or 143 genotype (Table 2). The frequency of OPPV positive animals was not significantly different between the 171QQ, QR, and RR genotypes as indicated by nominal and corrected p-values greater than 0.05 (Table 3) and equivalent odds ratios (Fig. 2). The 95% confidence intervals also indicate the range of potential effect sizes consistent with these data (Fig. 2). Also, the frequency of OPPV positive animals did not differ significantly between the 143HH and HR genotypes.
Table 2.
Number of ewes with (positive) or without (negative) detectable OPPV provirus among PRNP genotypes used for statistical comparison
| OPPV Provirus Status | % OPPV | ||
|---|---|---|---|
| PRNP genotype | negative | positive | positive |
| 171 QQ | 36 | 64 | 64.0 |
| 171 QR | 61 | 118 | 65.9 |
| 171 RR | 35 | 44 | 55.7 |
| 143 HH | 103 | 176 | 63.1 |
| 143 HR | 26 | 45 | 63.4 |
Table 3.
Significance level for effect of PRNP genotype upon frequency of animals with detectable OPPV provirus
| OPPV positive vs negative p-value | ||
|---|---|---|
| Genotype comparison | nominal | corrected |
| 171 QQ vs. QR | 0.23 | 0.90 |
| 171 QR vs. RR | 0.23 | 0.90 |
| 171 QQ vs. RR | 0.60 | 1.00 |
| 143 HH vs. RH | 0.78 | 1.00 |
P-values are before (nominal, left) and after (corrected, right) step-down Bonferroni multiple test correction
Figure 2.
Odds ratio and 95% confidence interval for effect of PRNP genotype upon frequency of OPPV positive animals.
OPPV provirus levels among PRNP genotypes
The levels of OPPV provirus were compared among the PRNP 171 and PRNP 143 genotypes to determine whether particular genotypes were associated with higher or lower provirus levels once a ewe became infected. Adjusted mean log-transformed provirus levels with 95% confidence interval were equivalent among codon 171 and among codon 143 genotypes (Fig. 3). Adjusted mean log-transformed provirus levels were not significantly different among the 171QQ, QR, and RR genotypes or among the 143HH and HR genotypes in which nominal and corrected p-values were greater than 0.05 (Table 4).
Figure 3.
Adjusted mean log10 provirus levels and 95% confidence interval among PRNP genotypes used for statistical comparison.
Table 4.
Significance level of OPPV proviral load levels between PRNP genotypes
| OPPV load p-value | ||
|---|---|---|
| Genotype comparison | nominal | Corrected |
| 171 QQ vs. QR | 0.07 | 0.27 |
| 171 QR vs. RR | 0.34 | 1.00 |
| 171 QQ vs. RR | 0.60 | 1.00 |
| 143 HH vs. RH | 0.27 | 1.00 |
p-values are before (nominal, left) and after (corrected, right) step-down Bonferroni multiple test correction
Discussion
The present study was performed to determine if a PRNP 171R selection program impacts the presence or magnitude of OPPV infection. Allelic variation in PRNP could affect OPPV status if PRNP variants produce changes in PrPc function or expression level relevant to OPPV, if PRNP is a pleiotropic gene, or if there are other molecules involved in prion pathogenesis that also affect OPPV pathogenesis. Alternatively, there may be nearby chromosomal regions affecting OPPV pathogenesis that are in linkage disequilibrium with certain PRNP alleles including, but not limited to, variants of PRNP promoter regions or PRNP homologues. However, the lack of association between PRNP genotype and OPPV status in this study indicates that the presence of a specific PRNP genotype does not influence the presence or magnitude of OPPV infection in this flock.
The study demonstrated that the frequency of sheep with detectable OPPV provirus was not related to the PRNP 171R (or 143R) allele in an Idaho ewe flock. This suggests that it is no more likely that a 171RR or 171QR sheep within a flock would become infected when compared to a 171QQ sheep. Likewise, the data suggest there is no difference in frequency of infection between the 143HH and 143HR sheep. Only ewes were sampled in this study so it is possible that introduction of rams could have a different affect, however it is unlikely considering that the frequency of OPPV in rams is equivalent, or perhaps lower than OPPV frequency in ewes [36,22].
Also, provirus levels in OPPV positive animals were not related to the PRNP 171R and 143R alleles. Thus, PRNP selection should not affect progression of disease once animals become infected with OPPV. A shift of flock genetics to a greater frequency of 171QR or 171RR sheep is unlikely to accelerate shedding or transmission of OPPV. In these sheep there also was no difference in provirus levels between animals of the 143 HH and 143HR genotypes, thus there are no documented cases where PRNP genotypes impact OPPV infection.
Recent studies have shown that factors such as breed and age are important for OPPV, therefore all analyses in this study accounted for breed, age and differences in how each breed handled OPPV with age. For example, Rambouillet ewes are less likely to be positive for OPPV provirus than Columbia ewes and Rambouillet ewes can also better control OPPV provirus levels than either Columbia or Polypay ewes [23,37]. Further, these breed differences can change over time as some breeds show increasing provirus levels with age while others do not [37]. However, all the analyses in this study accounted for age and breed in the association models so that these factors would not influence tests for association with PRNP genotype.
Interactions between retrovirus' and normal or abnormal prion protein have been previously observed. The current findings do not exclude the possibility that increases in ovine PrPc or CD230 expression could alter OPPV replication as observed in a human cell line where over-expression of human PrPc thwarted HIV-1 replication [32]. OPPV replicates in mammary macrophages and microglia and transmits via ewe milk [38-40] and PrPSc is found in macrophages of lymphoid follicles and microglia and transmits via ewe milk [41-44,31] thereby suggesting functional overlap between host proteins involved in both prion and lentivirus pathogenesis. Additional links between prion and retrovirus' are indicated by data showing that caprine arthritis-encephalitis virus (CAEV) aids PrPd accumulation in immortalized microglia in vitro [J Stanton, personal communication] and that scrapie infection increases MuLV expression and reciprocally MuLV accelerates scrapie pathogenesis [30].
This study is one of many examining PRNP selection effects. The PRNP 171RR genotype has no apparent effect on reproductive performance [45,46], ovulation rates and litter sizes [47], and only the Suffolk breed has lower lamb weaning weights [48]. Milk production and quality is not effected in Churra [49], East Friesian milk sheep [46] or Sardinian sheep and there are no significant changes in udder morphology [50]. Carcass and wool quality are not impaired [46,21] and 171R may positively affect average daily gain [51]. 171R has no effect on Salmonella resistance [21]. Finally, pedigree examination in Laxta Black Faced-type Navarra sheep showed no overall negative effect [52].
The present study taken together with previous investigations indicate that the correlated responses to PRNP 171R selection should be minimal. In total, ten different studies examining reproduction, meat, milk, fiber and infectious disease traits in a dozen different breeds found no overt negative effect from the PRNP 171R allele or 171RR genotype. Additional studies may supplement present and previous results by examining other breeds, genotypes, retrovirus strains, diseases, environmental or management conditions, or production traits. This investigation of a flock with endemic OPPV shows that the frequency of OPPV infection and level of OPPV provirus loads are not affected by the PRNP 171R allele (occurring either in the 171QR heterozygous or 171RR homozygous genotypes) and supports PRNP 171R selection as a component of Scrapie control programs.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RDH designed the study, performed sequence analysis, determined genotype distribution and frequencies, participated in statistical analysis, and drafted the manuscript. LHH participated in experimental design, developed and performed the RT-PCR assay, performed sequence analysis, and assisted in drafting the manuscript. SNW participated in experimental design, performed statistical analysis, and assisted in drafting the manuscript. KIOR participated in experimental design, performed sequence analysis, and provided editorial revisions to intellectual content. DPK participated in experimental design and provided editorial revisions to intellectual content. All authors read and approved the final manuscript.
Contributor Information
Robert D Harrington, Email: rdh@vetmed.wsu.edu.
Lynn M Herrmann-Hoesing, Email: lherrman@vetmed.wsu.edu.
Stephen N White, Email: swhite@vetmed.wsu.edu.
Katherine I O'Rourke, Email: korourke@vetmed.wsu.edu.
Donald P Knowles, Email: dknowles@vetmed.wsu.edu.
Acknowledgements
We are grateful to Liam Broughton, Lowell Kappmeyer, Linda Hamburg, Codie Hanke, and Marta Henrikkson for expert technical assistance. We thank the staff of the USDA-Agricultural Research Service National Sheep Experiment Station, Dubois, ID, USA for providing blood samples. This work was supported by USDA CRIS #5348-32000-025-00D.
References
- Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- Dickinson AG, Meikle VM, Fraser H. Identification of a gene which controls the incubation period of some strains of scrapie agent in mice. J Comp Pathol. 1968;78:293–299. doi: 10.1016/0021-9975(68)90005-4. [DOI] [PubMed] [Google Scholar]
- Bossers A, de Vries R, Smits MA. Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. J Virol. 2000;74:1407–1414. doi: 10.1128/JVI.74.3.1407-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossers A, Schreuder BE, Muileman IH, Belt PB, Smits MA. PrP genotype contributes to determining survival times of sheep with natural scrapie. J Gen Virol. 1996;77(Pt 10):2669–2673. doi: 10.1099/0022-1317-77-10-2669. [DOI] [PubMed] [Google Scholar]
- Hunter N, Foster JD, Goldmann W, Stear MJ, Hope J, Bostock C. Natural scrapie in a closed flock of Cheviot sheep occurs only in specific PrP genotypes. Arch Virol. 1996;141:809–824. doi: 10.1007/BF01718157. [DOI] [PubMed] [Google Scholar]
- O'Rourke KI, Holyoak GR, Clark WW, Mickelson JR, Wang S, Melco RP, Besser TE, Foote WC. PrP genotypes and experimental scrapie in orally inoculated Suffolk sheep in the United States. J Gen Virol. 1997;78(Pt 4):975–978. doi: 10.1099/0022-1317-78-4-975. [DOI] [PubMed] [Google Scholar]
- Westaway D, Zuliani V, Cooper CM, Da Costa M, Neuman S, Jenny AL, Detwiler L, Prusiner SB. Homozygosity for prion protein alleles encoding glutamine-171 renders sheep susceptible to natural scrapie. Genes Dev. 1994;8:959–969. doi: 10.1101/gad.8.8.959. [DOI] [PubMed] [Google Scholar]
- Acutis PL, Bossers A, Priem J, Riina MV, Peletto S, Mazza M, Casalone C, Forloni G, Ru G, Caramelli M. Identification of prion protein gene polymorphisms in goats from Italian scrapie outbreaks. J Gen Virol. 2006;87:1029–1033. doi: 10.1099/vir.0.81440-0. [DOI] [PubMed] [Google Scholar]
- Papasavva-Stylianou P, Kleanthous M, Toumazos P, Mavrikiou P, Loucaides P. Novel polymorphisms at codons 146 and 151 in the prion protein gene of Cyprus goats, and their association with natural scrapie. Vet J. 2007;173:459–462. doi: 10.1016/j.tvjl.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Vaccari G, Di Bari MA, Morelli L, Nonno R, Chiappini B, Antonucci G, Marcon S, Esposito E, Fazzi P, Palazzini N. et al. Identification of an allelic variant of the goat PrP gene associated with resistance to scrapie. J Gen Virol. 2006;87:1395–1402. doi: 10.1099/vir.0.81485-0. [DOI] [PubMed] [Google Scholar]
- Hamir AN, Gidlewski T, Spraker TR, Miller JM, Creekmore L, Crocheck M, Cline T, O'Rourke KI. Preliminary observations of genetic susceptibility of elk (Cervus elaphus nelsoni) to chronic wasting disease by experimental oral inoculation. J Vet Diagn Invest. 2006;18:110–114. doi: 10.1177/104063870601800118. [DOI] [PubMed] [Google Scholar]
- Johnson C, Johnson J, Clayton M, McKenzie D, Aiken J. Prion protein gene heterogeneity in free-ranging white-tailed deer within the chronic wasting disease affected region of Wisconsin. J Wildl Dis. 2003;39:576–581. doi: 10.7589/0090-3558-39.3.576. [DOI] [PubMed] [Google Scholar]
- O'Rourke KI, Besser TE, Miller MW, Cline TF, Spraker TR, Jenny AL, Wild MA, Zebarth GL, Williams ES. PrP genotypes of captive and free-ranging Rocky Mountain elk (Cervus elaphus nelsoni) with chronic wasting disease. J Gen Virol. 1999;80:2765–2769. doi: 10.1099/0022-1317-80-10-2765. [DOI] [PubMed] [Google Scholar]
- O'Rourke KI, Spraker TR, Hamburg LK, Besser TE, Brayton KA, Knowles DP. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J Gen Virol. 2004;85:1339–1346. doi: 10.1099/vir.0.79785-0. [DOI] [PubMed] [Google Scholar]
- Bishop MT, Hart P, Aitchison L, Baybutt HN, Plinston C, Thomson V, Tuzi NL, Head MW, Ironside JW, Will RG. et al. Predicting susceptibility and incubation time of human-to-human transmission of vCJD. Lancet Neurol. 2006;5:393–398. doi: 10.1016/S1474-4422(06)70413-6. [DOI] [PubMed] [Google Scholar]
- Cervenakova L, Goldfarb LG, Garruto R, Lee HS, Gajdusek DC, Brown P. Phenotype-genotype studies in kuru: implications for new variant Creutzfeldt-Jakob disease. Proc Natl Acad Sci USA. 1998;95:13239–13241. doi: 10.1073/pnas.95.22.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JE, Baybutt HN, Everington D, Will RG, Ironside JW, Manson JC. PRNP contains both intronic and upstream regulatory regions that may influence susceptibility to Creutzfeldt-Jakob disease. Gene. 2002;288:139–146. doi: 10.1016/S0378-1119(02)00466-3. [DOI] [PubMed] [Google Scholar]
- Zeidler M, Stewart G, Cousens SN, Estibeiro K, Will RG. Codon 129 genotype and new variant CJD. Lancet. 1997;350:668. doi: 10.1016/S0140-6736(05)63366-1. [DOI] [PubMed] [Google Scholar]
- O'Rourke KI. Ovine scrapie. New tools for control of an old disease. Vet Clin North Am Food Anim Pract. 2001;17:283–300. vi. [PubMed] [Google Scholar]
- USDA Phase II. Scrapie: Ovine Slaughter Surveillance Study 2002–2003. USDA: APHIS: VS, CEAH, National Animal Health Monitoring System. Fort Collins, CO; 2003. [Google Scholar]
- Vitezica ZG, Moreno CR, Lantier F, Lantier I, Schibler L, Roig A, Francois D, Bouix J, Allain D, Brunel JC. et al. Quantitative trait loci linked to PRNP gene controlling health and production traits in INRA 401 sheep. Genet Sel Evol. 2007;39:421–430. doi: 10.1051/gse:2007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutlip RC, Lehmkuhl HD, Sacks JM, Weaver AL. Seroprevalence of ovine progressive pneumonia virus in sheep in the United States as assessed by analyses of voluntarily submitted samples. Am J Vet Res. 1992;53:976–979. [PubMed] [Google Scholar]
- Herrmann-Hoesing LM, White SN, Lewis GS, Mousel MR, Knowles DP. Development and validation of an ovine progressive pneumonia virus quantitative PCR. Clin Vaccine Immunol. 2007;14:1274–1278. doi: 10.1128/CVI.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie SJ, Pearson LD, Zink MC, Bickle HM, Anderson BC, Marcom KA, DeMartini JC. Ovine lentivirus expression and disease. Virus replication, but not entry, is restricted to macrophages of specific tissues. Am J Pathol. 1995;146:250–263. [PMC free article] [PubMed] [Google Scholar]
- Arens M. Use of probes and amplification techniques for the diagnosis and prognosis of human immunodeficiency virus (HIV-1) infections. Diagn Microbiol Infect Dis. 1993;16:165–172. doi: 10.1016/0732-8893(93)90016-Z. [DOI] [PubMed] [Google Scholar]
- Verhofstede C, Reniers S, Van Wanzeele F, Plum J. Evaluation of proviral copy number and plasma RNA level as early indicators of progression in HIV-1 infection: correlation with virological and immunological markers of disease. AIDS. 1994;8:421–1427. doi: 10.1097/00002030-199410000-00008. [DOI] [PubMed] [Google Scholar]
- Vitone F, Gibellini D, Schiavone P, Re MC. Quantitative DNA proviral detection in HIV-1 patients treated with antiretroviral therapy. J Clin Virol. 2005;33:194–200. doi: 10.1016/j.jcv.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Brodie SJ, Marcom KA, Pearson LD, Anderson BC, de la Concha-Bermejillo A, Ellis JA, DeMartini JC. Effects of virus load in the pathogenesis of lentivirus-induced lymphoid interstitial pneumonia. J Infect Dis. 1992;166:531–541. doi: 10.1093/infdis/166.3.531. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Watt NJ, Hopkins J, Harkiss G, Woodall CJ. Quantitative analysis of maedi-visna virus DNA load in peripheral blood monocytes and alveolar macrophages. J Virol Methods. 2000;86:13–20. doi: 10.1016/S0166-0934(99)00169-X. [DOI] [PubMed] [Google Scholar]
- Lee KH, Jeong BH, Jin JK, Meeker HC, Kim JI, Carp RI, Kim YS. Scrapie infection activates the replication of ecotropic, xenotropic, and polytropic murine leukemia virus (MuLV) in brains and spinal cords of senescence-accelerated mice: implication of MuLV in progression of scrapie pathogenesis. Biochem Biophys Res Commun. 2006;349:122–130. doi: 10.1016/j.bbrc.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Ligios C, Sigurdson CJ, Santucciu C, Carcassola G, Manco G, Basagni M, Maestrale C, Cancedda MG, Madau L, Aguzzi A. PrPSc in mammary glands of sheep affected by scrapie and mastitis. Nat Med. 2005;11:1137–1138. doi: 10.1038/nm1105-1137. [DOI] [PubMed] [Google Scholar]
- Leblanc P, Baas D, Darlix JL. Analysis of the interactions between HIV-1 and the cellular prion protein in a human cell line. J Mol Biol. 2004;337:1035–1051. doi: 10.1016/j.jmb.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Schneider DA, Yan H, Fry LM, Alverson J, White SN, O'Rourke IK. Myenteric neurons of the ileum that express somatostatin are a target of prion neuroinvasion in an alimentary model of sheep scrapie. Acta Neuropathol. 2008;115:651–661. doi: 10.1007/s00401-008-0374-2. [DOI] [PubMed] [Google Scholar]
- Herrmann LM, Hotzel I, Cheevers WP, On Top KP, Lewis GS, Knowles D. Seven new ovine progressive pneumonia virus (OPPV) field isolates from Dubois Idaho sheep comprise part of OPPV clade II based on surface envelope glycoprotein (SU) sequences. Virus Res. 2004;102:215–220. doi: 10.1016/j.virusres.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective bonferroni test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- Arsenault J, Dubreuil P, Girard C, Simard C, Belanger D. Maedi-visna impact on productivity in Quebec sheep flocks (Canada) Prev Vet Med. 2003;59:125–137. doi: 10.1016/S0167-5877(03)00086-2. [DOI] [PubMed] [Google Scholar]
- Herrmann-Hoesing LM, White SN, Mousel MR, Lewis GS, Knowles DP. Ovine progressive pneumonia provirus levels associate with breed and Ovar-DRB1. Immunogenetics. 2008;60(12):749–758. doi: 10.1007/s00251-008-0328-9. [DOI] [PubMed] [Google Scholar]
- Carrozza ML, Mazzei M, Bandecchi P, Arispici M, Tolari F. In situ PCR-associated immunohistochemistry identifies cell types harbouring the Maedi-Visna virus genome in tissue sections of sheep infected naturally. J Virol Methods. 2003;107:121–127. doi: 10.1016/S0166-0934(02)00208-2. [DOI] [PubMed] [Google Scholar]
- Ebrahimi B, Allsopp TE, Fazakerley JK, Harkiss GD. Phenotypic characterisation and infection of ovine microglial cells with Maedi-Visna virus. J Neurovirol. 2000;6:320–328. doi: 10.3109/13550280009030758. [DOI] [PubMed] [Google Scholar]
- Herrmann-Hoesing LM, Palmer GH, Knowles DP. Evidence of proviral clearance following postpartum transmission of an ovine lentivirus. Virology. 2007;362:226–234. doi: 10.1016/j.virol.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Andreoletti O, Levavasseur E, Uro-Coste E, Tabouret G, Sarradin P, Delisle MB, Berthon P, Salvayre R, Schelcher F, Negre-Salvayre A. Astrocytes accumulate 4-hydroxynonenal adducts in murine scrapie and human Creutzfeldt-Jakob disease. Neurobiol Dis. 2002;11:386–393. doi: 10.1006/nbdi.2002.0558. [DOI] [PubMed] [Google Scholar]
- Caplazi P, O'Rourke K, Wolf C, Shaw D, Baszler TV. Biology of PrPsc accumulation in two natural scrapie-infected sheep flocks. J Vet Diagn Invest. 2004;16:489–496. doi: 10.1177/104063870401600601. [DOI] [PubMed] [Google Scholar]
- Herrmann LM, Cheevers WP, Davis WC, Knowles DP, O'Rourke KI. CD21-positive follicular dendritic cells: A possible source of PrP(Sc) in lymph node macrophages of scrapie-infected sheep. Am J Pathol. 2003;162:1075–1081. doi: 10.1016/S0002-9440(10)63904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konold T, Moore SJ, Bellworthy SJ, Simmons HA. Evidence of scrapie transmission via milk. BMC Vet Res. 2008;4:14. doi: 10.1186/1746-6148-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander BM, Stobart RH, Moss GE. Scrapie resistance and production traits in Rambouillet rams: Ram performance test 2002–2006. Res Vet Sci. 2008;85(2):345–348. doi: 10.1016/j.rvsc.2007.11.004. [DOI] [PubMed] [Google Scholar]
- De Vries F, Hamann H, Drogemuller C, Ganter M, Distl O. Analysis of associations between the prion protein genotypes and production traits in East Friesian milk sheep. J Dairy Sci. 2005;88:392–398. doi: 10.3168/jds.S0022-0302(05)72699-0. [DOI] [PubMed] [Google Scholar]
- Sweeney T, Hanrahan JP, O'Doherty E. Is there a relationship between prion protein genotype and ovulation rate and litter size in sheep? Anim Reprod Sci. 2007;101:153–157. doi: 10.1016/j.anireprosci.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Alexander BM, Stobart RH, Russell WC, O'Rourke KI, Lewis GS, Logan JR, Duncan JV, Moss GE. The incidence of genotypes at codon 171 of the prion protein gene (PRNP) in five breeds of sheep and production traits of ewes associated with those genotypes. J Anim Sci. 2005;83:455–459. doi: 10.2527/2005.832455x. [DOI] [PubMed] [Google Scholar]
- Alvarez L, Gutierrez-Gil B, San Primitivo F, de la Fuente LF, Arranz JJ. Influence of prion protein genotypes on milk production traits in Spanish Churra sheep. J Dairy Sci. 2006;89:1784–1791. doi: 10.3168/jds.S0022-0302(06)72247-0. [DOI] [PubMed] [Google Scholar]
- Salaris S, Casu S, Carta A. Investigating the relationship between the prion protein locus and udder morphology traits and milk yield in Sardinian sheep. J Anim Sci. 2007;85:2840–2845. doi: 10.2527/jas.2006-610. [DOI] [PubMed] [Google Scholar]
- Evoniuk JM, Berg PT, Johnson ML, Larson DM, Maddock TD, Stoltenow CL, Schauer CS, O'Rourke KI, Redmer DA. Associations between genotypes at codon 171 and 136 of the prion protein gene and production traits in market lambs. Am J Vet Res. 2007;68:1073–1078. doi: 10.2460/ajvr.68.10.1073. [DOI] [PubMed] [Google Scholar]
- Alfonso L, Parada A, Legarra A, Ugarte E, Arana A. The effects of selective breeding against scrapie susceptibility on the genetic variability of the Latxa Black-Faced sheep breed. Genet Sel Evol. 2006;38:495–511. doi: 10.1051/gse:2006017. [DOI] [PMC free article] [PubMed] [Google Scholar]