Abstract
To determine the genetic architecture of trout in Albania, 87 individuals were collected from 19 riverine and lacustrine sites in Albania, FYROM and Greece. All individuals were analyzed for sequence variation in the mtDNA control region. Among fourteen haplotypes detected, four previously unpublished haplotypes, bearing a close relationship to haplotypes of the Adriatic and marmoratus lineages of Salmo trutta, were revealed. Ten previously described haplotypes, characteristic of S. ohridanus, S. letnica and the Adriatic and Mediterranean lineages of S. trutta, were also detected. Haplotypes detected in this study were placed in a well supported branch of S. ohridanus, and a cluster of Mediterranean – Adriatic – marmoratus haplotypes, which were further delimited into three subdivisions of Mediterranean, marmoratus, and a previously non-described formation of four Adriatic haplotypes (Balkan cluster). Haplotypes of the Balkan cluster and the other Adriatic haplotypes, do not represent a contiguous haplotype lineage and appear not to be closely related, indicating independent arrivals into the Adriatic drainage and suggesting successive colonization events. Despite the presence of marmoratus haplotypes in Albania, no marbled phenotype was found, confirming previously reported findings that there is no association between this phenotype and marmoratus haplotypes.
Introduction
Major European peninsulas are known to have played a central role in the survival of animal and plants during ice-age maxima and have received a high degree of attention in terms of conservation of endemic taxa [1,2]. Compared with the Iberian and Italian peninsulas, the biodiversity and rich level of phenotypic variability present in the Balkan Peninsula have only recently been investigated by molecular techniques (e.g. [3-5]). As one of the 17 biodiversity hotspots of the world [6], this peninsula harbours numerous endemic taxa [4], including members of the genus Salmo (subsequently referred to as Balkan trout), which are especially diverse in this region. Many studies on the morphology and phenotypes of the fish of the Balkans were undertaken during the last century (e.g. [7,8]) and found high levels of endemism among Balkan trout. As a consequence, given the benefit of availability of modern molecular techniques, a number of recent studies have focused on revisiting Balkan trout taxonomy, population structure and demographic history [9-14]. However, much remains to be done, as the status of several nominal species and populations of Balkan trout remains unresolved, mainly as a result of the region's geographical, political and cultural isolation [15].
Considerable variation in external morphology of Balkan trout was reported in early studies [16,17], giving rise to many taxonomic units (see [18] for review). Recent molecular studies of trout from Bosnia-Herzegovina, Montenegro and FYROM [10,13,14] have confirmed this diversity. However, little clear association between phenotype and genotype has been found, and some well-established taxonomic groups, such as S. marmoratus [10], have been found not to be associated with detected genetic assemblages.
Several Salmo taxa have been reported to inhabit Albanian rivers and neighbouring drainages in FYROM and Greece. Examples include S. farioides, proposed by Karaman [17], and S. ohridanus, S. letnica, S. letnica lumi, S. trutta, S. macrostigma, S. peristericus, S. marmoratus and S. montenegrinus [19,18,21]. Unfortunately, confirmation of these observations and the continued existence of such trout in these waters, as well as their taxonomic status, remain uncertain, representing an absence from any comprehensive overview of Balkan trout demography, evolution and classification.
The data that do exist on trout in Albania are very scarce and mostly stem from an inventory of fishes undertaken in the country in the 1950s [22], or are restricted to certain areas (e.g. [21] on Lake Ohrid; [23] on the River Shkumbini). Rakaj [24] extended and brought up to date the work of Poljakov et al. [22] on Albanian ichthyofauna. He described trout from the rivers Shala and Valbona (Ohrid-Drin-Shkodra system; see also [25]) as well as from the lakes Shkodra and Ohrid, while trout have also been reported to exist within the rivers Bistrica [24], Cemit [24], Mati [20] and Shkumbini [23].
Very few genetic analyses of Albanian trout have been performed so far and all are restricted to lakes Ohrid and Prespa [26,13,27].
As inferred from several previous studies on Balkan trout [28,10], anthropogenically induced hybridisation, particularly with introduction of non-native trout lineages, has had a considerable impact on many indigenous trout stocks and has blurred the picture of the original genetic structure and phylogeography of Balkan trout. However, because of Albania's past political isolation and low level of economic development, it is probable that stocking with non-native strains of brown trout (e.g. Atlantic lineage) has not been performed here (I Wilson, personal communication). Therefore, despite any impact of over-fishing and intense poaching (authors' personal observations) on the population sizes of native trout, the present distribution and composition of trout in Albania may relatively faithfully reflect the natural situation, a rare situation for salmonid rivers in Europe given the widespread practice of stocking.
In the present survey, we analysed for the first time samples from a both extensive and intensive collection of trout from Albania and from some neighbouring drainages in FYROM and Greece (13 river basins altogether) along with S. ohridanus and S. letnica from Albanian waters of Lake Ohrid.
The main objective of this study was to determine the genetic architecture of Albanian trout from analysis of the mitochondrial DNA control region (mtDNA CR), and thus obtain phylogeographic information that could be compared with published data and make inferences on the historical demography and evolution of Balkan trout. We also looked for any indication of association between phenotype and mtDNA lineage.
Methods
Trout samples collection
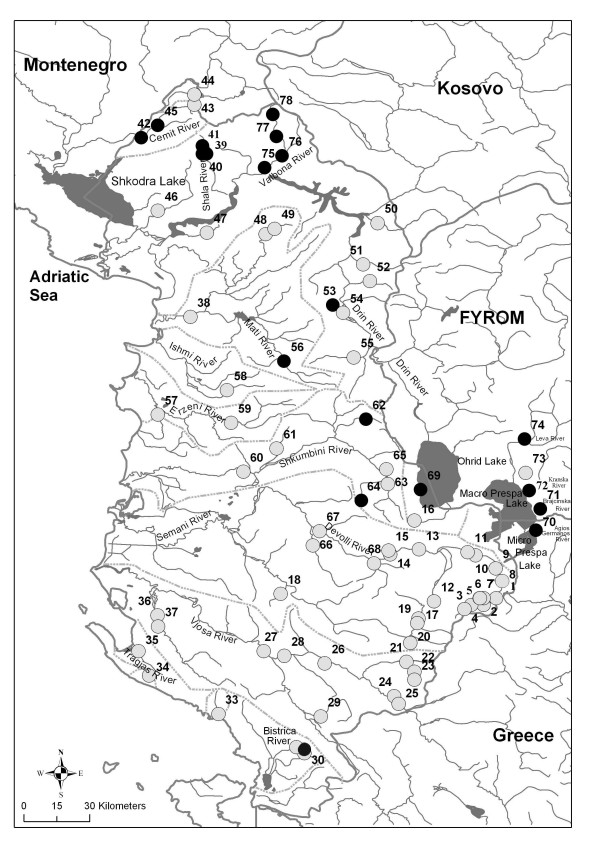
In 2005 and 2006, a total of 78 sampling sites were electrofished in rivers in Albania and the Megali Prespa basin in FYROM and Greece. The sampling in Albania (73 sampling sites) was performed not only to undertake a trout census in the country but also for the entire ichthyofauna. Locations were selected based upon observations published in the literature [23,22,20] and from local people. Emphasis was placed on both main water streams and isolated locations.
Among the sampled locations in Albania, trout were found at 15 of them (Fig. 1). Cake and Miho [23] reported trout at sites 61 and 63 (Fig. 1) in the River Shkumbini basin. However, during our sampling campaign, trout were not observed here, though they were found and sampled in two previously non-described locations within this catchment (62 and 64, Table 1). In addition, trout were observed for the first time in the River Mati catchment. On the other hand, they were not found at site 35 (on the River Tragjas), where local people report their existence.
Figure 1.
Map of sample sites. Sites where trout were found are marked with black (see Table 1); white spots are the sampling stations sampled without trout; dotted lines show the main river catchment.
Table 1.
Sampling locations (numbers as in Figure 1), sample size (N) and haplotype distribution of 14 mtDNA CR haplotypes resolved among 87 trout samples from Albania (AL), Former Yugoslav Republic of Macedonia (FYROM) and Greece
| Haplotype | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location (see, Figure 1) | Country | N | Taxon | Ad-AL1 | Ad-AL2 | Ad-AL3 | Ma-AL1 | Haplo1 | Haplo4 | Haplo5 | Haplo6 | ADcs11 | Haplo12 | Haplo14 | AdPrz | MEcs1 | ADcs1 |
| L. Ohrid (69) | AL | 5 | S. letnica | - | - | - | - | - | - | - | - | - | 5 | - | - | - | - |
| L. Ohrid (69) | AL | 5 | S. ohridanus | - | - | - | - | 1 | 1 | 2 | 1 | - | - | - | - | - | - |
| R. Cemit (42, 45) | AL | 3 | S. sp. | - | - | - | - | - | - | - | - | 3 | - | - | - | - | - |
| R. Seta (Drin) (53) | AL | 8 | S. sp. | - | - | 2 | - | - | - | - | - | - | - | 6 | - | - | - |
| R. Shala (Drin) (39) | AL | 4 | S. sp. | - | - | - | - | - | - | - | - | 3 | - | - | 1 | - | - |
| R. Zi (Shala, Drin) (40) | AL | 3 | S. sp. | - | - | - | - | - | - | - | - | 1 | - | - | 2 | - | - |
| R. Teth (Shala, Drin) (41) | AL | 3 | S. sp. | - | - | - | - | - | - | - | - | 3 | - | - | - | - | - |
| R. Valbona (Drin) (75–78) | AL | 11 | S. sp. | - | 3 | - | - | - | - | - | - | - | - | 1 | 1 | - | 6 |
| R. Mati (56) | AL | 5 | S. sp. | - | - | - | - | - | - | - | - | - | - | - | - | 5 | - |
| R. Shkumbini (64) | AL | 5 | S. sp. | 5 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| R. Qarishta (Skumbini) (62) | AL | 6 | S. sp. | - | - | - | - | - | - | - | - | 6 | - | - | - | - | - |
| R. Bistrica (31) | AL | 9 | S. sp. | - | - | - | 9 | - | - | - | - | - | - | - | - | - | - |
| R. Brajcino (Prespa) (71) | FYROM | 5 | S. peristericus | - | - | - | - | - | - | - | - | - | - | - | - | - | 5 |
| R. Kranska (Prespa) (72) | FYROM | 2 | S. peristericus | - | - | - | - | - | - | - | - | - | - | - | - | - | 2 |
| R. Leva (Prespa) (74) | FYROM | 5 | S. peristericus | - | - | - | - | - | - | - | - | - | - | - | - | - | 5 |
| R. Agios Germanos (Prespa) (70) | G | 8 | S. peristericus | - | - | - | - | - | - | - | - | - | - | - | - | - | 8 |
Trout were found at four of the five locations sampled in the Megali Prespa basin (in Greece and FYROM), including sites 70 and 71, where, according to the literature [9,29,17], they were expected to exist, and at sites 72 and 74. Trout were not observed at site 73.
The authors' general observation was that poaching for trout was very common in the area sampled, and where trout still exist, the observed densities were very low.
The 87 trout collected from 19 sites (Figure 1 and Table 1) were sorted among three species and one genus: S. ohridanus (5), S. letnica (5) and S. peristericus (20), and Salmo sp. (57). While the first two species are easily recognizable on the basis of both their distinct phenotype and specific native range (Lake Ohrid), the classification of S. peristericus was based upon its very restricted distribution (see [18]). The other specimens were phenotypically indefinable and were therefore assigned as Salmo sp. Mitochondrial DNA haplotypes detected in Albania, Greece and FYROM are reported in Table 1 and all the haplotypes used in phylogenetic analysis are listed in Table 2.
Table 2.
List of mtDNA CR haplotypes used for phylogeographic analysis and GenBank accession numbers
| Haplotype | Acc nb | Haplotype | Acc nb | Haplotype | Acc nb |
| Ad-AL1 | EU359770 | Ad12 | AY653216 | MEcs1 | AY836350 |
| Ad-AL2 | EU359768 | ADcs1 | AY836330 | MEcs10 | AY836359 |
| Ad-AL3 | EU359769 | ADcs11 | AY836340 | MEcs7 | AY836356 |
| AdRc | EU391632 | ADcs15 | AY836344 | ATcs1 | DQ841192 |
| AdN | DQ297172 | ADcs20 | AY836349 | AT11a | AY185578 |
| AdPe | DQ318126 | Haplo12 | AY926570 | ATs12 | AY836328 |
| AdPrz | DQ318129 | Haplo13 | AY926573 | Das1 | AY185568 |
| AdBoz | DQ318128 | Haplo14 | AY926571 | Da2 | AY185570 |
| AdTi | DQ318127 | Haplo15 | AY926572 | DaVl | DQ318123 |
| AdC1 | DQ381567 | Haplo16 | DQ381568 | Haplo1 | AY926564 |
| AdM1 | DQ381566 | Haplo17 | DQ381569 | Haplo4 | AY926561 |
| AdZ1 | DQ381565 | Haplo18 | DQ381570 | Haplo5 | AY926569 |
| ADs3 | AY260518 | Ma-AL1 | EU359771 | Haplo6 | AY926559 |
| Ad4 | AY260520 | MAcs1 | AY836365 | ||
| Ad11 | AY653218 | MA2 | AF321995 | ||
DNA amplification and sequencing
Total DNA was isolated from fin tissue preserved in 96% ethanol following the protocol of Medrano et al. [30]. The entire sequence of the mitochondrial DNA control region (mtDNA CR) was amplified by polymerase chain reaction (PCR) using primers 28RIBa [31] and HN20 [32]. Each 30 μL reaction included 1 μM of each primer, 0.2 μM dNTP, 1.5 μM MgCl2, 1 × PCR buffer, 1 U Taq polymerase (Applied Biosystems) and 100 ng of genomic DNA. The conditions for PCR were initial denaturation (95°C, 3 min) followed by 30 cycles of strand denaturation (94°C, 45 s), primer annealing (52°C, 45 s) and DNA extension (72°C, 2 min). All PCR amplifications were performed in a programmable thermocycler GeneAmp® PCR System 9700 (Applied Biosystems).
Amplified DNA fragments were run on a 1.5% gel and isolated from the gel using the QIAEX II gel Extraction Kit (QIAGEN).
The control region fragment between the tRNAPro gene and poly T-block of the amplified DNA (100 ng of purified PCR product) was sequenced using primer 28RIBa following ABI PRISM BigDye Terminator protocols (Applied Biosystems 3.1). The amplified DNA was salt-precipitated and analysed with an ABI PRISM 310 automated sequencer.
Data analysis
Sequences of the 5'-end of the mtDNA CR (ca. 561 bp) were aligned using the computer program ClustalX [33]. To assign individual haplotypes to trout species and lineages previously identified within the brown trout species complex, data were aligned against at least three haplotypes from each lineage (Me: Mediterranean; Ma: marmoratus; Da: Danubian; At: Atlantic), and compared to all known haplotypes found in trout samples across the Adriatic river system (Ad; Table 1).
Aligned haplotypes were imported into the program PAUP Version 4.0b10 [34] for phylogenetic analysis. Neighbour-Joining (NJ), maximum parsimony (MP), maximum likelihood (ML) and Bayesian analysis were used for phylogenetic reconstruction. For NJ, a Kimura 2-parameter model was chosen. For MP, insertions or deletions (indels) were included as a fifth character. A heuristic search (10 replicates) with Tree Bisection Reconnection (TBR) branch-swapping was employed to find the most parsimonious trees. For ML, a sequence evolution model was first chosen using the program Modeltest Version 3.7 [35] incorporated into PAUP. After choosing a model, a heuristic search (10 replicates) was used to estimate the most likely topology. Support values for the nodes were obtained with 1000 bootstrap replicates for MP, NJ, or ML analysis, whereby the fast stepwise addition method was used for ML. Bayesian analysis was performed with MrBayes version 3.1.2 [36] where posterior probabilities were obtained using the Markov chain Monte Carlo (MCMC) technique (Nst = 6, Rates = gamma, Ngen = 5,000,000, chains = 4).
Because of weak support for the Adriatic clade as a whole (see Results,) the genealogical relation of these haplotypes was also depicted using a 95% statistical parsimony network constructed from the 5'-end of mtDNA CR sequences using program TCS 1.3 [37]. Resolution of ambiguous loops in the TCS network was performed by comparing ML pair-wise distances of the haplotypes within a loop and identifying the most likely connections within it, reflected by the smallest pair-wise distances. ML pair-wise distances were computed under the model (HKY 85) using the program PAUP Version 4.0b10 [34].
Results
A total of 561 bp of the mtDNA CR was resolved in 87 individuals and compared with corresponding and already published sequences of various Salmo taxa.
In Lake Ohrid, five haplotypes, all previously described in Sušnik et al. [38] (marked with "Haplo"), were found, four of which were detected in S. ohridanus (Haplo 1, 4, 5 and 6) and one in S. letnica (Haplo 12). For the other samples, five already described haplotypes characteristic of the Adriatic (4) and Mediterranean (1) lineages of S. trutta were found. In addition, four previously unpublished haplotypes bearing close relation to others of the Adriatic (Ad-AL1 to 3) and marmoratus (Ma-AL1) were also detected.
Salmo peristericus from the FYROM part of the Prespa basin were fixed for haplotype ADcs1; this haplotype was also found in the River Valbona system in Albania (River Drin basin). Haplotype AdPrz was found in the rivers Valbona and Shala (also River Drin basin).
Haplo14, previously considered private for S. letnica in Lake Ohrid [13], was in this study found to exist also in trout in the Drin basin.
It is worth noting that all the samples from the River Bistrica were fixed for Ma-AL1, but none exhibited any phenotypic character state characteristic of S. marmoratus (field observations).
The highest level of genetic variation appeared to be in the Drin basin (haplotypes ADcs1, AdPrz and Haplo14) and the most common haplotype found in this study was ADcs1, found in Lake Prespa tributaries and the River Valbona.
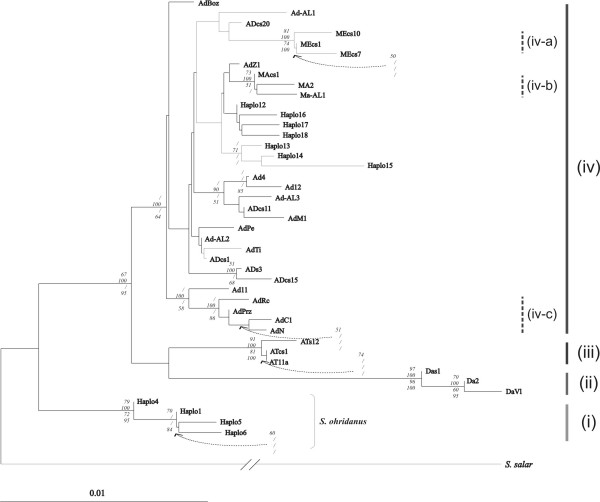
The phylogenetic organisation of the NJ distance tree clearly identified four well supported branches (Fig. 2): (i) S. ohridanus with Haplo1, 4, 5 and 6, (ii) the reference Danubian haplotypes, (iii) the reference Atlantic haplotypes, and (iv) a cluster of Mediterranean, Adriatic and marmoratus (ME-AD-MA) haplotypes exhibiting a very complex but poorly supported clade. All previously unreported haplotypes appeared in this clade.
Figure 2.
Neighbour-joining (NJ) tree for genus Salmo based on 561 bp of the 5'-end mtDNA control region and on a Kimura 2-parameter substitution model. In addition to haplotypes characteristic of Balkan trout from the Adriatic and Aegean drainages (Ad), three haplotypes representing Mediterranean (Me), Danubian (Da) and Atlantic (At) drainages were included in the analyses; haplotypes characteristic for Lake Ohrid S. letnica are marked with "Haplo"; bootstrap support values refer, from top to bottom, to NJ, maximum parsimony, maximum likelihood (HKY+I+G model, transition: transversion 2.6389; proportion of invariable sites (I) 0.6060; gamma distribution shape parameter 0.7375) and Bayesian methods; values <50 are marked with "/" or are not marked when there was no value above 50 in any of the analyses.
Character state phylogenetic (i.e., MP, ML and Bayesian) analyses revealed similar tree-topology with regard to the four main clades (Fig. 2) and provided a better resolution of the ME-AD-MA clade showing clear delimitation of three subdivisions: two already accepted groups of Mediterranean (iv-a) and marmoratus (iv-b) haplotypes [39,40], and (iv-c), a previously non-described formation of haplotypes AdRc, AdPrz, AdC1 and AdN (hereafter referred to as the Balkan cluster). The topology of the other Adriatic haplotypes remained largely unresolved.
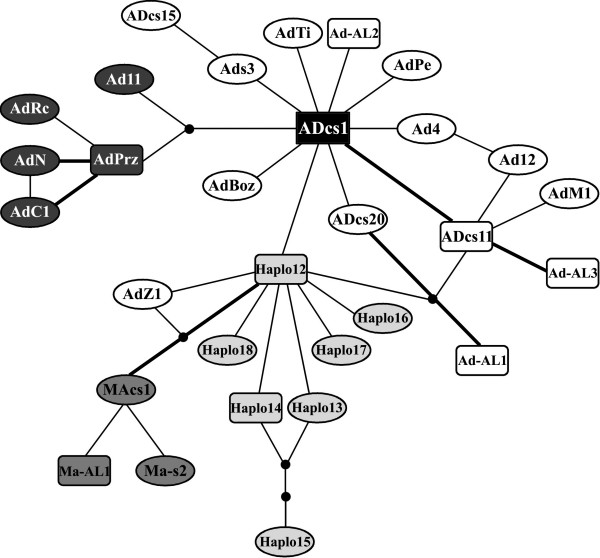
A network gathering the haplotypes found in this study and those previously published [40,13] is presented in Figure 3. Haplotype MEcs1 was found to exhibit several autapomorphies, which separated it considerably from other haplotypes and complicated the resolution of the network (data not shown). For this reason, this haplotype was excluded from further analysis. The general organization of the haplotype network obtained in this study featuring a multiple star-like structure with ADcs1 taking a central position was similar to the one reported by Cortey et al. [40] and confirmed in Sušnik et al. [13]. Those newly described are only one (Ad-AL2, Ad-AL3 and Ma-AL1) or two (Ad-AL1) mutation steps away from previously described haplotypes. AdPrz, a common haplotype in the southern Adriatic drainage [41], and AdRc and AdC1 from Lake Shkodra basin [13] and AdN from the River Neretva basin [10] form a separate group in the network, supporting the existence of the corresponding clade inferred from the phylogenetic tree (iv-c, Fig. 2). Interestingly, marmoratus haplotypes were found to be incorporated into the Adriatic clade network being apparently closely related to haplotypes predominantly detected in Ohrid trout (S. letnica).
Figure 3.
Haplotype network relating the Adriatic clade haplotypes found in Albanian trout populations (561 bp of the CR 5'-end) with previously published data [40,13]. Lines, regardless of length, represent single mutational events and link the haplotypes; black dots represent missing or theoretical haplotype; haplotypes found in Albania are in a square, those from Lake Ohrid are in light grey, those linked to S. farioides in black, while Ma haplotypes are in dark grey; most likely connections of the haplotypes within ambiguous loops, based on ML pair-wise distances are displayed with thick lines
Discussion
This study reveals for the first time the phylogenetic structure of trout populations in one of the last remaining incompletely explored regions of trout distribution in Europe. Drainages in Albania are linked to neighbouring systems in FYROM and Greece that also belong to the Adriatic river system as a whole. These rivers and lakes have been largely unmanaged with respect to stocking of non-native strains of trout, in contrast with most of the rest of Europe. However, stocking with trout from the same location has been practised over many years in Lake Ohrid, and in at least one tributary of Lake Prespa. Therefore, it was expected that non-native genetic signatures would not be detected and, indeed, this was the case: e.g. no haplotypes of Atlantic or Danubian brown trout phylogenetic lineages were found. Instead, Albanian trout populations are characterised by mtDNA haplotypes from the three other previously defined brown trout lineages [39]: Adriatic, Mediterranean and marmoratus. All of these three lineages are native to Mediterranean river systems. Moreover, Lake Ohrid contains the endemic species S. ohridanus (more closely related to S. obtusirostris of the Dalmatian river systems), with its unique haplotypes. Such pronounced genetic diversity places Albanian trout populations among the most variable in Europe. This finding is even more remarkable when one considers the very limited geographic distribution of these trout in Albania and the neighbouring area. Out of 78 locations sampled, we expected to find trout in at least 25 of them but were able to catch trout only in 19 sites. With few exceptions (sites 39,40,41) trout were always at very low density and it was difficult to catch more than five in several stations, probably as a result of heavy poaching by net, dynamite and chlorine. Consequently, although trout also exist in remote areas they are in danger of extinction.
Congruence between taxonomic group and mtDNA lineage
Lake Ohrid, the oldest lake in Europe, is shared between FYROM and Albania. According to historical data [21] and recent studies [11,38,13,27] at least two trout species, S. ohridanus and S. letnica, inhabit the lake and samples of both were included in this survey. Genetic analysis, including of mtDNA, has already been performed for these two taxa [38,13]. The results obtained in the present study corroborate the earlier findings, justifying the distinct taxonomic position of S. ohridanus in relation to its congeners, and supporting the species status of Ohrid trout S. letnica for conservation purposes [13]. The present study found a high frequency of Haplo14 in the Drin river system (previously reported only for Lake Ohrid), an unsurprising finding since until 1960 no dams existed along this river and Lake Ohrid was connected directly with rest of the system.
The second largest lake in the region, of Tertiary origin (>5 MY), is Lake Megali Prespa, whose tributaries are inhabited by S. peristericus [17]. During the Jurassic, lakes Prespa and Ohrid formed part of the Dassaretic lakes, which were linked with the Adriatic Sea. Trout from the River Agios Germanos, now a tributary of this lake after the stream was diverted from Lake Mikri Prespa between 1935 and 1945, have already been analysed at allozyme loci [42] and for mtDNA sequence variation [26]. On the basis of a diagnostic allele at CK-2*, this population was first thought to be distinct from other populations of brown trout in Greece. However, based on subsequent partial mtDNA CR and cyt b gene sequence analysis, it was then placed back in the S. trutta complex. All the populations of S. peristericus examined in the present study were fixed for the haplotype ADcs1, which distinguished them from all trout populations surveyed here apart from the population from the River Valbona (Drin basin), where this haplotype was also found. A recent hypothesis concerning demographic patterns of the Adriatic lineage [40,14] considers ADcs1 as the central haplotype (Fig. 3): it is the most common haplotype in the Iberian Peninsula, where the Adriatic lineage is thought to have originated [40]. The plesiomorphic state of the haplotype ADcs1 and its presence in S. peristericus indicates its ancestry within the Adriatic lineage and does not support the recognition of this taxon as a separate species. Nevertheless, given that S. peristericus is distinct morphologically from all other Balkan trout [9] and restricted geographically to Lake Megali Prespa basin, we stress the importance of this taxon as a unit that needs conserving.
No marbling phenotype characteristic of marble trout was observed in any of the individual trout caught in the present study, even though S. marmoratus has been reported to be present in the rivers Valbona and Drin [19,24,25]. On the other hand, the marmoratus haplotype (Ma-A1) was detected in southern Albania, in the River Bistrica where Rakaj [20] has described the local form as S. peristericus. As no marbling was observed in the trout from this river, this supports the view that the marmoratus mtDNA lineage and the marbling phenotype are not linked: previous reports have described the existence of marmoratus haplotypes in many populations of phenotypically brown trout across the Mediterranean river basins, including rivers in Dalmatia [39], central Italy [43], Greece [26] and Corsica (unpublished data). The marbling phenotype is only characteristic of this lineage in its north Adriatic range, where the phenotype was first described [44,45].
Phylogeographic considerations
Much effort has been put in resolving salmonid genetic structure and phylogeographic signals in the Adriatic drainage [41,10,43,38,14]. To update the overall current picture, genetic data on samples from Albania and neighbouring freshwater systems in FYROM and Greece can now be incorporated into previously obtained data and used to make inferences on trout phylogeography and historical demography in the western Balkans and supplement the current knowledge concerning the rest of the Adriatic drainage.
The presence of four evolutionary lineage haplotypes (of S. ohridanus, S. marmoratus, and Adriatic and Mediterranean S. trutta) in Albania points to a complicated demographic history and rich diversity of trout populations in the country. The lineages AD and ME, both reported here as present in Albania, have been studied and reviewed thoroughly by Cortey et al. [40] who suggested that they had originated in the Iberian peninsula some 150,000 years ago, with haplotypes ADcs1 and MEcs1 as the most ancestral, respectively. It is thought that these lineages expanded together from west to east across the Mediterranean basin during the extreme Pleistocene glacial maxima and therefore would have reached the western Balkans relatively recently. In Albania, the ME haplotype (MEcs1) was found in a single river basin (River Mati), and was the only haplotype present there. Indications of a patchwork distribution of ME haplotypes have already been reported for the western Mediterranean drainages [40], the Aegean and Adriatic drainages in Greece [26] and in central Italy [43]. As Albanian rivers represent the limit of the geographical range of the ME lineage, it seemed likely that here the concentration of such haplotypes would be low and that they would be very sensitive to stochastic events (gene flow, bottlenecks, etc.). Such events appear to have been particularly intense in the Balkan Peninsula during the Pleistocene [4] and could be the main reason for both the present geographical limitations of distribution and local fixation of the MEcs1 haplotype.
A complex and particularly fuzzy phylogenetic relationship among AD haplotypes, already observed by Cortey et al. [40] and Sušnik et al. [13], was noticed in this study, and only the so-called Balkan cluster (haplotypes AdN, AdRc, AdC1 and AdPrz) was well resolved, with a bootstrap value of 100 per cent in MP and 86 per cent in Bayesian. This cluster corresponds to the AdN-AdPrz cluster previously described by Marić et al. [41] and Razpet et al. [10] for the rivers Neretva and Prizrenska Bistrica. In those studies, the distribution of haplotypes corresponded well with the distribution of the questionable taxon S. farioides (from rivers Krka (Croatia), Neretva (Bosnia and Herzegovina) and Prizrenska Bistrica (Kosova), tributaries of Lake Shkodra (Montenegro) and of the River Drin and Lake Ohrid) [17,9]. Two additional haplotypes constituting the Balkan cluster and used here as reference haplotypes, AdRc and AdC1, also originate from the S. farioides range (Lake Shkodra tributaries; see [13]) and additionally support the proposed haplotype-species association. In this study, the haplotype AdPrz was found in the River Drin basin, the eastern limit of the range of S. farioides, and close to the type location of Prizren, which shares the same water shed. Thus, the results from this study support the congruence of the distribution of the Balkan haplotype cluster and the range of S. farioides. As reported previously [10,13], haplotypes of the Balkan cluster, including AdPrz, on the one hand and other Ad haplotypes on the other hand, do not represent a contiguous haplotype lineage (see Fig. 3) and appear not to be closely related, indicating independent arrivals into the Adriatic drainage and suggesting successive colonization events.
The data referring to the distribution of the marmoratus haplotype in Albania do not contribute much to resolving contradictory notions about the centre of origin and demographic patterns of marmoratus lineage (c.f., [46,39,48]). The newly described Ma-AL1 haplotype, recorded for a previously non-surveyed location in Albania, broadens the known genetic diversity of the marmoratus lineage, and highlights its extensive but patchy distribution, as observed across a broad stretch of Mediterranean river systems [26,39,43].
Balkan trout are composed of a genetic mosaic of haplotypes, related to most of the other trout lineages of the Mediterranean area analysed and reported in other studies. However, due to a complexity of past migrations, colonisations and extinctions, as well as that of many other organisms [4], the Balkans has been considered a hotspot of trout biodiversity. The region's unique mix of habitats and topography has created a peninsula rich in endemism, and ironically its isolation (both physical and political) has helped to conserve a complex structure of trout populations, particularly in Albania. This first investigation of a little explored area has revealed a glimpse into a partly understandable and partly fuzzy web of relationships.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AS participated in the study design and coordination and drafted the manuscript. SM carried out the molecular genetic studies and prepared the sequence alignment. PB participated in the design and coordination of the study and in writing the manuscript. AJC conceived the study, succeeded in finding funding, participated in its design and coordination, and helped to draft the manuscript. SSh organized the logistic for the fieldwork, participated with the collection of data and helped to draft the manuscript. SS carried out phylogenetic analyses and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Aleš Snoj, Email: ales.snoj@bfro.uni-lj.si.
Saša Marić, Email: sasa@bf.bio.bg.ac.yu.
Patrick Berrebi, Email: patrick.berrebi@univ-montp2.fr.
Alain J Crivelli, Email: a.crivelli@tourduvalat.org.
Spase Shumka, Email: sprespa@yahoo.co.uk.
Simona Sušnik, Email: simona.susnik@bfro.uni-lj.si.
Acknowledgements
We thank I Koutseri and Dr S Petkovski for organizing sampling campaigns in Greece and FYROM, and all the people who helped in the field. Many thanks go to I Wilson whose constructive comments helped to improve the manuscript and A Sandoz for drawing the map of Albania.
This study was funded by a grant awarded by the French Embassy in Tirana, Albania, to Dr AJ Crivelli and Dr M Lutz, and by the Ministry of Science and Environment Protection of the Republic of Serbia (Grant No. ON 143040).
References
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc Lond. 1996;58:247–276. [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF. Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Culver DC, Christman MC, Sket B, Trontelj P. Sampling adequacy in an extreme environment: species richness patterns in Slovenian caves. Biodivers Conserv. 2004;13:1209–1229. doi: 10.1023/B:BIOC.0000018153.49280.89. [DOI] [Google Scholar]
- Kryštufek B, Reed JM. In: Balkan biodiversity pattern and process in the European hotspot. Griffiths HI, Kryštufek B, Reed JM, editor. Dordrecht: Kluwer Academic Publishers; 2004. Pattern and process in Balkan biodiversity – an overview; pp. 203–217. [Google Scholar]
- Trontelj P, Goricki S, Polak S, Verovnik R, Zaksek V, Sket B. Age estimates for some subterranean taxa and lineages in the Dinaric Karst. Acta Carsologica. 2007;36:183–189. [Google Scholar]
- Conservation International. 2004. http://www.conservation.org/
- Karaman S. Salmonidi Balkana. Bull Soc Scient Skoplje. 1926;2:253–268. [Google Scholar]
- Sorić V. Salmonids in the Ohrid-Drim-Skadar system. Acta Soc Zool Bohemoslov. 1990;54:305–319. [Google Scholar]
- Delling B. PhD thesis. Stockholm University; 2003. Species diversity and phylogeny of Salmo with emphasis on southern trouts (Teleostei, Salmonidae) [Google Scholar]
- Razpet A, Sušnik S, Jug T, Snoj A. Genetic variation among trout in the River Neretva basin, Bosnia and Herzegovina. J Fish Biol. 2007;70:94–110. doi: 10.1111/j.1095-8649.2007.01392.x. [DOI] [Google Scholar]
- Sell J, Spirkovski Z. Mitochondrial DNA differentiation between two forms of trout Salmo letnica, endemic to the Balkan Lake Ohrid, reflects their reproductive isolation. Mol Ecol. 2004;13:3633–3644. doi: 10.1111/j.1365-294X.2004.02362.x. [DOI] [PubMed] [Google Scholar]
- Snoj A, Melkiè E, Sušnik S, Muhamedagić S, Dovè P. DNA phylogeny supports revised classification of Salmothymus obtusirostris. Biol J Linn Soc Lond. 2002;77:399–411. doi: 10.1046/j.1095-8312.2002.00130.x. [DOI] [Google Scholar]
- Sušnik S, Snoj A, Wilson I, Mrdak D, Weiss S. Historical demography of brown trout (Salmo trutta) in the Adriatic drainage including the putative S. letnica endemic to Lake Ohrid. Mol Phylogenet Evol. 2007;44:63–76. doi: 10.1016/j.ympev.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Sušnik S, Weiss S, Tomljanović T, Delling B, Treer T, Snoj A. Reticulate evolution: ancient introgression of the Adriatic brown trout mtDNA in softmouth trout Salmo obtusirostris (Teleostei: Salmonidae) Biol J Linn Soc Lond. 2007;90:139–152. doi: 10.1111/j.1095-8312.2007.00717.x. [DOI] [Google Scholar]
- Smith KG, Darwall WRT. The status and distribution of freshwater fish endemic to the Mediterranean Basin. IUCN Gland and Cambridge; 2006. [Google Scholar]
- Heckel J. Fortsetzung des im Julihefte 1851 enthaltenen Berichtes über eine, auf Kosten der kais. Akademie der Wissenschaften unternommene, ichtyologische Reise. Sitzungsberichte der Mathematisch-Naturwissenschaftlichen Classe der kaiserlichen Akademie der Wissenschaften. 1852;8:347–390. [Google Scholar]
- Karaman S. Beitrag sur kenntnis der susswasserfishe Jugoslaviens. Bull Soc Scient Skoplje. 1937;18:130–139. [Google Scholar]
- Kottelat M, Freyhof J. Handbook of European freshwater fishes. Cornol: Publications Kottelat; 2007. [Google Scholar]
- Karaman S. The Radika River trouts. Folia Balcanica. 1957;1:57–70. [Google Scholar]
- Rakaj N, Flloko A. Conservation status of freshwater fish of Albania. Biol Conserv. 1995;72:195–199. doi: 10.1016/0006-3207(94)00082-2. [DOI] [Google Scholar]
- Stanković S. In: Monographiae Biologicae 9. Junk W, editor. Den Haag; 1960. The Balkan Lake Ohrid and its Living World. [Google Scholar]
- Poljakov GD, Filipi ND, Basho K. Peshqit e Shqipërisë. Universiteti Shtetëror i Tiranës; 1958. [Google Scholar]
- Cake A, Miho A. Ihtiofauna e lumit Shkumbin. Tiranë: Monografi; 1999. [Google Scholar]
- Rakaj N. Iktiofauna E Shqipërisë. Tiranë: Shtëpia Botuese "Libri Universitar"; 1995. [Google Scholar]
- Schöffmann J. Zur gegenwätigen Situation der Marmorierten Forelle (Salmo marmoratus Cuvier, 1817) in Albanien, ihrem südlichsten Verbreitungsraum. Österreichs Fisherei. 1994;47:132–136. [Google Scholar]
- Apostolidis AP, Triantaphyllidis C, Kouvatsi A, Economidis PS. Mitochondrial DNA sequence variation and phylogeography among Salmo trutta L. (Greek brown trout) populations. Mol Ecol. 1997;6:531–542. doi: 10.1046/j.1365-294X.1997.d01-176.x. [DOI] [PubMed] [Google Scholar]
- Wilson I. Low genetic variability in the summer Koran (Salmo letnica aestivalis Stefanovic) of Lake Ohrid. Albanian J Nat Tech Sci. 2004;16:3–13. [Google Scholar]
- Jug T, Berrebi P, Snoj A. Distribution of non-native trout in Slovenia and their introgression with native trout populations as observed through microsatellite DNA analysis. Biol Conserv. 2005;123:381–388. doi: 10.1016/j.biocon.2004.11.022. [DOI] [Google Scholar]
- Karakousis Y, Triantaphyllidis C, Economidis PS. Morphological variability among seven populations of brown trout, Salmo trutta L., in Greece. J Fish Biol. 1991;38:807–817. doi: 10.1111/j.1095-8649.1991.tb03620.x. [DOI] [Google Scholar]
- Medrano FJ, Aasen E, Sharrow L. DNA extraction from nucleated red blood cells. Biotechniques. 1990;8:43. [PubMed] [Google Scholar]
- Snoj A, Jug T, Melkiè E, Sušnik S, Pohar J, Dovè P, Budihna N. Mitochondrial and microsatellite DNA analysis of marble trout in Slovenia. Quaderni ETP. 2000;29:5–11. [Google Scholar]
- Bernatchez L, Danzmann RG. Congruence in control-region sequence and restriction-site variation in mitochondrial DNA of brook charr (Salvelinus fontinalis Mitchill) Mol Biol Evol. 1993;10:1002–1014. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4637–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland: Sinauer Associates; 2000. [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Sušnik S, Knizhin I, Snoj A, Weiss S. Genetic and morphological characterization of a Lake Ohrid endemic, Salmo (Acantholingua) ohridanus with a comparison to sympatric Salmo trutta. J Fish Biol. 2006;67:2–23. doi: 10.1111/j.0022-1112.2006.00902.x. [DOI] [Google Scholar]
- Bernatchez L. The evolutionary history of brown trout (Salmo trutta L.) inferred from phylogeographic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution. 2001;55:351–379. doi: 10.1111/j.0014-3820.2001.tb01300.x. [DOI] [PubMed] [Google Scholar]
- Cortey M, Pla C, Garcia-Marin JL. Historical biogeography of Mediterranean Trout. The role of allopatry and dispersal events. Mol Phylogenet Evol. 2004;33:831–844. doi: 10.1016/j.ympev.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Marić S, Sušnik S, Simonović P, Snoj A. Phylogeographic study of brown trout from Serbia, based on mitochondrial DNA control region analysis. Genet Sel Evol. 2006;38:411–430. doi: 10.1051/gse:2006012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakousis Y, Triantaphyllidis C. Genetic structure and differentiation among Greek brown trout (Salmo trutta L.) populations. Heredity. 1990;64:297–304. doi: 10.1038/hdy.1990.37. [DOI] [PubMed] [Google Scholar]
- Splendiani A, Giovannotti M, Cerioni PN, Caniglia ML, Caputo V. Phylogeographic inferences on the native brown trout mtDNA variation in central Italy. Ital J Zool (Modena) 2006;73:179–189. doi: 10.1080/11250000600679751. [DOI] [Google Scholar]
- Cuvier G. Le règne animal, distribué d'après son organisation, pour servir de base à l'histoire naturelle des animaux et d'introducion à l'anatomie comparée. Paris: Déterville; 1829. [Google Scholar]
- Heckel JJ, Kner R. Die Süsswasserfische der Österreichen Monarchie mit Rücksicht auf der angrenzenden Länder. Leipzig: Engelmann; 1858. [Google Scholar]
- Antunes A, Templeton AR, Guyomard R, Alexandrino P. The role of nuclear genes in intraspecific evolutionary inference: Genealogy of the transferrin gene in the brown trout. Mol Biol Evol. 2002;19:1272–1287. doi: 10.1093/oxfordjournals.molbev.a004188. [DOI] [PubMed] [Google Scholar]
- Berrebi P, Povž M, Jesenšek D, Cattaneo-Berrebi G, Crivelli AJ. The genetic diversity of native, stocked and hybrid populations of marble trout in the Soèa river, Slovenia. Heredity. 2000;85:277–287. doi: 10.1046/j.1365-2540.2000.00753.x. [DOI] [PubMed] [Google Scholar]
- Giuffra E, Bernatchez L, Guyomard R. Mitochondrial control region and protein-coding genes sequence variation among phenotypic forms of brown trout Salmo trutta from northern Italy. Mol Ecol. 1994;3:161–171. doi: 10.1111/j.1365-294X.1994.tb00117.x. [DOI] [PubMed] [Google Scholar]