Abstract
Background
The INK4b-ARF-INK4a tumour suppressor locus controls the balance between progenitor cell renewal and cancer. In this study, we investigated how higher-order chromatin structure modulates differential expression of the human INK4b-ARF-INK4a locus during progenitor cell differentiation, cellular ageing and senescence of cancer cells.
Results
We found that INK4b and INK4a, but not ARF, are upregulated following the differentiation of haematopoietic progenitor cells, in ageing fibroblasts and in senescing malignant rhabdoid tumour cells. To investigate the underlying molecular mechanism we analysed binding of polycomb group (PcG) repressive complexes (PRCs) and the spatial organization of the INK4b-ARF-INK4a locus. In agreement with differential derepression, PcG protein binding across the locus is discontinuous. As we described earlier, PcG repressors bind the INK4a promoter, but not ARF. Here, we identified a second peak of PcG binding that is located ~3 kb upstream of the INK4b promoter. During progenitor cell differentiation and ageing, PcG silencer EZH2 attenuates, causing loss of PRC binding and transcriptional activation of INK4b and INK4a. The expression pattern of the locus is reflected by its organization in space. In the repressed state, the PRC-binding regions are in close proximity, while the intervening chromatin harbouring ARF loops out. Down regulation of EZH2 causes release of the ~35 kb repressive chromatin loop and induction of both INK4a and INK4b, whereas ARF expression remains unaltered.
Conclusion
PcG silencers bind and coordinately regulate INK4b and INK4a, but not ARF, during a variety of physiological processes. Developmentally regulated EZH2 levels are one of the factors that can determine the higher order chromatin structure and expression pattern of the INK4b-ARF-INK4a locus, coupling human progenitor cell differentiation to proliferation control. Our results revealed a chromatin looping mechanism of long-range control and argue against models involving homogeneous spreading of PcG silencers across the INK4b-ARF-INK4a locus.
Background
Development and homeostasis require the coordinate regulation of cell proliferation and differentiation. The INK4b-ARF-INK4a tumor suppressor locus (Figure 1A) plays a central role in controlling the equilibrium between progenitor cell renewal and cancer risk [1-8]. This locus encodes three cell cycle inhibitory proteins: p15INK4b, p14ARF and p16INK4a [3,8]. p15INK4b and p16INK4a are closely related proteins and both act on the Rb-pathway through the inhibition of the proliferation-promoting cyclin-dependent kinases CDK4 and CDK6. p14ARF is structurally and functionally unrelated to p15INK4b or p16INK4a and works primarily through activation of the p53 pathway. A large number of studies have suggested a role for the INK4b-ARF-INK4a locus in cancer suppression and promotion of ageing. p16INK4a, in particular, has been implicated in balancing the need for tissue renewal and the risk of tumourigenesis [1-8]. Perhaps not surprisingly, the regulation of the INK4b-ARF-INK4a locus is highly complex. INK4b-ARF-INK4a expression is controlled by various signal transduction pathways and patterns of expression vary depending on physiological circumstances. Coordinated regulation of the whole locus, as well as differential gene expression, has been described [3]. Unfortunately, regulation of INK4b has received significantly less attention than that of ARF and INK4a.
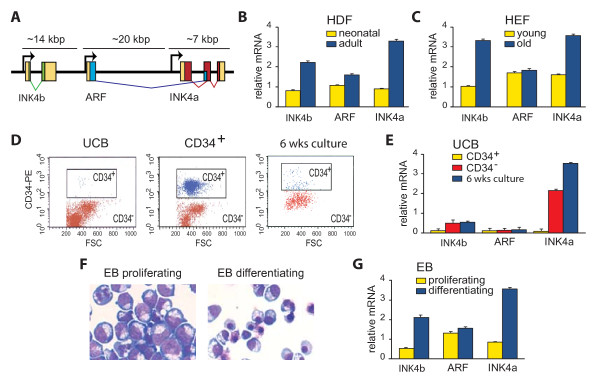
Figure 1.
Expression of the INK4b-ARF-INKa locus during cellular ageing and differentiation. (A) Organization of the human INK4b-ARF-INK4a locus (not drawn to scale), encoding three distinct proteins, p15INK4b, p14ARF and p16INK4a. The untranslated regions (yellow boxes), the coding sequences of p15INK4b (green), p14ARF (blue) and p16INK4a (red) are indicated. (B) INK4b and INK4a, but not ARF, are upregulated in ageing human diploid fibroblasts (HDFs). RT-qPCR analysis of INK4b-ARF-INK4a expression in neonatal (yellow bar) versus adult (blue bar) HDFs. Bar graphs represent the mean of three independent biological replicate experiments, each analyzed in triplicate by RT-qPCR. mRNA levels are expressed relative to Gapdh. Error bars represent standard error of the mean. (C) INK4b and INK4a are selectively upregulated in ageing human embryonic fibroblasts (HEFs). Comparison of INK4b-ARF-INK4a expression in HEF cells (TIG3) at a low passage doubling (PDL 26, yellow) and high PDL (PDL 64, blue). (D) Flow cytometrical analysis of umbilical cord blood cells showing forward scatter (FSC) on the x-axes and CD34 staining on the y-axes. The immature CD34+ cells (blue) and mature CD34- cells (red) were sorted (left hand panel). Isolated CD34+ cells were reanalysed (middle panel; red dots represent 65% of the population). Following 6 weeks of culture these cells stained negative for CD34 (less than ~15%; right hand panel). (E) INK4b-ARF-INK4a expression in CD34+ (yellow) cells, CD34- (red) cells, and the progeny of seeded CD34+ cells after 6 weeks of culture (blue). (F) Erythroblasts (EB) kept under proliferation conditions for 2 days following induction of differentiation towards erythrocytes. Cells were cytocentrifuged onto glass slides and stained for haemoglobin (brown colour) and with standard cytologic dyes. (G) Expression of INK4b-ARF-INK4a in proliferating (yellow) erythroblst cells and cells differentiating towards erythrocytes (blue).
The polycomb group (PcG) silencers form an important class of transcriptional corepressors that control the expression of the INK4b-ARF-INK4a locus [9]. This was first suggested by the finding that the PcG protein, BMI1, promotes oncogenesis in mice through the silencing of INK4a [10]. Since then, other PcG proteins have also been implicated in the silencing of the INK4b-ARF-INK4a locus, including the histone H3 lysine 27 (H3K27) methyltransferase EZH2 [9-22]. However, in contrast to INK4a and ARF, the role of PcG proteins in INK4b expression control has not been extensively studied.
PcG proteins function as part of larger multi-protein assemblages, referred to as polycomb repressive complexes (PRCs) [23-25]. One major class is formed by PRC1-like complexes, which include assemblages harbouring BMI1. PRC1 class complexes have been implicated in chromatin compaction and histone H2A ubiquitylation. The second major class is formed by PRC2-like complexes, harbouring histone H3K27 methylases such as EZH2. However, it is important to stress that there is likely to be a great variety of PRCs and additional enzymatic activities. For example, PRC1 subunits are also part of alternate assemblages such as Drosophila dRAF [26] and its mammalian relatives [27,28]. The dRAF complex, harbouring the BMI1 homolog PSC, dRING and the demethylase dKDM2, ubiquitylates histone H2A and demethylates histone H3K36 during gene silencing [26]. Very recently, an essential role for N-acetylglucosamination in PcG repression was established [29]. Importantly, PRCs have also been implicated in the higher-order chromatin organization through looping [30,31].
Although the developmental roles of many PcG proteins await further analysis, recent research has emphasized their importance in dynamic gene control during the differentiation of precursor cells, in cancer and cellular senescence [9-22,32-36]. In particular, several of these studies have shown the importance of EZH2 for the dynamic regulation of gene silencing, orchestrating the differentiation of progenitor cells.
Here, we have addressed the role of PcG silencers in the regulation of the human INK4b-ARF-INK4a locus during the differentiation of progenitor cells, cellular ageing and cellular senescence of cancer cells. During these diverse physiological processes INK4b and INK4a were coordinately induced, whereas ARF remained unaltered. In order to investigate the underlying mechanisms, we analysed the spatial organization of the INK4b-ARF-INK4a locus. Our results revealed long-range control through chromatin looping rather than 'blanket' spreading of PcG proteins across the whole INK4b-ARF-INK4a locus. We conclude that PcG proteins control the higher-order chromatin conformation of the INK4b-ARF-INK4a locus, providing a molecular mechanism for coupling cell differentiation to proliferation control.
Results
Selective induction of INK4b and INK4a during cellular ageing, differentiation and senescence
We compared the expression of the human INK4b-ARF-INK4a locus in neonatal and adult human diploid fibroblasts (HDFs). The proliferation rate of adult HDFs is ~2-3 times slower than that of neonatal HDFs. In order to monitor INK4b-ARF-INK4a expression, we extracted RNA, which was followed by reverse transcription and real-time quantitative polymerase chain reaction (RT-qPCR) with gene-selective primers (Figure 1B). Compared to neonatal cells (yellow), the expression of INK4a and INK4b, but not of ARF, in adult HDFs (blue) is significantly higher. We also compared human embryonic fibroblasts (HEFs) with a low passage number (PDL 26) with old cells (PDL 64). We observed a gradual, up to two- to threefold, increase in doubling time as the passage number became higher. INK4a and INK4b are clearly upregulated in HEFs with a high passage number, whereas ARF expression remained unchanged (Figure 1C). We conclude that, in these untransformed human diploid cells, INK4a and INK4b, but not ARF, are coordinately upregulated during ageing.
Next, we examined the expression of the INK4b-ARF-INK4a locus in human cells with a broad versus restricted potential for differentiation. We sorted CD34+ and CD34- cells isolated from human umbilical cord blood (UCB; Figure 1D). CD34+ UCB cells comprise quiescent stem cells, but they mainly represent the transiently amplifying compartment of multipotent and early myeloid progenitors [37]. In contrast, the CD34- fraction contains committed cells in late stages of differentiation. A fluorescence activated cell sorting analysis of the isolated CD34+ cells revealed a purity of about 65% (Figure 1D, middle panel). We cultured the purified CD34+ cells for 6 weeks, after which the majority of cells (~85%) matured to CD34- cells, representing mainly postreplicative erythroblasts and granulocytes. An expression analysis of CD34+ (yellow) and CD34- cells (red) immediately following isolation, and CD34+ derived cells cultured for 6 weeks (blue), demonstrated up-regulation of INK4b and very strong INK4a induction during differentiation (Figure 1E). In contrast, ARF expression remained unaltered.
As an alternative way to study the effects of cell differentiation, we cultured erythroblasts (EB) from human fetal liver. After 7 days, EBs were either kept under proliferating conditions or induced to differentiation towards erythrocytes for 2 days (Figure 1F). Again, the differentiation was accompanied by selective activation of INK4a and INK4b (Figure 1G).
As a model for senescence in cancer cells, we used MON human malignant rhabdoid tumour (MRT) cells. MRTs are caused by the loss of the hSNF5 subunit of the SWI/SNF chromatin remodelling complex [38]. Re-expression of hSNF5 in MRT cells restores SWI/SNF recruitment to INK4b and INK4a, causing the eviction of PRCs and a loss of silencing [20]. Consequently, these cells first undergo a G1/S cell cycle arrest and later become senescent [20,39]. hSNF5-induced cellular senescence of MRT cells is p16INK4a-dependent [20,39]. We suspect that the hSNF5-mediated senescence of MRT cells might be due to the inability of oncogenic stress signalling to activate INK4a expression in the absence of hSNF5. In summary, we identified a number of diverse physiological processes in which human INK4a and INK4b are coordinately induced, while ARF expression remained unaltered.
EZH2 is down-regulated during progenitor cell differentiation
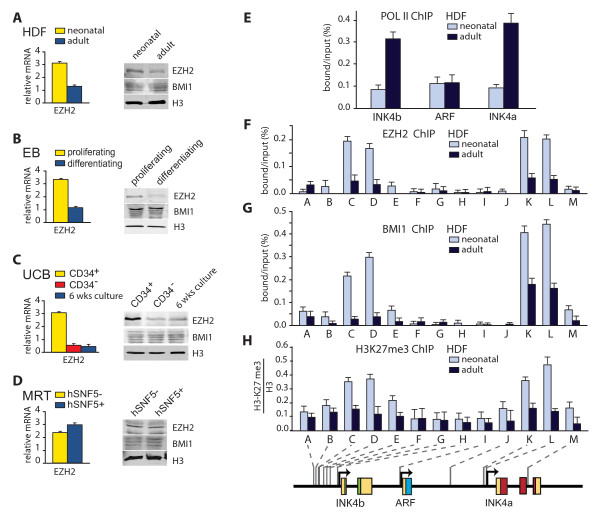
Because the PcG silencers EZH2 and BMI1 play important roles in the repression of the INK4b-ARF-INK4a locus, we investigated their expression in young versus adult HDFs and during cellular differentiation. RT-qPCR and Western immunoblotting revealed lower EZH2 levels in adult HDFs compared to the neonatal cells. In contrast, the BMI1 levels were comparable (Figure 2A). When we compared proliferating EBs with differentiating cells we again observed a reduction of EZH2 levels, but not of BMI1 (Figure 2B). Likewise, UCB CD34+ progenitor cells expressed much higher levels of EZH2 than mature CD34- cells (Figure 2C). Upon culture, CD34+ cells matured and, concomitantly, EZH2 expression was strongly attenuated, whereas BMI1 levels remained stable (Figure 2C). However, re-expression of hSNF5 in MRT cells mediates INK4a and INK4b induction without affecting EZH2 or BMI1 levels [20]. We conclude that, in differentiating haematopoietic progenitor cells, the expression of the PRC2 subunit EZH2 wanes. In contrast, levels of the PRC1 subunit BMI1 remained constant.
Figure 2.
EZH2 attenuation during progenitor cell differentiation. (A-D) PRC2 subunit EZH2, but not the PRC1 subunit BMI1, is down-regulated during cellular ageing and differentiation. The expression of EZH2 was analysed by RT-qPCR and Western immunoblotting in: (A) neonatal and adult human diploid fibroblasts (HDFs); (B) proliferating and differentiating erythroblasts, (C) immature CD34+ progenitors and mature CD34- cells isolated directly from umbilical cord blood and CD34+ cells that differentiated and lost CD34 expression following 6 weeks of culture; (D) malignant rhabdoid tumour cells that either lack or express hSNF5. For characterization of these cells see Figure 1. In parallel, BMI1 and histone H3 levels were determined. (E) Selective RNA POL II recruitment to INK4a and INK4b, but not ARF, in ageing HDFs. Chromatin immunoprecipitation (ChIP)-quantitative polymerase chain reaction (qPCR) analysis of RNA POL II binding to the INK4b-ARF-INK4a locus in neonatal (light blue) and adult (dark blue) HDFs. The following primer sets were used: H (INK4b), I (ARF) and L (INK4a). All ChIP data presented in this study are the result of at least three biological replicates. Background levels were determined using antibodies directed against GST. The abundance of specific DNA sequences in the immunoprecipitates was analysed by qPCR and corrected for the independently determined amplification curves for each primer set. ChIP signal levels for each region are presented as percentage of input chromatin. Bar graphs represent the mean of three independent experiments, each analysed in triplicate by qPCR. Error bars represent standard error of the mean.(F) PRCs bind INK4a and INK4b in neonatal but not in adult HDFs. ChIP-qPCR analysis revealed highly localized binding of EZH2 to the INK4a promoter region (primer sets K and L) and an area ~3 kb upstream of the INK4b promoter (primer sets C and D) in neonatal HDFs. In adult HDFs, EZH2 protein binding is strongly reduced. Analysis was as described above. (G) Following waning of EZH2, BMI1 binding to INK4a and INK4b is reduced significantly. ChIP-qPCR analysis of BMI1 binding to the INK4b-ARF-INK4a locus in neonatal and adult HDFs. The positions of the amplified regions (A-M) of the INK4b-ARF-INK4a locus are indicated at the bottom. (H) ChIPs using antibodies directed against histone H3K37 me3 revealed increased H3K27 methylation at- and around the PRC binding sequences upstream of INK4b and at the INK4a promoter. H3K27 me3 ChIPs were normalized against histone H3.
In order to determine the effects of EZH2 down-regulation on chromatin occupancy at the INK4b-ARF-INK4a locus, we used chromatin immunoprecipitations (ChIPs) monitored by qPCR. Comparing neonatal and adult HDFs revealed an increased recruitment of RNA polymerase II (RNA POLII) to INK4a and INK4b in adult cells, consistent with their enhanced transcription (Figure 2E). Next, we established the pattern of EZH2 and BMI1 binding, and the relative level of histone H3K27 me3 (Figure 2F-H). PRC binding at the INK4a promoter is already well established. We performed a detailed analysis of EZH2 and BMI1 binding to the INK4b upstream region and used a selection of primers targeting INK4a and ARF we published earlier [20] to serve as a reference. In addition to the INK4a promoter region (primer sets K and L), we identified a second peak of PRC binding to an area ~3 kb upstream of the INK4b promoter (primer sets C and D). Outside these two domains, binding of EZH2 and BMI1 across the INK4b-ARF-INK4a locus was low. H3K27 me3 levels follow EZH2 binding but are spread over a larger area (Figure 2H). Although only EZH2 was down-regulated in adult HDFs, both EZH2 and BMI1 occupancy at INK4a and INK4b was strongly reduced in these cells (Figure 2F and 2G). This observation agrees well with earlier studies showing that EZH2 can facilitate binding of other PcG proteins [23-25,40]. Like EZH2 binding, H3K27 me3 levels were strongly reduced in adult HDFs compared to neonatal cells (Figure 2H). In senescing MRT cells a distinct mechanism operates. hSNF5 re-expression does not affect EZH2 expression (Figure 2D), but enables SWI/SNF recruitment to INK4a and INK4b, leading to PRC eviction [20].
EZH2 is required for coordinate silencing of INK4a and INK4b
Our results showed that attenuation of EZH2 is accompanied by a loss of PRC1 and PRC2-binding, recruitment of RNA POL II and induction of INK4a and INK4b. ARF expression remained unaffected and is not co-regulated with INK4a and INK4b in the human cells studied here. These observations suggest a critical role for EZH2 in the selective regulation of INK4a and INK4b expression, during ageing and differentiation. To test whether EZH2 is indeed required for the silencing of INK4a and INK4b, we used a shRNA strategy to attenuate its levels in neonatal HDFs. Cells were transduced with lentiviruses expressing either shRNAs targeting EZH2 mRNA (EZH2 KD) or a scrambled control. Three days following the transduction, EZH2 levels were effectively reduced in cells treated with the appropriate shRNA (Figure 3A). In contrast, BMI1 levels were unaffected. Loss of EZH2 caused a strong induction of INK4a and INK4b but not of ARF (Figure 3B). RNA POL II was selectively recruited to the INK4a and INK4b loci, as revealed by ChIP-qPCR (Figure 3C). Depletion of EZH2 leads to its expected disappearance from INK4a and INK4b, but also causes loss of BMI1 binding (Figure 3D and 3E). Similar results were obtained in MRT cells (Additional file 1, Figure S2). Taken together, these results suggest that EZH2 attenuation is sufficient for the dissociation of PcG silencers and induction of INK4a and INK4b.
Figure 3.
Loss of EZH2 causes derepression of INK4a and INK4b. (A) Neonatal human diploid fibroblasts were transduced with lentiviruses expressing either shRNAs targeting E(z)h2 mRNA (EZH2 KD) or a scrambled control. Three days following transduction, EZH2 levels were analysed by real-time quantitative polymerase chain reaction (RT-qPCR) and Western immunoblotting, as described above. The band corresponding to EZH2 is indicated with an arrowhead. In parallel, BMI1 and histone H3 levels were determined. (B) Loss of EZH2 causes transcriptional activation of INK4b and INK4a. Seventy-two hours following transduction, relative expression levels of INK4b, ARF and INK4a were determined by RT-qPCR of isolated mRNA. (C) EZH2 depletion leads to RNA POL II recruitment to INK4a and INK4b, as determined by chromatin immunoprecipitation (ChIP)-qPCR. (D-E) EZH2 depletion causes loss of PRCs from INK4a and INK4b, as revealed by ChIP-qPCR using antibodies directed against EZH2 (D) or BMI1 (E). All analyses were as described in the legend to Figures 1 and 2.
A chromatin loop, linking the repressed INK4a and INK4b, is released during induced expression
We wondered whether, because of their coordinate regulation by PcG silencers, INK4a and INK4b might be in close physical proximity, in spite of their being over 35 kb apart. In order to investigate the three-dimensional conformation of the INK4b-ARF-INK4a locus, we used chromatin conformation capture (3C) technology in combination with qPCR [41,42]. We first compared the INK4b-ARF-INK4a locus higher-order chromatin structure in neonatal and adult HDFs (Figure 4A). Cells were cross-linked with formaldehyde, followed by chromatin isolation and restriction digestion with EcoRI. Our preliminary analysis yielded 10 suitable EcoRI fragments across almost 70 kb of DNA encompassing the INK4b-ARF-INK4a locus (see Methods). Samples were ligated under conditions that favour the union of DNA fragments that are physically connected and qPCR across junctions was used to determine the relative cross-linking frequency between restriction fragments. All 3C data presented here are the result of three independent biological replicate experiments. The 'constant' primer and the TaqMan probe (grey bar) were designed in the EcoRI fragment ~4 kb to 2 kb upstream of INK4b, harbouring the PRC-binding sequences. Plotting of the ligation frequencies to this 'bait' fragment revealed a clear peak at fragment 9 overlapping the INK4a promoter proximal region. These experiments were complimented by 3C analysis using a bait fragment near the INK4a promoter. Now, we observed a peak at fragment 2, encompassing the PRC-binding domain upstream of the INK4b promoter (Figure 4B). We conclude that, in neonatal HDFs, the repressed INK4b-ARF-INK4a locus has a looped structure. The PRC-bound regions upstream of INK4b and proximal to the INK4a promoter are close in nuclear space, whereas the ~35 kb of intervening DNA, including the ARF promoter, loops out.
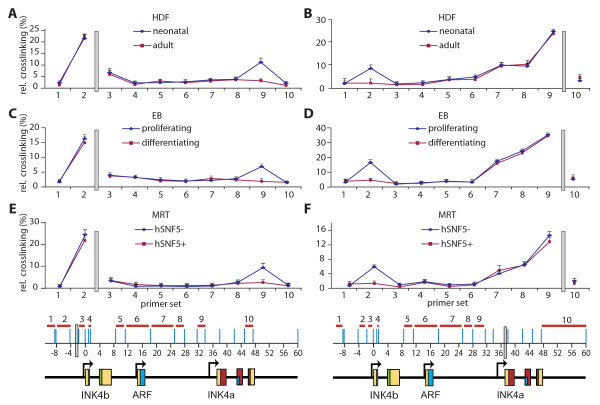
Figure 4.
Regulated chromatin looping between INK4a and INK4b. (A-F) Chromatin conformation capture - quantitative polymerase chain reaction (qPCR) analyses of long-range interactions at the INK4b-ARF-INK4a locus. Locus-wide relative cross-linking frequencies of EcoRI fragments 1 to 10 are plotted. The constant primer and the TaqMan probe (grey bar) were either in the EcoRI fragment -4 to -2 kb upstream of INK4b (A, C and E) or overlapping the INK4a promoter (B, D and F). The locations within the INK4b-ARF-INK4a genomic region of the EcoRI sites (blue lines), bait fragment (harbouring the grey bar, indicating the TaqMan probe) and suitable candidate EcoRI fragments (numbered red segments) are shown at the bottom. The relative cross-linking frequency of each fragment is plotted, with the fragment numbers indicated on the X-axis. (A, B) Neonatal (blue) and adult (red) human diploid fibroblasts. (C, D) Proliferating- (blue) and differentiating (red) eryhtroblasts. (E, F) Proliferating MRT cells lacking hSNF5 (blue) and senescing MON cells expressing hSNF5 (red). Each point represents the mean of three independent biological replicates, each analysed in triplicate by qPCR. Standard deviations are depicted by error bars.
In adult HDFs, the higher-order chromatin conformation of the INK4b-ARF-INK4a locus is dramatically different. 3C-qPCR analyses revealed a loss of long-range interaction between INK4a and INK4b, suggesting the locus adopted a linear conformation (Figure 4A-B). In order to study the effects of progenitor cell differentiation, we undertook a similar 3C analysis of the chromatin structure in proliferating and differentiating ERs (Figure 4C and 4D). Our results revealed the presence of a chromatin loop between INK4a and INK4b in proliferating cells, which is similar to that in neonatal HDFs. When ERs were induced to differentiate towards erythrocytes, the silent chromatin loop dissolved and INK4a and INK4b became de-repressed. Finally, we compared proliferating MRT cells with senescing cells after hSNF5 expression (Figure 4E and 4F). Following hSNF5 expression and PRC removal, the repressive chromatin loop is released and the INK4b-ARF-INK4a locus assumes a linear conformation.
Together, our results showed that, in neonatal HDFs, haematopoietic progenitor cells and MRT cancer cells, the repressed INK4b-ARF-INK4a locus assumes a looped conformation. The ~35 kb chromatin loop links the PRC-binding regions of INK4a and INK4b, whilst excluding ARF. Concomitant with the increased transcription of INK4a and INK4b in adult HDFs, following differentiation or senescence, the chromatin loop dissolves. The loss of looping is concomitant with the loss of PRC-binding, which, in HDFs and differentiating EBs, is caused by attenuation of EZH2. In MRT cells driven towards senescence, EZH2 levels remain stable, but the PRCs are removed by SWI/SNF action. This duly leads to a release of the repressive loop and gene activation, again suggesting that PRC-binding is required for looping.
PRC-binding is required for looping between INK4a and INK4b
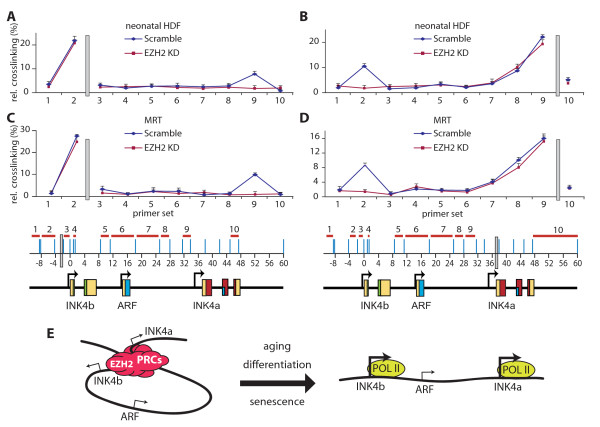
To test whether EZH2 is crucial for loop formation, we transduced neonatal HDFs with lentiviruses expressing either a shRNA targeting EZH2 mRNA or a scrambled control. As shown above, depletion of EZH2 leads to a loss of PRC binding to INK4a and INK4b upstream regions (Figure 3). 3C-qPCR analyses revealed that a loss of EZH2 also results in the release of the repressive chromatin loop that links INK4a and INK4b (Figure 5A and 5B). As shown above, the restoration of SWI/SNF targeting in MRT cells provides an alternate mechanism of PRC removal and loop release (Figure 4E and 4F; Additional file 1, Figure S1). However, EZH2 depletion in the absence of hSNF5 expression, also led to loss of EZH2 and BMI1 binding (Additional file 1, Figure S2) and loss of looping (Figure 5C and 5D). Taken together, these results show that PRC binding is critical for chromatin looping at the INK4b-ARF-INK4a locus. When PRC binding is lost, due either to diminished EZH2 levels or because of SWI/SNF action in MRT cells, the repressive chromatin loop is released concomitant with INK4a and INK4b induction (Figure 5E).
Figure 5.
EZH2 is required for looping between INK4a and INK4b. (A-D) Chromatin conformation capture-quantitative polymerase chain reaction (PCR) analysis on neonatal human diploid fibroblasts (A, B) and malignant rhabdoid tumour (MRT) cells (C, D) transduced with lentiviruses expressing either shRNAs targeting E(z)h2 mRNA (red) or a scrambled control (blue). Procedures were as described in the legend to Figure 4. (E) Parsimonious model for the coordinate regulation of human INK4a and INK4b by PcG repressive complexes (PRCs). In multipotent progenitor cells, young fibroblasts and highly proliferating MRT cancer cells, PRCs simultaneously bind the INK4a promoter and the INK4b upstream promoter region. This creates a looped chromatin structure that links the silenced INK4b and INK4a, but excludes ARF. EZH2 is critical for PRC binding and silencing. During progenitor cell differentiation and ageing, EZH2 attenuates, causing reduced locus occupancy of PRCs and release of the repressive loop. Concomitantly, RNA POL II is recruited selectively to INK4b and INK4a, but not to ARF, leading to gene transcription. MRT cells contain a crippled SWI/SNF complex, due to loss of the hSNF5 tumor suppressor subunit. hSNF5 re-expression in these cells restores SWI/SNF targeting to INK4a and INK4b, causing PRC eviction and release of the repressive loop. For details see discussion.
Discussion
PcG proteins control the balance between differentiation and proliferation
Our results and those of others [18,34,36,40,43], emphasize a crucial role for EZH2 in orchestrating progenitor cell differentiation and proliferation control. Ezhkova et al. [18] observed derepression of INK4a and INK4b due to EZH2 down-regulation, controlling the balance between the proliferative basal layer of progenitor cells and non-proliferating differentiated epidermal cells. These results in the mouse epidermal lineage are highly reminiscent of our findings in human haematopoietic progenitor cells. The histone H3K36 demethylase JHDM1b/KDM2b, a mammalian homolog of the dRAF signature subunit dKDM2, binds and regulates INK4b [19]. Reminiscent of the results described here, He et al. [19] observed that a knockdown of JHDM1b/KDM2b or RING1b in primary mouse embryo fibroblasts causes induction of INK4b and INK4a but not of ARF or the p53-pathway. Consequently, these cells undergo arrest and cellular senescence, for which p15INK4b, it turns out, are critical. In response to oncogenic RAS-signalling in human fibroblasts, the H3K27 me3 demethylase JMDJ3/KDM6B activates INK4a, but not ARF [11,13]. In mouse embryo fibroblasts, JMJD3/KDM6B activates both INK4a and ARF, possibly reflecting a difference in INK4/ARF regulation between mice and man [3,44]. However, as mentioned above, other studies have also presented induction of INK4a and INK4b, but not ARF in mouse cells (see, for example, [18] and [19]). Here, we identified EZH2- and BMI1-binding sequences upstream of the INK4b promoter, providing additional evidence for the control of INK4b by PcG repression. Finally, we emphasize that, under different physiological conditions, different combinations of INK4b-ARF-INK4a expression will be relevant. See, for example, the many examples of co-induction of ARF and INK4a [3,7,8]. Related to this point, it is easy to imagine that, in those situations, the INK4b-ARF-INK4a locus might assume a more distinct conformation than that described here.
Regulation of INK4a and INK4b by EZH2 might be a conserved mechanism required to balance progenitor cell proliferation and differentiation. Unfortunately, this coupling might have sinister consequences when control of EZH2 expression is lost [40]. EZH2 is over-expressed in a variety of tumours, potentially blocking the tumour suppression function of INK4b-ARF-INK4a. In these cells, EZH2 seems to promote de-differentiation and uncontrolled proliferation. Notably, the expression level of EZH2 in early murine haematopoietic cells correlates with their expansion potential. EZH2 overexpression in early haematopoietic progenitors led to a loss of their repopulating ability [43]. Together with early studies in Drosophila, these observations emphasize the importance of PcG protein regulation and dosage.
Following the hSNF5 expression in MRT cells, which triggers the cellular senescence programme, EZH2 levels do not change. In these cells, the SWI/SNF chromatin remodeller is crippled due to a loss of hSNF5 and, therefore, unable to evict PRCs from INK4a and INK4b regulatory elements [20]. Following hSNF5 expression, SWI/SNF is able to remove PRCs from INK4a and INK4b, causing the release of the repressive chromatin loop and gene expression. Thus, a loss of function of a remodeller that counteracts PcG silencing may have similar consequences as EZH2 over-expression. Together with the earlier identification of SWI/SNF as a suppressor of PcG mutations in flies [23,45], these findings emphasize the evolutionary conservation of this regulatory antagonism.
Higher-order chromatin structure and gene expression control
Several studies have implicated higher-order chromatin structure in the regulation of complex multi-gene loci [30,31,46-51]. Typically, models explaining gene control at a distance invoke either spreading of factors along a chromatin fibre or long-range protein-protein interactions that cause looping-out of the intervening chromatin. Two earlier studies proposed a continuous spreading across the whole INK4b-ARF-INK4a locus of either heterochromatinization [52] or PcG proteins [15]. However, the ChIP data in the latter study actually showed a clear peak of PRC binding at the INK4a promoter, which tapers off and is near background at the ARF gene. This pattern of binding at INK4a is reminiscent of what we observed [20]. For the cell types and physiological processes studied here, we favour a discontinuous looping mechanism of INK4b-ARF-INK4a locus control over models invoking blanket spreading of silencers. First, there are two distinct peaks of PRC binding: first, ~3 kb upstream of the INK4b promoter and, second, at the INK4a promoter. Outside these domains, PRC binding is near background levels and we detected no significant PRC-binding at ARF. We note that the PRC-binding peak upstream of INK4b, albeit that it is nearby, does not coincide with the RD regulatory region identified by Gonzalez et al. [52]. In addition, INK4a and INK4b are coordinately derepressed during cell differentiation, ageing and senescence, whereas ARF remained unaffected. Finally, the PRC-bound INK4a and INK4b regulatory regions are linked in nuclear space. Loss of PRC-binding causes the release of chromatin looping and induction of INK4a and INK4b.
Our findings for the INK4b-ARF-INK4a locus dove-tail well with other studies revealing PcG-mediated chromatin looping [30,31]. Chromatin looping is a consequence of the association of proteins bound to two or more distal regulatory elements (see, for example, [50,51,53-55] and references therein). When two genes are brought into the same microenvironment, such as the PRC bound INK4a and INK4b, it facilitates coordinated regulation. The looping-out of an intervening gene, such as ARF, provides a physical separation which allows independent regulation. Thus, in our view, the looped-out chromatin per se is neither active nor repressive. Rather, transcriptional consequences are determined locally, by repressors or activators that can alter the local chromatin status. Here, we presented an example where the higher-order chromatin conformation of a human multi-gene locus reflects its differential pattern of gene expression during diverse physiological processes.
Conclusion
Two classic problems in the study of transcription are the mechanism of long-range gene control and the regulation of multi-gene loci. Here, we studied how PcG silencers control expression of the human INK4b-ARF-INK4a tumour suppressor locus. We concentrated on a variety of physiological processes during which INK4a and INK4b are coordinately upregulated, whereas ARF expression remains unaltered. In agreement with differential regulation, our ChIP analysis revealed non-homogeneous PRC binding to the INK4b-ARF-INK4a locus. In addition to the INK4a promoter, we identified a PRC-binding sequence ~3 kb upstream of the INK4b promoter. PRC binding to these two regions mediates the formation of a ~35 kb chromatin loop, linking INK4b and INK4a but excluding ARF. EZH2 attenuation causes the release of the repressive loop and upregulation of INK4a and INK4b. Thus, EZH2 levels determine the higher-order chromatin structure and expression pattern of the INK4b-ARF-INK4a locus, coupling human progenitor cell differentiation to proliferation control. Our findings revealed a looping mechanism of INK4b and INK4a control, but are difficult to reconcile with models invoking the continuous spreading of PcG silencers.
Methods
Cell isolation, cell culture and lentiviral procedures
UCB was collected in 10 ml Hanks+Hepes (H+H) with 1% heparin by nursing staff of the Department of Obstetrics and Gynecology at the Sint Franciscus Hospital, Rotterdam, The Netherlands, following informed consent of the mothers. Mononuclear cells were isolated by Ficoll density centrifugation (Lymphoprep, Nycomed Pharma, Oslo, Norway). The cell suspension was washed twice with Hanks Balanced Salt Solution (HBSS, Gibco, Breda, The Netherlands) and CD34+ cell were isolated using the indirect CD34+Microbead kit (Miltenyi Biotec, Germany). The purity of these CD34+ cells was 65-70% as determined by flow cytometry. CD34+ cells were cultured at a density of 1-3 × 104/ml in serum free enriched DMEM as previously described [56]. Medium was supplemented with 100 ng/ml stem cell factor (SCF), 100 ng/ml Flt3-L, and 20 ng/ml trombopoietin. Cells were cultured for 6 weeks and the medium was refreshed every week. In order to establish EB cultures, human fetal liver tissue was obtained from elective abortions of patients who had previously signed informed consent forms giving permission for them to be used for research studies (protocol approved by Erasmus MC medical ethical committee). Fetal livers of 12-18 week human embryos were dissected and passed through a 70-μm nylon mesh. EBs were cultivated at a density of 1-2 × 106 cells/ml in serum-free medium (StemSpan; Stem Cell Technologies, BC, Canada) enriched with lipids lipids (40 ng/ml cholesterol-rich lipid mix; Sigma) and supplemented with erythropoietin (2 U/ml, a gift of Orthobiotech, Tilburg, The Netherlands), dexamethasone (1 μM; Sigma, MO, USA) SCF (50 ng/ml, supernatant of CHO producer cells) [57]. The EB culture was expanded by daily partial medium changes and the addition of fresh factors, keeping cell density between 1.5-2 × 106 cells/ml. Proliferation kinetics and size distribution of the cell populations were monitored daily using an electronic cell counter (CASY-1, Schärfe-System, Reutlingen, Germany). In order to induce terminal differentiation EBs were washed and reseeded at 1.5-2 × 106 cells/ml in lipid-enriched StemSpan supplemented with Epo (5 U/ml) and iron-loaded transferrin (1 mg/ml; SCIPAC Ltd, Kent, UK) [57]. Differentiating EBs were maintained at 2-3 × 106 cells/ml and harvested 48 h after induction. Cell morphology was analysed in cytospins stained with histological dyes and neutral benzidine [58], using an OlympusBx40 microscope (40× objective, NA 0.65), an OlympusDp50 CCD camera and Viewfinder Lite 1.0 acquisition software. Tissue culture of HDFs and MON MRT cells was performed according to standard protocols. Adult HDFs (Cascade Biologics, Oregon, USA, CAT No. C-004-5C), were isolated from adult human fore skin and have the potential for ~12 population doublings. Neonatal HDFs (Cascade Biologics, CAT No. C-004-5C), were isolated from neonatal foreskin and have the potential for ~16 population doublings. For our experiments, we used adult and neonatal HDFs of comparable, early passages. TIG3 cells were obtained from the Health Science Research Resource Bank (Osaka, Japan, http://cellbank.nibio.go.jp/celldata/jcrb0506.htm). HDF, MON and TIG3 cells were grown in DMEM supplemented with 10% (v/v) FCS. The hSNF5 expressing lentiviral vector has been described [39]. High titre vector stocks were produced in 293T cells by co-transfection of transfer vector constructs with packaging constructs using standard transfection procedures [59]. To deplete EZH2, cells were transduced with lentiviruses expressing shRNA directed against EZH2 (Clone TRCN0000040073 and TRCN0000040075; Expression Arrest™-The RNAi consortium (TRC) Human shRNA library purchased from Open Biosystems, USA) for 3 days. In a control experiment, the cells were transduced with scramble, non-targeting lentiviruses.
Cell extracts and western blotting
Cell extracts were prepared in RIPA buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 1% NaDOC, 0.1% SDS) and protein concentration determined. ~50 μg of extract was resolved SDS-PAGE, and transferred to 0.45 μm nitrocellulose membrane. Haematopoetic progenitor cells were lysed directly in an equal volume of 2× SDS loading buffer (1 × Tris.Cl pH6.8, 20% glycerol, 4% SDS, O.2 M DTT, 0.001% bromophenol blue). After transfer, membranes were blocked for 1 h in T-PBS with 5% milk and 0.1% Tween20 prior to incubation with primary (overnight) and secondary (1 h) antibodies. Primary antibodies: SUZ12 (Abcam, ab12073), BMI1 (Abcam; ab14389), EZH2 (Santa Cruz; Sc-25383) and Histone H3 (Abcam; ab1791). Western blots were developed with the ECL detection kit (PIERCE) or visualized with the IRDye 680/800 CW (LI-COR) and ODYSSEY Infrared Imageing System according to the supplier's instructions.
RT-qPCR and ChIP-qPCR assays
Total RNA was extracted from cells using the SV Total RNA Isolation System (Promega, WI, USA). cDNA was synthesized from 1 μg of total RNA using random hexamers and Superscript™ II RNase H-Reverse Transcriptase (Invitrogen). Quantitative real-time PCR (MyIQ, Bio Rad) was performed using SYBR Green I. PCR primers were designed using Beacon designer (Premier Biosoft). The qPCR Core Kit (Invitrogen) was used with 400 nM of each primer under the following cycling conditions: 3 min. at 95°C followed by 40 cycles of 10 s at 95°C and 45 s at 60°C. Gapdh was used as an endogenous reference for normalization. Enrichment of specific sequences was calculated using the comparative CT method [60]. ChIPs were performed essentially as described by the Upstate protocol http://www.upstate.com. Cross-linked chromatin was prepared from ~2 × 107 cells. Cells were treated with 1% formaldehyde for 20 min at room temperature. Chromatin isolation, sonication yielding fragments of 300-600 bp and immunoprecipitations were performed according to protocol. The following antibodies were used: BMI1 (Abcam; ab14389), EZH2 (Santa Cruz; Sc-25383), POL II (Santa Cruz; Sc-899), Histone H3 (Abcam; ab1791) and H3-K27 me3 (Upstate; 07-449). The abundance of specific DNA sequences in the immunoprecipitates was determined by qPCR and corrected for the independently determined amplification curves of each primer set. ChIPs using species and isotype-matched immunoglobins directed against an unrelated protein (GST) were used to determine background levels analysed by qPCR as described above. Enrichment of specific DNA sequences was calculated using the comparative CT method [60]. ChIP levels for each region are presented as percentage of input chromatin. H3K27 me3 ChIPs were normalized against histone H3. All data presented are the result of at least three biological replicate experiments. PCR primer sequences are provided in Additional file 1, Table S1. Statistical analysis was performed using R software http://www.r-project.org/.
Chromatin conformation capture assay
The 3C-qPCR assay was performed as described [61]. Formaldehyde-fixed nuclei, prepared from ~107 cells, were digested with EcoRI overnight, followed by ligation with T4 DNA ligase at 16°C for 4 h. Cross-links were reversed and DNA was purified. The PCR control template, primer efficiency and ligation efficiency were determined by digesting and ligating a BAC clone, which encompassed the entire INK4b-ARF-INK4a locus, as previously described [61]. To correct for differences in the quality and quantity of templates, ligation frequencies between the fragments were normalized to a fragment in the human Ercc3 locus. Sample purity and digestion efficiency has been carefully assessed as described [61]. Quantification of ligated products was performed by real-time qPCR with a Taqman probe corresponding to a sequence within a DNA fragment overlapping the PRC-binding region of either INK4b or INK4a. Our analysis yielded 10 suitable EcoR1 fragments covering the INK4b-ARF-INK4a locus. The primers and probe sequences are listed in Additional file 1, Table S2. The amplification conditions used in 3C assays are available on request. Cross-linking frequencies were calculated as previously described [51,61].
Abbreviations
ChIP: chromatin immunoprecipitation; EB: erythroblast; HDF: human diploid fibroblast; HEF: human embryonic fibroblasts; MRT: malignant rhabdoid tumour; PcG: polycomb group; PCR: polymerase chain reaction; PRC: PcG repressive complex; RT-qPCR: real-time quantitative PCR; SCF: stem cell factor; UCB: umbilical cord blood; 3C: chromatin conformation capture; H3K27: H3 lysine 27.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SKK and CPV conceived and designed the study and wrote the manuscript. SKK and PSK performed most of the experiments. EF, FP and MvL participated in the experiments involving haematopoietic cells and the writing of the parts describing them. All authors have read and approved the manuscript
Supplementary Material
Supplementary tables and figures.
Contributor Information
Sima Kheradmand Kia, Email: kheradmandkia@erasmusmc.nl.
Parham Solaimani Kartalaei, Email: p.solaimani@gmail.com.
Elnaz Farahbakhshian, Email: e.farahbakhshian@erasmusmc.nl.
Farzin Pourfarzad, Email: f.pourfarzad@erasmusmc.nl.
Marieke von Lindern, Email: m.vonlindern@erasmusmc.nl.
C Peter Verrijzer, Email: c.verrijzer@erasmusmc.nl.
Acknowledgements
We thank Wouter de Laat and Petra Klous for their advice on 3C and Sjaak Philipsen and Daan Noordermeer for their critical reading of the manuscript. This work was supported by grants from the Dutch Cancer Society KWF EMCR2006-3583 and the Dutch government BSIK 03038, SCDD to CPV.
References
- Beausejour CM, Campisi J. Ageing: balancing regeneration and cancer. Nature. 2006;443:404–405. doi: 10.1038/nature05221. [DOI] [PubMed] [Google Scholar]
- Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE. The Regulation of INK4/ARF in cancer and ageing. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13(1):77–83. doi: 10.1016/S0959-437X(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Jacobs JJL, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PAC, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27 me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agherbi H, Gaussmann-Wenger A, Verthuy C, Chasson L, Serrano M, Djabali M. Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PLoS One. 2009;4:e5622. doi: 10.1371/journal.pone.0005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenführ M, Maertens G, Banck M, Zhou MM, Walsh MJ, Peters G, Gil J. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Martinez-Leal JF, Rizzo S, Martinez D, Hudson D, Visakorpi T, Peters G, Carnero A, Beach D, Gil J. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene. 2005;24:5543–5551. doi: 10.1038/sj.onc.1208735. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine J-C. et al. The Polycomb group proteins bind throughout the INK4a-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gu X, Su I, Bottino R, Contreras J, Tarakhovsky A, Kim S. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, Rappsilber J, Helin K, Hansen KH. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. Embo J. 2007;26:1637–1648. doi: 10.1038/sj.emboj.7601632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15. Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradmand Kia S, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF Mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. MCB. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, El Messaoudi-Aubert S, Racek T, Stock JK, Nicholls J, Rodriguez-Niedenführ M, Gil J, Peters G. Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS One. 2009;4:e6380. doi: 10.1371/journal.pone.0006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo A, Olthof S, Han L, Vellenga E, de Haan G, Schuringa JJ. Repression of BMI1 in normal and leukemic human CD34+ cells impairs self-renewal and induces apoptosis. Blood. 2009;114:1498–1505. doi: 10.1182/blood-2009-03-209734. [DOI] [PubMed] [Google Scholar]
- Müller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. curr Opin Genet Dev. 2009;19:150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome Regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers J, Verrijzer CP. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 2008;22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C, Sánchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Gambetta MC, Oktaba K, Muller J. Essential Role of the glycosyltransferase Sxc/Ogt in polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Tiwari VK, McGarvey KM, Licchesi JDF, Ohm JE, Herman JG, Sch, uuml, beler D, Baylin SB. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6(12):2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen AM, Evers B, Prasad AA, Tanger E, Cornelissen-Steijger P, Jonkers J, van Lohuizen M. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr Biol. 2008;18:1094–1099. doi: 10.1016/j.cub.2008.06.070. [DOI] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Bruggeman S, Hulsman D, Tanger E, Buckle TBM, Zevenhoven J, van Tellingen O, van Lohuizen M. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12:328–341. doi: 10.1016/j.ccr.2007.08.032. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons G, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U, Karlsson G, Karlsson S. Signaling pathways governing stem-cell fate. Blood. 2008;111:492–503. doi: 10.1182/blood-2007-07-075168. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Oruetxebarria I, Venturini F, Kekarainen T, Houweling A, Zuijderduijn LM, Mohd-Sarip A, Vries RG, Hoeben RC, Verrijzer CP. p16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. J Biol Chem. 2004;279:3807–3816. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- Simon J, Lange C. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E, Dontje B, de Haan G. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107:2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G. An INKlination for epigenetic control of senescence. Nat Struct Mol Biol. 2008;15:1133–1134. doi: 10.1038/nsmb1108-1133. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. Brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-E. [DOI] [PubMed] [Google Scholar]
- Cleard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- de Laat W, Klous P, Kooren J, Noordermeer D, Palstra R, Simonis M, Splinter E, Grosveld F, James JB. Current Topics in Developmental Biology. Vol. 82. Academic Press; 2008. 3-Dimensional organization of gene expression in erythroid cells; pp. 117–139. full_text. [DOI] [PubMed] [Google Scholar]
- Madan Babu M, Janga SC, de Santiago I, Pombo A. Eukaryotic gene regulation in three dimensions and its impact on genome evolution. Curr Opin Genet Dev. 2008;18(6):571–582. doi: 10.1016/j.gde.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. Making connections: boundaries and insulators in Drosophila. Curr Opin Genet Dev. 2007;17(5):394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra R-J, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Molecular Cell. 2002;10:1453–1465. doi: 10.1016/S1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Klatt P, Delgado S, Conde E, Lopez-Rios F, Sanchez-Cespedes M, Mendez J, Antequera F, Serrano M. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature. 2006;440:702–706. doi: 10.1038/nature04585. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T, Katsani KR, Verrijzer CP. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 2002;21:1775–1781. doi: 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrascheck M, Escher D, Mahmoudi T, Verrijzer CP, Schaffner W, Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucl Acids Res. 2005;33:3743–3750. doi: 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Merchav S, Wagemaker G. Detection of murine bone marrow granulocyte/macrophage progenitor cells (GM-CFU) in serum-free cultures stimulated with purified M-CSF or GM-CSF. Int J Cell Cloning. 1984;2:356–367. doi: 10.1002/stem.5530020604. [DOI] [PubMed] [Google Scholar]
- Leberbauer C, Boulme F, Unfried G, Huber J, Beug H, Mullner EW. Different steroids co-regulate long-term expansion versus terminal differentiation in primary human erythroid progenitors. Blood. 2005;105:85–94. doi: 10.1182/blood-2004-03-1002. [DOI] [PubMed] [Google Scholar]
- Beug H, Palmieri S, Freudenstein C, Zentgraf H, Graf T. Hormone-dependent terminal differentiation in vitro of chicken erythroleukemia cells transformed by ts mutants of avian erythroblastosis virus. Cell. 1982;28:907–919. doi: 10.1016/0092-8674(82)90070-8. [DOI] [PubMed] [Google Scholar]
- Follenzi A, Sabatino G, Lombardo A, Boccaccio C, Naldini L. Efficient Gene delivery and targeted expression to hepatocytes in vivo by improved lentiviral vectors. Hum Gene Ther. 2002;13:243–260. doi: 10.1089/10430340252769770. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta]CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2(7):1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables and figures.