Abstract
To detect rare epigenetic effects associated with assisted reproduction, it is necessary to monitor methylation patterns of developmentally important genes in a few germ cells and individual embryos. Bisulfite treatment degrades DNA and reduces its complexity, rendering methylation analysis from small amounts of DNA extremely challenging. Here we describe a simple approach that allows determining the parent-specific methylation patterns of multiple genes in individual early embryos. Limiting dilution (LD) of bisulfite-treated DNA is combined with independent multiplex PCRs of single DNA target molecules to avoid amplification bias. Using this approach, we compared the methylation status of three imprinted (H19, Snrpn and Igf2r) and one pluripotency-related gene (Oct4) in three different groups of single mouse two-cell embryos. Standard in vitro fertilization of superovulated oocytes and the use of in vitro matured oocytes were not associated with significantly increased rates of stochastic single CpG methylation errors and epimutations (allele methylation errors), when compared with the in vivo produced controls. Similarly, we compared the methylation patterns of two imprinted genes (H19 and Snrpn) in individual mouse 16-cell embryos produced in vivo from superovulated and non-superovulated oocytes and did not observe major between-group differences. Using bovine oocytes and polar bodies as a model, we demonstrate that LD even allows the methylation analysis of multiple genes in single cells.
Key words: assisted reproduction, bisulfite (pyro)sequencing, bovine, DNA methylation, early embryo, imprinted genes, mouse, oocyte, single cell analysis
Introduction
Epidemiological studies in humans and experimental evidence in animal models indicate that assisted reproductive technologies (ARTs) interfere with epigenetic reprogramming of developmentally important, in particular imprinted genes during gametogenesis and early embryogenesis and may thus be associated with abnormal development.1–5 Genomic imprinting constitutes the parental allele-specific silencing of alleles by differential methylation of cis-regulatory regions (imprinting control centers). It plays an essential role in mammalian development.6,7 Because the most dramatic epigenetic changes occur during germ cell and early embryo development, these are vulnerable time windows for environmental influences on the epigenome of an organism. Following their entry in the genital ridge, mouse primordial germ cells undergo genome-wide demethylation. Parental germline-specific methylation patterns are then established de novo during spermatogenesis and oogenesis, respectively.8 Maternally imprinted genes acquire their methylation marks in a temporally coordinated manner during follicle development and oocyte growth.9,10 In mouse zygotes and early embryos genome-wide demethylation waves erase most of the germline methylation patterns, followed by de novo establishment of somatic methylation patterns around the time of implantation.11,12 Although a larger number of non-imprinted CpG islands within active transcription units may undergo incomplete demethylation during early embryogenesis, only the estimated 100–200 imprinted genes among approximately 25,000 mammalian genes fully escape this methylation reprogramming after fertilization and maintain their germline-specific methylation patterns.13,14
In order to assess the possible effects of different ARTs, including in vitro maturation (IVM) of oocytes, ovarian stimulation, in vitro fertilization (IVF), and/or culture of embryos, on gametogenesis and embryogenesis, it is necessary to analyze the methylation profiles of individual oocytes and early embryos. Methylation data from an apparently homogenous cell population, i.e., a pool of oocytes or blastomeres, provide an incomplete picture, in particular when there is an amplification bias. Epimutations affecting the physiological methylation patterns may be considerably more frequent than somatic DNA mutations, creating an enormous epigenetic variability.15,16 It becomes increasingly clear that an “average cell” with a representative methylation profile does not exist. Moreover, rare events (“needle in a haystack”) are masked in pools of oocytes or embryos. Bisulfite sequencing still is the gold standard for methylation analysis.17 However, most protocols for methylation analysis of imprinted and other non-imprinted, developmentally important genes require larger numbers of cells, i.e., pools of approximately 100 oocytes9,10,18,19 or individual blastocysts with 70–100 cells each20,21 for bisulfite treatment of DNA, followed by amplification, cloning and (pyro)sequencing of differentially methylated regions (DMRs). The degradation and low complexity of bisulfite-converted DNA are a serious challenge for the methylation analysis of small amounts of DNA from a single cell or early embryo. The preferential amplification of either methylated or unmethylated DNA molecules or the stochastic amplification of a single or only few molecules in the starting sample can yield results that are not representative for the studied sample.22 Currently, there are no reliable methods for whole genome amplification of bisulfite-converted DNA in the picogram range.23 Here, we present a simple limiting dilution method which allows one to analyze the methylation patterns of multiple genes in single oocytes or embryos.
Results
Principle of limiting dilution bisulfite (pyro)sequencing.
To avoid amplication bias and to recover as many DNA molecules as possible from the starting sample, we have developed a novel method (Fig. 1) based on limiting dilution24 of bisulfite-treated DNA from individual cells and/or early embryos. The sequestration of individual DNA molecules from the starting sample in separate PCR reactions reduces the methylation status of the targeted chromosomal region to a binary state: an analyzed CpG site on a single DNA molecule is either methylated or not methylated. Each generated bisulfite sequence (amplicon) represents an individual DNA molecule in the starting sample. Multiplex nested PCR allows one to simultaneously analyze several genes in individual early embryos and oocytes.
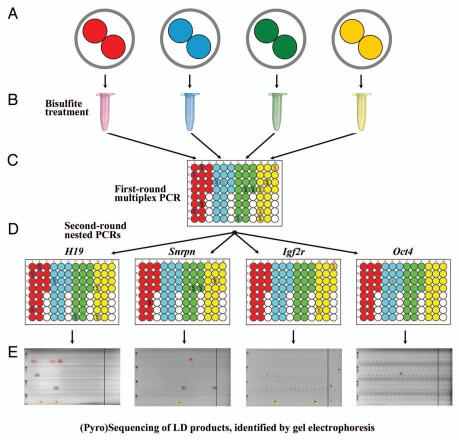
Figure 1.
Principle of limiting dilution bisulfite (pyro)sequencing. (A) Individual two-cell embryos (indicated by different colors) without contaminating sperm or cumulus cells are transferred into separate reaction tubes. (B) DNA isolation and bisulfite conversion. (C) First-round multiplex PCR: the bisulfite-treated DNA of individual embryos (note the color code) is diluted and distributed across 20 wells of a microtiter plate. Four negative controls (without template) are added for each LD (embryo). Wells containing a template for any of the studied gene are marked with an ideogramatic DNA molecule. (D) Second-round nested PCRs of the studied genes are performed in new microtiter plates using one microliter each of the corresponding first-round PCR product as template. (E) The second-round PCR products are visualized on agarose gels. Lanes on the right side of the vertical line present the negative controls. Gene-specific LD products of a particular embryo (encircled in different colors) are analyzed by bisulfite sequencing or pyrosequencing. Please note that the negative control lanes of the Igf2r plate contain a few unspecific PCR products.
LD analysis of three imprinted and one pluripotency gene in individual mouse two-cell embryos.
We have compared the parent-specific methylation patterns of one paternally methylated (H19) and two maternally methylated (Snrpn and Igf2r) imprinted genes in individual mouse two-cell embryos of the following experimental groups: (i) naturally fertilized in vivo produced embryos from in vivo grown and matured oocytes in unstimulated cycles (NFU group), (ii) in vitro fertilized embryos derived from in vivo grown and matured superovulated oocytes (IVF group) and (iii) in vitro fertilized embryos derived from preantral oocytes that were grown and matured in vitro for 13 days (IVC group). To discriminate between maternal and paternal alleles of imprinted genes, the embryos were hybrids from Mus musculus (C57BL/6J x CBA/Ca) females and M. musculus castaneus (CAST/Ei) males. The pluripotency-related gene Oct4 was included in the multiplex assay to detect contamination with sperm and/or somatic cells. Oct4 is unmethylated in early embryos, but highly methylated in sperm, and cumulus and other somatic cells of the follicle or oviduct.
Bisulfite-treated DNA of a single two-cell embryo was diluted to a final volume of 200 µl and evenly distributed into 20 wells of a microtiter plate (Fig. 1). Two-cell embryos with fully replicated chromosomes are endowed with 8 double-stranded (ds) DNA molecules (alleles) of each studied gene. According to a Poisson distribution, most wells do not contain a DNA target molecule, some wells contain a single DNA molecule and very rarely a well may contain two target molecules. Because bisulfite-treated DNA is heavily degraded, the number of wells containing an amplifiable DNA template is always markedly lower than the number of DNA molecules in the starting sample. Four water controls were added to each PCR assay to exclude amplification products caused by environmental DNA contamination. Nested PCR was performed with a first-round multiplex assay using a mixture of outer primers for the four target genes. For each gene, a second round singleplex PCR was performed in a separate plate using 1 µl of the multiplex PCR product as a template for gene-specific inner primers. The second-round PCR products (5 µl each) of the four plates (for H19, Snrpn, Igf2r and Oct4) were run on agarose gels to identify wells with a gene-specific amplification product, which was then analyzed by bisulfite pyrosequencing. The remaining 20 µl of second-round PCR product were sufficient for two pyrosequencing reactions per positive well. Strain-specific SNPs in the second-round H19, Snrpn and Igf2r amplicons were used to determine the parental origin of the analyzed allele in addition to its methylation status.
Table 1 summarizes the methylation results in the three analyzed two-cell embryo groups. A total of 26 naturally fertilized control embryos were studied. Figure 2 presents the methylation patterns of the recovered alleles in each analyzed embryo. Two embryos, NFU21 and NFU22, exhibited abnormally methylated alleles, i.e., all or most (at least 75%) CpGs on a given DNA molecule were aberrantly methylated, indicative of epimutations. NFU21 showed one normally methylated maternal Igf2r allele and one completely unmethylated maternal Igf2r allele with a paternal methylation imprint. This indicates a mosaic Igf2r epimutation. NFU22 was endowed with two abnormally methylated maternal H19 alleles with a paternal methylation imprint, consistent with an H19 epimutation in a non-mosaic state. A few alleles showed single CpG errors, referring to aberrantly methylated CpGs in an overall correctly methylated allele. Single CpG faults most likely represent stochastic methylation errors without functional implications, or alternatively incomplete bisulfite conversion events or amplification errors. In NFU8 and NFU16, one maternal Snrpn allele each displayed a demethylated CpG surrounded by 8 methylated CpGs. NFU7 and NFU10 showed one paternal Snrpn allele each with a methylated CpG and 8 unmethylated CpGs. The average rate of single CpG errors in the four studied genes was 1.1% (4/358). In 26 IVF embryos (Fig. 3) we did not find a single epimutation in the 138 alleles analyzed. Single CpG errors were also rare (12/619 or 2%). Of 18 analyzed IVC embryos (Fig. 4), two showed epimutations. IVC3 displayed an aberrantly methylated maternal H19 allele in addition to two unmethylated H19 alleles. IVC18 displayed one aberrantly methylated maternal H19 allele (mosaic state) and one aberrantly demethylated maternal Igf2r allele. Previously, it had been shown that methylation abnormalities can occur in multiple imprinted genes within the same embryo.21 Although the rate of single CpG errors (8/227 or 3.5%) was somewhat higher than in the two other groups, there were no significant differences (χ2 tests) in the number of single CpG errors or epimutations between the NF, IFV and IVC groups. Interestingly, the maternal alleles of imprinted genes showed a higher number of epimutations (6/131; 5%) and single CpG errors (15/616; 2.4%) than paternal alleles (0/73; 0% and 6/442; 1.4%, respectively). However again, these differences were not statistically significant.
Table 1.
Summary of methylation results in mouse two-cell embryos
| Embryo groups | H19 | Snrpn | Igf2r | Oct4a | |
| NFU | Number of embryos analyzed | 26 | 26 | 26 | 26 |
| Number of recovered maternal alleles per embryo | 0.6 | 0.4 | 0.4 | 0.4 | |
| paternal alleles per embryo | 0.3 | 0.4 | 0.1 | 0.4 | |
| Number (percentage) of abnormal maternal alleles | 2/16 (13%) | 0/10 (0%) | 1/10 (10%) | 0/10.5 (0%) | |
| abnormal paternal alleles | 0/8 (0%) | 0/11 (0%) | 0/3 (0%) | 0/10.5 (0%) | |
| Number (percentage) of maternal single CpG errorsb | 0/56 (0%) | 2/64 (3%) | 0/54 (0%) | 0/21 (0%) | |
| paternal single CpG errorsb | 0/29 (0%) | 2/95 (2%) | 0/18 (0%) | 0/21 (0%) | |
| IVF | Number of embryos analyzed | 26 | 26 | 26 | 26 |
| Number of recovered maternal alleles per embryo | 0.9 | 0.7 | 0.5 | 0.8 | |
| paternal alleles per embryo | 0.7 | 0.8 | 0.3 | 0.8 | |
| Number (percentage) of abnormal maternal alleles | 0/24 (0%) | 0/18 (0%) | 0/12 (0%) | 0/19.5 (0%) | |
| abnormal paternal alleles | 0/17 (0%) | 0/20 (0%) | 0/8 (0%) | 0/19.5 (0%) | |
| Number (percentage) of maternal single CpG errorsb | 0/93 (0%) | 6/117 (5%) | 1/60 (2%) | 1/39 (3%) | |
| paternal single CpG errorsb | 3/58 (5%) | 0/173 (0%) | 0/40 (0%) | 1/39 (3%) | |
| IVC | Number of embryos analyzed | 18 | 18 | 18 | 18 |
| Number of recovered maternal alleles per embryo | 1.2 | 0.6 | 0.4 | 0.4 | |
| paternal alleles per embryo | 0.3 | 0.1 | - | 0.4 | |
| Number (percentage) of abnormal maternal alleles | 2/22 (9%) | 0/11 (0%) | 1/8 (13%) | 0/6.5 (0%) | |
| abnormal paternal alleles | 0/5 (0%) | 0/1 (0%) | - | 0/6.5 (0%) | |
| Number (percentage) of maternal single CpG errorsb | 6/77 (8%) | 0/57 (0%) | 0/38 (0%) | 0.5/13 (4%) | |
| paternal single CpG errorsb | 1/20 (5%) | 0/9 (0%) | - | 0.5/26 (4%) |
Because paternal and maternal alleles could not be distinguished, an equal number of paternal and maternal alleles was assumed to calculate the number of recovered alleles, abnormal alleles and single CpG errors, respectively.
Abnormal alleles excluded.
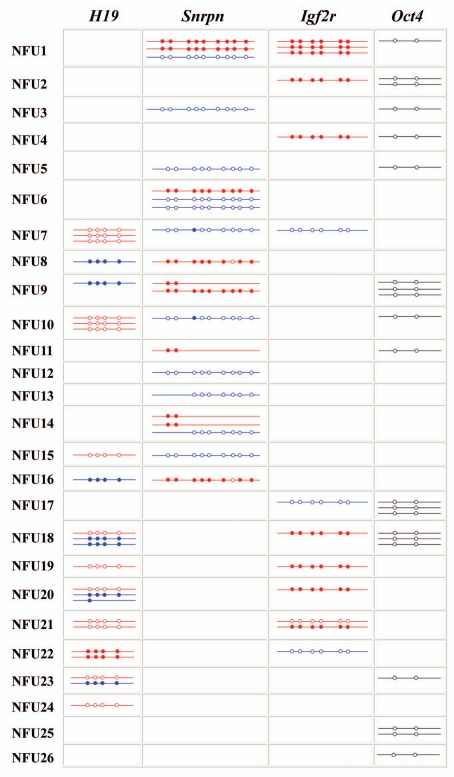
Figure 2.
Methylation patterns of H19, Snrpn, Igf2r and Oct4 in 26 naturally fertilized (NFU) mouse (M. musculus x M. musculus castaneus) two-cell embryos. Each line indicates an individual allele (LD product). Each box displays different alleles that were recovered from a single embryo (the embryo number is indicated on the left side). Maternal alleles of imprinted genes are highlighted in red and paternal alleles in blue. Oct4 alleles are marked in black, because the parental origin was not determined. Open circles represent unmethylated CpG sites and filled circles methylated CpGs. The missing CpGs on some alleles could not be analyzed due to low sequence quality. Note the abnormal methylation patterns of one maternal Igf2r allele in NFU21 and the two abnormally methylated H19 alleles in NFU22. NF7U, NFU8, NFU10 and NFU16 display one Snrpn allele each with a single CpG methylation error.
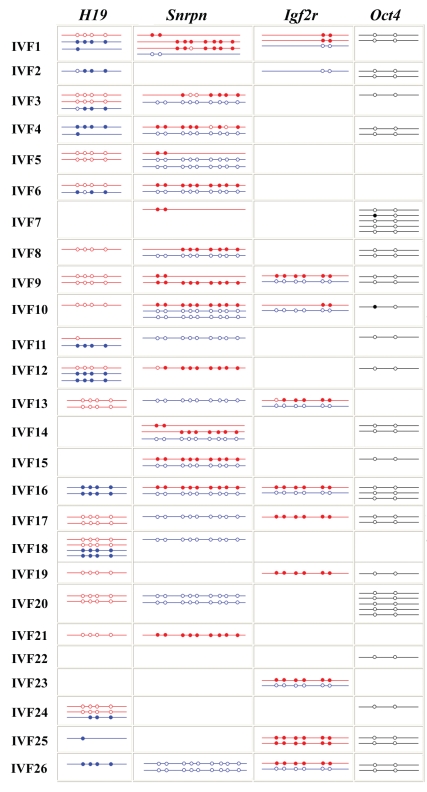
Figure 3.
Methylation patterns of H19, Snrpn, Igf2r and Oct4 in 26 in vitro fertilized (IVF) mouse (M. musculus x M. musculus castaneus) two-cell embryos. Each line indicates an individual allele (LD product). Each box displays different alleles that were recovered from a single embryo. Maternal alleles of imprinted genes are highlighted in red and paternal alleles in blue. Oct4 alleles are marked in black. Open circles represent unmethylated CpG sites and filled circles methylated CpGs.
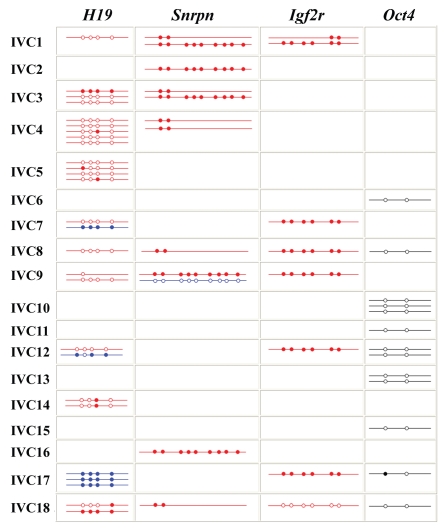
Figure 4.
Methylation patterns of H19, Snrpn, Igf2r and Oct4 in 18 mouse (M. musculus x M. musculus castaneus) two-cell embryos from in vitro cultured (IVC) preantral oocytes. Each line indicates an individual allele (LD product). Maternal alleles of imprinted genes are highlighted in red and paternal alleles in blue. Oct4 alleles are marked in black. Open circles represent unmethylated CpG sites and filled circles methylated CpGs.
Efficiency and robustness of LD bisulfite pyrosequencing.
On average one allele of a given gene was recovered from a single two-cell embryo (Table 1). Since unreplicated two-cell embryos are endowed with four and replicated embryos with eight ds DNA target molecules, the recovery rate was 13–25%. Because bisulfite-treated DNA is heavily degraded, it is not surprising that the recovery rate is dependent on amplicon size. From 70 embryos, we obtained 92 alleles for H19 (176 bp amplicon size), 78 for Oct4 (197 bp), 71 for Snrpn (264 bp) and 41 for Igf2r (384 bp). In the three studied imprinted genes, in which the parental alleles could be discriminated, we always obtained more maternal than paternal alleles. In the NFU group we had 36 maternal versus 22 paternal alleles, in the IVF group 54 versus 45, and in the IVC group 41 versus 6. This preferential amplification was not dependent on the methylation status; it was evident for the paternally methylated H19 gene (62 maternal versus 30 paternal alleles) as well as for the maternally methylated Snrpn (39 versus 32) and Igf2r (30 versus 11) genes.
In total, we obtained methylation patterns of 282 alleles representing 1,232 CpGs. Essentially all (>99.9%) analyzed CpGs exhibited methylation values of <20%, as expected for unmethylated sites, or >80%, typical for methylated sites. Theoretically, the methylation levels scored for individual CpGs should all be either 0% or 100%. However, the actually measured methylation values also depend on the sequence context and other factors. The thresholds for methylation typing were empirically determined in previous studies in references 25 and 26, by pyrosequencing a large number of oocyte DNA samples with homogeneous methylation status such as bisulfite converted LD products or cloned PCR products of completely methylated or completely demethylated DMRs. The methylation difference between duplicate measurements of the same LD products was around 1% (data not shown). This demonstrates that pyrosequencing is a robust technique for methylation typing of CpGs on individual DNA molecules (Yes/No answer). Of the three studied imprinted genes, we obtained 131 maternal and 73 paternal alleles. There was only a single LD product (of Igf2r in embryo IVF26), which contained two different parental alleles. Collectively, these results suggest that the vast majority of LD products represents a single DNA molecule in the starting sample. None of 78 analyzed Oct4 alleles was fully methylated. This shows that the studied samples were not contaminated with sperm and/or somatic cell DNA.
LD analysis of two oppositely imprinted genes in individual mouse 16-cell embryos.
The most likely explanation for the excess of maternal LD products in two-cell embryos is the presence of the second polar body representing one additional maternal allele. To test this hypothesis, we performed an LD analysis of the paternally methylated H19 and the maternally methylated Snrpn gene in mouse 16-cell embryos, where the second polar body is already degraded. Because each single embryo contains 32 (unreplicated) to 64 (fully replicated) ds DNA molecules of a target gene, the bisulfite-converted DNA of a single embryo was distributed into 90-wells of a microtiter plate. To study the effects of superovulation on the embryonic epigenome, we compared the methylation imprints of in vivo produced embryos from naturally fertilized oocytes of unstimulated cycles (NFU group) versus stimulated cycles (NFS). For H19, we obtained 41 maternal and 37 paternal alleles from 9 individual NFU and 52 maternal and 35 paternal alleles from 10 NFS embryos (Fig. 5 and Table 2). We observed one NFU and two NFS embryos with abnormally demethylated paternal H19 alleles in a mosaic state and one NFS embryo with a single abnormally methylated maternal H19 allele. Alleles with at least 75% abnormally (de)methylated CpGs were considered as epimutations. For Snrpn, we recovered 26 maternal and 37 paternal alleles from 11 NFU and 30 maternal and 42 paternal alleles from 10 NFS embryos. One embryo in the NFU group showed a single paternal Snrpn allele and two embryos in the NFS group a single maternal Snrpn allele with epimutations. These differences are not statistically significant. The number of stochastic methylation errors was approximately 2% in both the paternal and the maternal allele in both groups of embryos. Altogether, we recovered 149 maternal (67 NFU and 82 NFS) alleles and 151 paternal (74 NFU and 77 NFS) alleles, which strongly argues against a parent-specific amplification bias of our assay. On average, we recovered 7.6 alleles per gene and embryo, which corresponds to 12–24% of all DNA target molecules in the starting sample, similar to the recovery rate in two-cell embryos.
Figure 5.
Methylation patterns of H19 (left side) and Snrpn (right side) in in vivo produced mouse (M. musculus x M. musculus castaneus) 16-cell embryos from unstimulated (NFU group) versus superovulated matings (NFS group). Each line indicates an individual allele (LD product). Maternal alleles (highlighted in red) and paternal alleles (blue) from the same embryo are grouped together. Open circles represent unmethylated CpG sites and filled circles methylated CpGs.
Table 2.
Summary of methylation results in mouse 16-cell embryos
| Embryo groups | H19 | Snrpn | |
| NFU | Number of embryos analyzed | 9 | 11 |
| Number of recovered maternal alleles per embryo | 4.6 | 2.4 | |
| paternal alleles per embryo | 4.1 | 3.4 | |
| Number (percentage) of abnormal maternal alleles | 0/41 (0%) | 0/26 (0%) | |
| abnormal paternal alleles | 1/37 (3%) | 1/37 (3%) | |
| Number (percentage) of maternal single CpG errorsa | 4/155 (3%) | 4/181 (2%) | |
| paternal single CpG errorsa | 11/123 (9%) | 0/305 (0%) | |
| NFS | Number of embryos analyzed | 10 | 10 |
| Number of recovered maternal alleles per embryo | 5.2 | 3.0 | |
| paternal alleles per embryo | 3.5 | 4.2 | |
| Number (percentage) of abnormal maternal alleles | 1/52 (2%) | 2/30 (7%) | |
| abnormal paternal alleles | 4/35 (11%) | 0/42 (0%) | |
| Number (percentage) of maternal single CpG errorsa | 0/193 (0%) | 8/204 (4%) | |
| paternal single CpG errorsa | 9/113 (8%) | 2/339 (1%) |
Abnormal alleles excluded.
LD analysis of individual bovine oocytes and polar bodies.
To demonstrate the efficiency of LD in single cell methylation analysis, we analyzed three imprinted genes (H19, SNRPN and PEG3) and the pluripotency-related gene OCT4 in single bovine oocytes and their corresponding first polar bodies. Both oocyte and first polar body are endowed with two ds DNA molecules for each target gene. Figure 6 presents the methylation patterns of eight IVM oocytes and their first polar bodies. The imprinted genes were analyzed by direct bisulfite sequencing of the LD products to examine a larger number of CpG sites (20 for H19, 30 for SNRPN and 18 for PEG3), OCT4 was analyzed by bisulfite pyrosequencing. OCT4 alleles were obtained from all eight analyzed oocytes and from two polar bodies. With the exception of a single CpG error in one OCT4 allele of oocyte no. 4, all CpGs were unmethylated, as expected for oocytes. Therefore, we can largely exclude somatic cell DNA contamination. In total, we recovered 13 (81%) of the 16 OCT4 molecules in the eight oocytes and 3 (19%) of the 16 in the first polar bodies. For H19, SNRPN and PEG3, we obtained 3 (19%), 5 (31%) and 2 (13%) alleles, respectively, from oocytes and none, 1 (6%) and 2 (13%), respectively, from polar bodies. Thus, the average recovery rate (for all four studied genes) of single-cell LD methylation analysis is 34% (22/64) in oocytes and 5/64 (8%) in polar bodies. Due to this remarkably high efficiency relatively few cells are needed to obtain a representative view on the methylation status and its variation in a particular cell type. The lower recovery rate in polar bodies, compared to oocytes can be explained by DNA degradation, which starts as soon as the polar body is extruded. Similar to mouse embryo IVC18, the bovine oocyte no. 4 displayed epimutations in both the H19 and SNRPN gene, whereas PEG3 imprinting and OCT4 promoter methylation were normal in both oocyte and polar body. The rate of single CpG errors was 1% (3/292), similar to those in mouse two- and 16-cell embryos.
Figure 6.
Methylation patterns of H19, SNRPN, PEG3 and OCT4 in bovine IVM oocytes and first polar bodies. Each line indicates an individual allele (LD product). Each box displays alleles that were recovered from a single oocyte and/or the corresponding polar body (the oocyte number is indicated on the left side). Maternal alleles of imprinted genes are highlighted in red. OCT4 alleles are marked in black. Open circles represent unmethylated CpG sites and filled circles methylated CpGs. CpG site 17 (gray dot) of bovine PEG3 represents a C/T SNP and was excluded from methylation analysis. Please note the abnormally methylated H19 and SNRPN alleles in oocyte no. 4.
Discussion
Technical aspects.
Bisulfite sequencing is difficult for methylation analysis of single cells and early embryos. Previous studies used either pools of oocytes9,10,18,19 or advanced blastocyst stages.20,21 However, bisulfite sequencing has been adapted to agarose-embedded cells to reduce DNA degradation, which allowed studying one DMR in single cells or embryos.27–29 Recently, a restriction-enzyme-based microfluidic assay for single cell methylation analysis of one target gene has been described in reference 30. Although this assay may be useful for screening one or two CpG sites of a promoter in individual cells, it is not able to resolve the methylated and the unmethylated allele in the same cell. An added advantage of bisulfite sequencing is the analysis of multiple (up to several dozen) adjacent CpG sites in the same amplicon, which is necessary to distinguish stochastic single CpG methylation errors from aberrant methylation of the entire allele. It is generally assumed that the density of methylated CpGs in a cis-regulatory region rather than individual CpGs turn a gene “on” or “off”.31,32 Limiting dilution combined with bisulfite pyrosequencing or direct sequencing, as described here, gives the methylation patterns of multiple genes in bisulfite-treated DNA of single cells or embryos without embedding.
We have developed two different multiplex assays, each with four genes, for studying imprinted gene methylation in the murine25 and bovine model,26 respectively. Outer primers of different studied genes must have similar melting temperatures. The primers were then optimized in a stepwise manner, gene by gene to avoid mispriming. Due to the low complexity of bisulfite-converted DNA that is depleted of C (sense) or G (antisense strand) nucleotides, the primers exhibit reduced specificity. The final validation was made by wet experiments. Development of a multiplex assay for four genes takes two to four weeks. Since each gene-specific second-round PCR requires 1 µl first-round PCR product, our LD assay potentially allows multiplex methylation analysis of up to 25 genes in single cells or embryos. Although multiplexing of ten or more bisulfite primer pairs is difficult, multiplex compatibility is an important advantage of our LD approach, compared to other protocols.
LD relies on independent PCRs of single DNA target molecules and, thus prevents stochastic amplification of a single or a few molecules in the starting sample. On average, we recovered one allele of a target gene in the multiplex from a single two-cell embryo and seven to eight alleles from a 16-cell embryo. For many embryos, we obtained alleles of multiple genes and both paternal and maternal alleles. By analyzing each embryo individually, we can largely exclude artifacts due to somatic cell contamination.
Effects of different ARTs on imprinted gene methylation in early embryos.
One reassuring finding of our study is that none of 26 IVF two-cell embryos from superovulated oocytes showed an epimutation (in 54 maternal and 40 paternal alleles analyzed), whereas 2 of 26 naturally fertilized non-superovulated control embryos displayed an epimutation in H19 and Igf2r, respectively (in 36 maternal and 22 paternal alleles). Obviously, epimutations also occur at a low rate in non-ART embryos and standard IVF protocols do not appear to drastically increase this rate, at least in the mouse model. One of 18 IVC two-cell embryos derived from in vitro grown and matured preantral oocytes showed an epimutation in H19 and another one epimutations in both H19 and Igf2r (in 41 maternal and 6 paternal alleles studied). Although this rate is somewhat higher than in the NF and IVF groups, this difference is not statistically significant.
Most epimutations were observed in a mosaic state. For example, in embryo NFU21 one of two recovered maternal Igf2r alleles and in embryo IVC3 one of three maternal H19 alleles displayed a paternal methylation imprint. This is consistent with the idea that most epimutations in early embryos are due to failures in the maintenance of the methylation patterns, rather than failures in imprint establishment.
When we compared the H19 and Snrpn methylation patterns of in vivo produced 16-cell embryos, we observed a higher number of alleles with epimutations in superovulated embryos (7/159; 4.5%) than in non-superovulated controls (2/141; 1.5%), however again this difference was not statistically significant. Three epimutations were found on maternal alleles and six on paternal alleles, always in a mosaic state. Taken together, we did neither observe dramatic effects of IVF and IVC on methylation imprints in mouse two-cell embryos nor a major impact of superovulation on methylation imprints in 16-cell embryos. However, this does not necessarily contradict previous studies suggesting epigenetic abnormalities after ART manipulations in mouse embryos, fetuses and/or placentae.20,21,33
Our main goal was to demonstrate the feasibility of imprinted gene methylation analysis in a few cells. To this end, we analyzed individual two-cell embryos, which were cultured for only 24–36 h, and 16-cell embryos, which were produced in vivo. ART transfers in human infertility treatment and in vitro production of animals are typically done with blastocyst embryos and at the earliest with eight-cell stages. In the mouse model, different embryo culture conditions, in particular the addition of serum to embryo culture medium can lead to aberrant imprinted gene methylation and expression.34,35 Loss of imprinting appears to occur after the mouse two-cell stage and prior to the blastocyst stage and can persist in a tissue-specific manner, with placental tissues being more sensitive to perturbations than the embryo itself.36 ART-induced epigenetic changes may also depend on the genotypic background. Mouse embryos derived from a B6 female and a castaneus male, as used in our study, appear to be more resistant to methylation changes than Cast x B6 crosses.34
Another special feature of our study is that we distinguished between single CpG methylation errors, which are most likely stochastic errors without pathological consequences and true epimutations (allele methylation errors), which can be expected to interfere with imprinted gene regulation. Allele errors are rare events and with current methods it is almost impossible to analyze a sufficient number of alleles in early embryos in order to detect minor between-group differences. For example, the power for χ2 tests (1 df) to find significant (p < 0.05) differences in the incidence of epimutations between 16-cell embryos from superovulated versus non-superovulated oocytes (Table 2) is 15% for the maternal and 31% for the paternal H19 alleles and 27% and 19%, respectively, for the maternal and paternal Snrpn alleles. Correspondingly, the required sample sizes for a power of 80% were 916 maternal and 267 paternal H19 and 245 maternal and 540 paternal Snrpn alleles. In contrast, when all aberrantly methylated CpGs regardless whether they represent single CpG errors or epimutations, are added up, it is much easier to reach statistical significance. When adding up all aberrantly hypomethylated CpG sites on the maternal Snrpn allele, there is a significant (p < 0.001) difference between superovulated 16-cell embryos (24/220; 11%) and non-superovulated controls (4/181; 2%). If we distinguish between single CpG errors and epimutations, we have 4% (8/204) single CpG errors and two epimutations at the maternal Snrpn allele in superovulated embryos and 2% (4/181) single CpG errors and no epimutation in controls, which is not a significant difference.
On average, we observed approximately 3% epimutations in both mouse two-cell embryos (6 of 204 alleles) and 16 cell-embryos (9 of 300 alleles), without dramatic differences between ART and non-ART groups. Nevertheless, this rate is much higher than the incidence of imprinting disorders in human new-born populations. Although epigenetic mechanisms and DNA methylation patterns of imprinted genes are not fully conserved among mammalian species and one has to be careful to extrapolate from animal models to the human situation, it is tempting to speculate that epimutations that mainly occur in a mosaic state in early embryos contribute to the high rate of implantation failure and/or pregnancy loss in mammals. Previously, we have shown that approximately 10% of human spontaneous abortions and stillbirths display methylation abnormalities in multiple imprinted genes.37
Materials and Methods
Mouse embryos.
To obtain naturally fertilized (NFU) embryos, 3–6 months-old C57BL/6J x CBA/Ca females were housed together with a 5–12-month old CAST/Ei male. Two-cell embryos were flushed 24 h after mating from female oviducts and transferred to Leibovitz L-15 medium (PAN, Aidenbach, Germany). After washing four-times in 50 µl droplets of PBS, single embryos without contaminating cumulus cells or sperms (by visual inspection) were transferred into 10 µl PBS each and frozen at −80°C until further analysis. The 26 naturally fertilized (NFU) embryos studied here were obtained from five different matings.
For superovulation, 3–6-months-old C57BL/6J x CBA/Ca females were injected with 7.5 IU PMSG (Intervet, Unterschleißheim, Germany) and 48 hours later with 7.5 IU hCG (Merck Serono, Darmstadt, Germany). Cumulus oocyte complexes (COC) with competent metaphase II oocytes were harvested 14 h after superovulation from oviducts and transferred to 100 µl α-MEM glutamax medium (Invitrogen, Karlsruhe, Germany) supplemented with 3 mg/ml BSA. Pools of about 20–30 COCs were in vitro fertilized with 1 × 105 capacitated sperms from 5–12-months old CAST/Ei males. The remaining cumulus cells and sperms were removed after 4–6 h. Oocytes were washed twice with PBS and incubated for 36 h at 37°C in 100 µl IVF medium covered with embryo oil. Two-cell embryos were collected 24–36 hours after IVF and washed four times. Single embryos were frozen in 10 µl PBS each. The 26 analyzed IVF embryos were obtained from nine superovulated females and three different IVF attempts.
Preantral follicles were isolated from 12–14-day-old C57BL/6J x CBA/Ca females and cultured, as described previously in reference 25. Culture medium consisted of α-MEM glutamax with 5% FCS, 5 µg/ml insulin, 5 µg/ml transferrin, 5 ng/ml sodium selenite (Sigma-Aldrich, Munich, Germany), and 10 mIU/ml recombinant FSH. In vitro folliculo- and oogenesis was performed for 12 days and on every fourth day 20 µl of medium was replenished; 10 mIU/ml LH (kindly donated by Merck Serono) was added once at the beginning of culture. In vitro ovulation was stimulated by 1.5 IU/ml recombinant hCG (donated by Merck Serono) and 5 ng/ml recombinant EGF (Promega, Mannheim, Germany). After 18 hours of maturation, COC containing competent metaphase II oocytes were harvested and used for IVF. Eighteen in vitro grown (IVC) and fertilized two-cell embryos were obtained from ten in vitro follicle cultures and five different IVFs.
To obtain early morula-stage embryos, 6–8-week old superovulated (for protocol, see above) or non-superovulated C57BL6/J females were mated with Cast/Ei males. Embryos were flushed from the oviducts 60 h after fertilization. Only “high quality” 16-cell embryos (by visual inspection) were used for further analysis. Embryos with developmental delay or morphological signs of fragmentation were discarded.
Bovine oocytes.
Oocytes were collected from abattoir ovaries and in vitro matured in TCM199 medium (Sigma-Aldrich), as described previously in reference 26. Cumulus cells were removed by enzymatic treatment with 0.1% hyaluronidase and subsequent vortexing. To prepare the oocytes for physical manipulation, they were washed three times in TCMair and then incubated in TCMair containing 7.5 µg/ml cytochalasin B (Sigma-Aldrich) for 10 min at 39°C. The extruded first polar body was visualized by inverted light microscopy and removed using microneedles (with 20–25 µm diameter). Oocytes were washed again three times in PBS containing 0.1% polyvinyl pyrrolidone. The oocyte and the corresponding polar body were transferred into separate tubes and frozen at −80°C until further use.
Primer design.
PCR and sequencing primers (Table 3) were designed using the Pyrosequencing Assay Design Software (Biotage, Uppsala, Sweden). To discriminate between maternal (M. musculus) and paternal (M. musculus castaneus) alleles, primers were chosen from regions that contain a strain-specific SNP. The four studied mouse genes (H19, Snrpn, Igf2r and Oct4) as well as bovine OCT4 were analyzed by bisulfite pyrosequencing, which can quantify the methylation levels of a limited number (1–7) of CpGs located within 30–50 bp 3′ from the sequencing primer. Three imprinted bovine genes (H19, SNRPN and PEG3) were analyzed by direct sequencing of bisulfite-treated DNA. This allows one to look at 20–30 CpGs within an amplicon. Addition of M13 sequence and a G or C rich tag containing at least 50% cytosines (forward primer) or guanines (reverse primer) to the nested PCR primers for direct sequencing improved the bisulfite sequence quality,26,38 by increasing the binding specificity during the Sanger sequencing reaction.
Table 3.
Genes and primers for bisulfite (pyro)sequencing
| Gene/repeat | Primer | Sequence (5′-3′) | Amplicon length (bp) | Chromosomal localization (bp)a | Number of CpGs | Strain-specific SNP |
| mH19 | outer forward | AAA TTT TAA TTT TGG TTG TTT TTG G | MMU 7 | |||
| outer reverse | AAT CAA TTA AAA AAA TAA TAA AAC CC | 292 | 149, 766, 893-149, 767, 185 | |||
| nested forward | TGG TTG TTT TTG GAA TAT AAT GTT | |||||
| nested reverse | bAAA AAC AAA ACA CCT ATA CCC TTC | 176 | 149, 766, 905-149, 767, 072 | |||
| pyrosequencing 1 | TTT AAG ATG ATA GTT ATT AG | 4 | ||||
| pyrosequencing 2 | TTG GTT TAT GGG GTT | 1 | rs338202061 | |||
| mIgf2r | outer forward | GGT ATT TTG AGG GTG TAA ATT GTA | MMU 17 | |||
| outer reverse | AAC CCT AAC ACA ACT AAA CAA CAT | 422 | 12, 935, 356-12, 935, 778 | |||
| nested forward | GAA GGG TTT TGT GAT TAG GGT TAA | |||||
| nested reverse | bAAC CCT AAC ACA ACT AAA CAA CAT | 384 | 12, 935, 394-12, 935, 778 | |||
| pyrosequencing 1 | GTT GTA AGA GAG GTA AGT TT | 4 | ||||
| pyrosequencing 2 | AAA GGG TTG GAT TTT TAG | 2 | rs46625914 | |||
| mSnrpn | outer forward | TTG GTA GTT GTT TTT TGG TAG GAT | MMU 7 | |||
| outer reverse | ATA AAC CCA AAT CTA AAA TAT TTT AAT CA | 295 | 67, 149, 848-67, 150, 143 | |||
| nested forward | bTTG GTA GTT GTT TTT TGG TAG GAT | |||||
| nested reverse | TAA AAT ACA CTT TCA CTA CTA AAA TCC AC | 264 | 67, 149, 879-67, 150, 143 | |||
| pyrosequencing 1 | TCC CAA ACA ATA ACT A | 2 | rs46036463 | |||
| pyrosequencing 2 | ACT CCC TCT CCT CTC TAC | 7 | ||||
| mOct4 | outer forward | TTG AGT GGG TTG TAA GGA TAG G | MMU 17 | |||
| outer reverse | AAA AAA TTT CAC CTC TCC CTC C | 329 | 35, 642, 662-35, 642, 991 | |||
| nested forward | GTA GGG GTG AGA GGA TTT TGA A | |||||
| nested reverse | bCCA CCC TCT AAC CTT AAC CTC T | 197 | 35, 642, 766-35, 642, 963 | |||
| pyrosequencing | GTT TGG AAG ATA TAG GTA GA | 2 | ||||
| bH19 | outer forward | GAG GGG TAT TGA GAG GTT G | BTA 29 | |||
| outer reverse | CAA ACA TAA AAA TCC CTC AAT ATC CC | 279 | 51, 358, 501-51, 358, 779 | |||
| inner forward | cAGA GGT TGT GGG TGT GGA GAT A | |||||
| inner reverse | dTCC TCT CCC ACC TTC AAC AA | 230 | 51, 358, 512-51, 358, 741 | 20 | ||
| bSNRPN | outer forward | GGG GTG GGG TAG ATA TTA TTT T | NW_001501801.1 (BTA 21) | |||
| outer reverse | AAA AAA AAA AAA TAT TAC CCA CCA CAC | 299 | 23, 984-24, 282 | |||
| inner forward | cGGT TTT TTT GTT TGA GAG AG | |||||
| inner reverse | dAAA AAA AAA AAA TAT TAC CCA C | 276 | 23, 984-24, 259 | 30 | ||
| bPEG3 | outer forward | GAT ATG TTT ATT TTT GGT TGT TGG | BTA18 | |||
| outer reverse | ACC CTA ATC CCA AAC TCC AAC TAA CC | 280 | 64, 374, 680-64, 374, 959 | |||
| inner forward | cGTG TGG GGG TAT TAG AGT TTG T | |||||
| inner reverse | dACC CTA ATC CCA AAC TCC A | 235 | 64, 374, 680-64, 374, 914 | 19 | ||
| bOCT4 | outer/inner forward | TGG AGA GGG GTT TTG AAG AA | BTA 23 | |||
| outer reverse | CAA AAA TCT CCA CCC AAA CC | 180 | 27, 905, 312-27, 905, 491 | |||
| inner reverse | bTCC AAA CCC CAA ACT CCT AAA CT | 156 | 27, 905, 312-27, 905, 467 | |||
| pyrosequencing | GGA GTT GGA AGT GAA GGT | 2 |
According to Ensembl version 61 of the mouse genome and NCBI release Btau 4.0 of the bovine genome, respectively.
Biotinylated.
Tagged with M13 (italics) and stuffer sequence (bold) TGT AAA ACG ACG GCC AGT CCA CTC ACT CAC CCA CCC.
Tagged with M13 (italics) and stuffer sequence (bold) CAG GAA ACA GCT ATG ACC GGG TGG GAG GTG GGA GGG.
Bisulfite conversion.
DNA from individual mouse embryos and bovine oocytes, respectively, was extracted and bisulfite converted using the EZ DNA Methylation Direct Kit (Zymo Research, Orange, CA USA), which is particularly suited for small amounts of DNA. Briefly, 1 µl of proteinase K (20 µg/µl) and 10 µl of 2x digestion buffer were added to the tube containing a single embryo or oocyte. After incubation for 20 min at 50°C, 130 µl of bisulfite conversion mix were added. Proteinase K digestion breaks down the proteins associated with the DNA freeing it for the conversion reaction, which was performed in a thermal cycler at 98°C for 8 min and 64°C for 3.5 h. Then the DNA was cleaned with a spin column and eluted in 10 µl elution buffer.
Limiting dilution (LD) bisulfite pyrosequencing of mouse embryos.
For the LD analysis of two-cell embryos, the bisulfite-treated DNA was diluted to a final volume of 200 µl. Aliquots with 10 µl template each were distributed into 20 wells of a microtiter plate; four negative controls containing no template were added to each LD. For 16-cell embryos, the 10 µl bisulfite-converted DNA of a single morula were diluted 1:90 to a final volume of 900 µl and distributed into 90 wells of a PCR microtiter plate; six water controls were added. The optimum dilution was determined empirically. To ensure that most LD products represent a single DNA molecules, the number of wells should be larger than the maximum number of DNA molecules in the starting sample and after LD analysis at least half of the wells should not contain a PCR product.
First-round multiplex PCR was performed in 25 µl reactions, each well containing 2.5 µl AmpliTaq Gold buffer and 0.25 µl AmpliTaq Gold polymerase (Applied Biosystems, Darmstadt, Germany), 4 µl MgCl2, 0.5 µl dNTPs and 0.3 µl (10 pmol/µl) of each outer primer of the studied genes (Table 3). PCR was carried out with an initial denaturation step at 95°C for 9 min, 35 cycles of 94°C for 20 sec, 52°C for 30 sec and 72°C for 1 min, and a final extension step at 72°C for 10 min. In a second round, nested singleplex PCRs for each of the studied genes were performed using 1 µl of the first-round multiplex PCR product as a template. The reaction volume consisted of 2.5 µl 10x buffer and 0.2 µl Fast Start Taq polymerase (Roche, Mannheim, Germany), 0.5 µl dNTPs, 1.25 µl (10 pmol/µl) of each forward and reverse primer and 18.3 µl PCR grade water. Cycling conditions were as follows: H19, 95°C for 5 min, 35 cycles of 95°C for 30 sec, 57°C for 30 sec and 72°C for 1 min and a final extension at 72°C for 7 min; Igf2r, 95°C for 5 min, 37 cycles of 95°C for 30 sec, 57°C for 30 sec and 72°C for 1 min, and a final extension at 72°C for 7 min; Snrpn, 95°C for 5 min, 29 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min, and a final extension step; Oct4, 95°C for 5 min, 38 cycles of 95°C for 30 sec, 57°C for 30 sec, and 72°C for 1 min, and then 72°C for 7 min. The singleplex PCRs (5 µl each) were run on an agarose gel to visualize reactions yielding a product representing a single DNA molecule (allele) in the starting sample. Pyrosequencing was performed on a Pyromark Q96MD system with the PyroGold SQA reagent kit (Qiagen, Hilden, Germany). The Pyromark Q-CpG software was used to analyze the methylation levels as well as the strain specific SNPs.
Direct bisulfite sequencing of bovine oocytes and polar bodies.
The bisulfite-treated DNA of a single oocyte or first polar body was distributed in 5 wells each of a microtiter plate. Three water controls were also run. Multiplex PCR with a mixture of outer primers for the four studied genes (Table 3), followed by gene-specific single PCRs with nested inner primers was used to amplify individual alleles. FastStart Taq Polymerase (Roche Diagnostics) was used in both first and second round PCR. In general, PCR amplifications were carried out in 25 µl reactions (see above) with an initial denaturation step at 95°C for 4 min, 30–35 cycles of 95°C for 30 sec, primer-specific annealing temperature for 30 sec, 72°C for 45 sec, and a final extension step at 72°C for 5 min. Multiplex PCR was performed with an annealing temperature of 54°C for 35 cycles. Gene-specific nested PCR of H19 was performed with 65°C annealing temperature and 32 cycles; SNRPN, PEG3 and OCT4 were amplified at 57°C annealing temperature for 30 cycles. Direct bisulfite sequencing of the three imprinted genes was performed on an ABI 3130xl automated sequencer. OCT4 was analyzed by bisulfite pyrosequencing, as described above.
Outlook
Although ARTs are widely applied in animals and humans, their epigenetic effects in individual germ cells and early embryos have not been systematically analyzed. Improving human infertility treatment and the in vitro production of embryos in farm animals requires a better knowledge about the natural epigenetic variability in early embryos and the extrinsic variations due to manipulations of germ cells and/or embryos. Our LD assay provides the possibility to analyze the methylation patterns of multiple genes in minute amounts of DNA. Our results on bovine oocytes demonstrate that LD can efficiently recover alleles from single bisulfite-treated cells. The LD protocol described here is robust and should be valuable in various kinds of tissues, including somatic cell nuclear transfer derived embryos as well as stem cells and cancer cells of various types.
Acknowledgments
This work was supported by grants of the German Research Foundation (PAK 86 and FOR 1041). We thank Dr. E. Schneider and Dr. C.J. Scholz for help with statistical analyses.
Abbreviations
- ART
assisted reproductive technology
- COC
cumulus oocyte complex
- DMR
differentially methylated region
- ds
double-stranded
- IVC
in vitro culture
- IVF
in vitro fertilization
- IVM
in vitro maturation
- LD
limiting dilution
- NFS
natural fertilization from stimulated cycles
- NFU
natural fertilization from unstimulated cycles
- PCR
polymerase chain reaction
- SNP
single nucleotide polymorphism
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Horsthemke B, Ludwig M. Assisted reproduction: the epigenetic perspective. Hum Reprod Update. 2005;11:473–482. doi: 10.1093/humupd/dmi022. [DOI] [PubMed] [Google Scholar]
- 2.Wrenzycki C, Herrmann D, Lucas-Hahn A, Gebert C, Korsawe K, Lemme E, et al. Epigenetic reprogramming throughout preimplantation development and consequences for assisted reproductive technologies. Birth Defects Res C Embryo Today. 2005;75:1–9. doi: 10.1002/bdrc.20035. [DOI] [PubMed] [Google Scholar]
- 3.Haaf T. Methylation dynamics in the early mammalian embryo: implications of genome reprogramming defects for development. Curr Top Microbiol Immunol. 2006;310:13–22. doi: 10.1007/3-540-31181-5_2. [DOI] [PubMed] [Google Scholar]
- 4.Amor DJ, Halliday J. A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum Reprod. 2008;23:2826–2834. doi: 10.1093/humrep/den310. [DOI] [PubMed] [Google Scholar]
- 5.Grace KS, Sinclair KD. Assisted reproductive technology, epigenetics and long-term health: a developmental time bomb still ticking. Semin Reprod Med. 2009;27:409–416. doi: 10.1055/s-0029-1237429. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson-Smith AC, Surani MA. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293:1086–1089. doi: 10.1126/science.1064020. [DOI] [PubMed] [Google Scholar]
- 7.Kelsey G. Genomic imprinting—roles and regulation in development. Endocr Dev. 2007;12:99–112. doi: 10.1159/000109637. [DOI] [PubMed] [Google Scholar]
- 8.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 9.Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79:530–538. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- 10.Hiura H, Obata Y, Komiyama J, Shirai M, Kono T. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells. 2006;11:353–361. doi: 10.1111/j.1365-2443.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- 11.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 12.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 13.Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 14.Smallwood SA, Tomizawa SI, Krueger F, Ruf N, Carli N, Segonds-Pichon A, et al. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet. 2011;43:811–814. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goyal R, Reinhardt R, Jeltsch A. Accuracy of DNA methylation pattern preservation by the Dnmt1 methyltransferase. Nucleic Acids Res. 2006;34:1182–1188. doi: 10.1093/nar/gkl002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider E, Pliushch G, el Hajj N, Galetzka D, Puhl A, Schorsch M, et al. Spatial, temporal and interindividual epigenetic variation of functionally important DNA methylation patterns. Nucleic Acids Res. 2010;38:3880–3890. doi: 10.1093/nar/gkq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olek A, Oswald J, Walter J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 1996;24:5064–5066. doi: 10.1093/nar/24.24.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anckaert E, Romero S, Adriaenssens T, Smitz J. Effects of low methyl donor levels in culture medium during mouse follicle culture on oocyte imprinting establishment. Biol Reprod. 2010;83:377–386. doi: 10.1095/biolreprod.109.082164. [DOI] [PubMed] [Google Scholar]
- 19.Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, et al. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–820. doi: 10.1242/dev.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fauque P, Jouannet P, Lesaffre C, Ripoche MA, Dandolo L, Vaiman D, et al. Assisted reproductive technology affects developmental kinetics, H19 imprinting control region methylation and H19 gene expression in individual mouse embryos. BMC Dev Biol. 2007;7:116. doi: 10.1186/1471-213X-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet. 2010;19:36–51. doi: 10.1093/hmg/ddp465. [DOI] [PubMed] [Google Scholar]
- 22.Warnecke PM, Mann JR, Frommer M, Clark SJ. Bisulfite sequencing in preimplantation embryos: DNA methylation profile of the upstream region of the mouse imprinted H19 gene. Genomics. 1998;51:182–190. doi: 10.1006/geno.1998.5371. [DOI] [PubMed] [Google Scholar]
- 23.Mill J, Petronis A. Profiling DNA methylation from small amounts of genomic DNA starting material: efficient sodium bisulfite conversion and subsequent whole-genome amplification. Methods Mol Biol. 2009;507:371–381. doi: 10.1007/978-1-59745-522-0_27. [DOI] [PubMed] [Google Scholar]
- 24.Daser A, Thangavelu M, Pannell R, Forster A, Sparrow L, Chung G, et al. Interrogation of genomes by molecular copy-number counting (MCC) Nat Methods. 2006;3:447–453. doi: 10.1038/nmeth880. [DOI] [PubMed] [Google Scholar]
- 25.Trapphoff T, el Hajj N, Zechner U, Haaf T, Eichenlaub-Ritter U. DNA integrity, growth pattern, spindle formation, chromosomal constitution and imprinting patterns of oocytes from vitrified pre-antral follicles. Hum Reprod. 2010;25:3025–3042. doi: 10.1093/humrep/deq278. [DOI] [PubMed] [Google Scholar]
- 26.Heinzmann J, Hansmann T, Zechner U, Haaf T, Niemann H. Epigenetic profile of developmentally important genes in bovine oocytes. Mol Reprod Dev. 2011;78:188–201. doi: 10.1002/mrd.21281. [DOI] [PubMed] [Google Scholar]
- 27.Geuns E, de Rycke M, van Steirteghem A, Liebaers I. Methylation imprints of the imprint control region of the SNRPN-gene in human gametes and preimplantation embryos. Hum Mol Genet. 2003;12:2873–2879. doi: 10.1093/hmg/ddg315. [DOI] [PubMed] [Google Scholar]
- 28.Geuns E, Hilven P, van Steirteghem A, Liebaers I, de Rycke M. Methylation analysis of KvDMR1 in human oocytes. J Med Genet. 2007;44:144–147. doi: 10.1136/jmg.2006.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geuns E, de Temmerman N, Hilven P, van Steirteghem A, Liebaers I, de Rycke M. Methylation analysis of the intergenic differentially methylated region of DLK1-GTL2 in human. Eur J Hum Genet. 2007;15:352–361. doi: 10.1038/sj.ejhg.5201759. [DOI] [PubMed] [Google Scholar]
- 30.Kantlehner M, Kirchner R, Hartmann P, Ellwart JW, Alunni-Fabbroni M, Schumacher A. A high-throughput DNA methylation analysis of a single cell. Nucleic Acids Res. 2011;39:44. doi: 10.1093/nar/gkq1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sontag LB, Lorincz MC, Georg Luebeck E. Dynamics, stability and inheritance of somatic DNA methylation imprints. J Theor Biol. 2006;242:890–899. doi: 10.1016/j.jtbi.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 33.Fortier AL, Lopes FL, Darricarrere N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet. 2008;17:1653–1665. doi: 10.1093/hmg/ddn055. [DOI] [PubMed] [Google Scholar]
- 34.Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- 35.Khosla S, Dean W, Brown D, Reik W, Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001;64:918–926. doi: 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- 36.Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, Bartolomei MS. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131:3727–3735. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- 37.Pliushch G, Schneider E, Weise D, el Hajj N, Tresch A, Seidmann L, et al. Extreme methylation values of imprinted genes in human abortions and stillbirths. Am J Pathol. 2010;176:1084–1090. doi: 10.2353/ajpath.2010.090764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han W, Cauchi S, Herman JG, Spivack SD. DNA methylation mapping by tag-modified bisulfite genomic sequencing. Anal Biochem. 2006;355:50–61. doi: 10.1016/j.ab.2006.05.010. [DOI] [PubMed] [Google Scholar]