Abstract
HA22-LR is a recombinant immunotoxin for the treatment of B-cell malignancies that contains the Fv portion of an anti-CD22 antibody fused to a functional portion of Pseudomonas exotoxin A. In the present study, we attempted to improve this molecule. First, we produced a single-chain version of HA22-LR (scdsFv-HA22-LR) in which a peptide linker was introduced between the disulfide-linked light and heavy chains to enable production via single fermentation. No difference in cytotoxic activity was observed between scdsFv-HA22-LR and prototype HA22-LR. Next, we attempted to increase the affinity of scdsFv-HA22-LR by using alanine scanning mutagenesis of complementarity determining regions (CDRs) to assess the specific contribution of each CDR residue to the antigen binding. We found that mutation of asparagine 34 in VLCDR1, which is located at the VL/VH interface, to alanine (N34A) caused a substantial increase in affinity and activity. Estimated KD values measured by fluorescence-activated cell sorting were lowered by 10-fold: 0.056 nM in the N34A mutant compared to 0.58 nM in wild type (WT). Cell viability assays of CD22-positive B-cell lymphoma and leukemia cell lines showed that the N34A mutant had increased cytotoxicity ranging from ∼2 (HAL-1, IC50(WT): 2.37 ± 0.62 ng/ml, IC50(N34A): 1.32 ± 0.41 ng/ml) to 10 (SUDHL-6, IC50(WT): 0.47 ± 0.090 ng/ml, IC50(N34A): 0.048 ± 0.018 ng/ml)-fold compared to WT immunotoxin. The present study suggests that the N34A mutant of scdsFv-HA22-LR could have important consequences in a clinical setting.
Key words: immunotoxin, HA22, affinity-maturation, alanine scan, VH/VL interface
Introduction
The binding of antibodies to specific antigens on cancer cells has prompted their use as targeted therapies for cancer.1 The Food and Drug Administration has approved 30 antibody-based therapies, and it is expected that many more will follow.2 Immunotoxins are a category of immunoconjugates in which antibodies are joined to protein toxins. They exploit the precision of antibodies and the lethality of protein toxins to target and kill cancer cells expressing specific cell surface proteins. Any tumor-associated cell-surface antigen is a potential target for immunotoxins as long as it is not expressed on essential normal cells. A variety of plant, fungal and bacterial toxins have been adapted for use with immunotoxins, including ricin, diphtheria toxin and Pseudomonas exotoxin A (PE).3,4
Our approach to targeted therapy is to genetically fuse the variable fragment (Fv) of a tumor-reactive antibody to a portion of PE. PE-based immunotoxins are currently in clinical studies for the treatment of lymphomas and leukemias, as well as solid tumors.5,6 A Phase 1 study of the anti-CD25 immunotoxin LMB-2 (anti-TacFv-PE38) showed a 23% response rate in patients with hematologic malignancies refractory to standard chemotherapy.7 Also, a Phase 1 study of the anti-mesothelin immunotoxin SS1P demonstrated minor but encouraging responses for treating solid tumors in patients with mesothelioma or ovarian cancer who had failed standard therapies.6
We have focused much of our recent efforts in targeting CD22 on B cell malignancies. The recombinant immunotoxin BL22 contains the Fv fragment of an anti-CD22 mAb fused to a 38 kDa fragment of PE.8 In Phase 1 and Phase 2 clinical studies, BL22 was highly active in hairy cell leukemia (HCL) despite prior purine analog treatment and resistance.9 Patients with chronic lymphocytic leukemia (CLL), however, had poorer response to BL22, which we attribute to much lower expression of CD22 on CLL compared with HCL cells.5 To enable more immunotoxin to bind to and enter cells, rather than disassociating from the antigen, the off-rate of BL22 was decreased by mutagenesis of the third complementary determining region (CDR) of the heavy chain (VHCDR3).10 The resulting immunotoxin, HA22 (moxetumomab pasudotox), contains 3 amino acid mutations; 100Ser-100aSer-100bTyr in Fv of BL22 were changed to 100Thr-100aHis-100bTrp.10 The mutant immunotoxin bound CD22 with a 10-fold higher affinity due to a slower off-rate. It has significantly improved cytotoxicity and is undergoing Phase 1 testing in HCL, CLL, non-Hodgkin lymphoma and acute lymphoblastic leukemia in children NCT00659425.11
To achieve more productive intracellular trafficking and less immunogenicity, the proteolytic susceptibility of the PE38 portion of HA22 was modified.12 The new immunotoxin, HA22-LR, has a deletion of most of domain II of PE. HA22-LR has the same activity as HA22, but has two remarkable and unexpected properties. One is that it kills CLL cells from patients much more effectively than HA22. The other is that it has much less toxicity to mice, which suggests it should have fewer side effects in patients than HA22.
Because of the clinical benefits obtained with HA22, we decided to further improve this molecule by increasing its affinity and consequently its activity. Although much effort has been put into improving the affinity of HA22 Fv by mutating several mutational “hot spot” residues of CDRs, the improvements were relatively small.13 In the present study, we exploited a different strategy of affinity maturation in which the functional contributions to binding of individual CDR residues was precisely assessed by alanine scanning mutagenesis.14 An additional problem with HA22 is that it requires two separate fermentations to make the protein, because the variable domain of light chain (VL) and heavy chain (VH)-PE38 portions are produced separately and then assembled into one protein by redox shuffling and protein renaturation.15 In the current paper, we describe the production of an anti-CD22 Fv in which the light and heavy chains are connected by both a peptide linker and a disulfide bond to stabilize the immunotoxin to make production easier.16 The resulting immunotoxin called HA22 (scds)Fv-LR was used for the mutational analysis (Fig. 1).
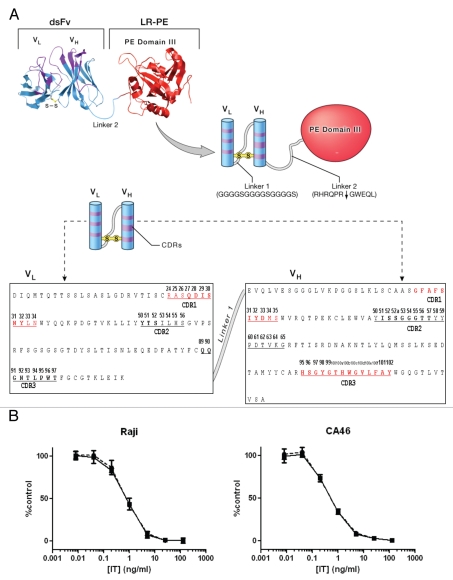
Figure 1.
(A) The structure of scdsFv version of HA22LR. The 2-chain disulfide- and peptide-(linker 1) linked Fv of an antibody targeting CD22 is combined with the domain III of native PE by the linker which bears Furin-cleavage site (linker 2) to create an immunotoxin. The Fv portion sequence is shown. Residue numbering is based on the Kabat numbering scheme.22 The CDR regions are defined according to Kabat et al. (underlined) and IMGT23 (boldface). Alanine-scanned CDR residues are shown in red. (B) Activities of dsFv-form (circles and dotted line) and scdsFv-form (squares and solid line) of HA22-LR on CD22-positive cells. The cytotoxicity was measured by WST-8 in triplicate 3 times. Typical cytotoxic curves are shown. Data are expressed as the mean ± SD. IT, immunotoxin.
Results
Generation of anti-CD22 immunotoxin, scdsFv-HA22-LR (single chain-, disulfide bond-Fv-HA22-LR).
We made the scdsFv form of HA22-LR by introducing a peptide-linker between light chain and heavy chain of Fv (Fig. 1A). This protein was made from the newest generation of PE-based immunotoxins, which consist of a disulfide-linked Fv (dsFv) of a mAb attached to domain III of PE by a linking peptide, which bears a furin-cleavage site. Introduction of a peptide linker between the VH and VL enabled us to produce the immunotoxin in a single fermentation. This is particularly advantageous for large-scale production of clinical-grade protein, which requires high homogeneity of product and the best curtailment of the cost.16 The purity of scFv-HA22-LR protein was over 90%. Cytotoxic activities of conventional dsFv-form and newly generated scdsFv-form of HA22-LR were compared in WST-8 cell viability assays using Raji and CA46 cell lines (Fig. 1B). No difference in cytotoxic activity was observed between scdsFv-HA22-LR and dsFv-HA22-LR [IC50(scdsFv): 0.84 ± 0.29 ng/ml (Raji), 0.49 ± 0.10 ng/ml (CA46), IC50(dsFv): 0.75 ± 0.01 ng/ml (Raji), 0.43 ± 0.07 ng/ml (CA46)].
Next, to further improve this molecule, we attempted an affinity maturation of scdsFv-HA22-LR. We exploited the alanine scan strategy to identify functionally important residues of the Fv. It is very likely that VHCDR3 region is the primary antigen binding site of HA22 Fv because VHCDR3 is the primary antigen binding site in most antibodies,17,18 and because our previous studies using a phage-based in vitro evolution method10,13 showed that Thr100-His100a-Trp100b in VHCDR3 is the only region whose modification drastically altered the affinity of HA22 Fv. Also, insertion of Gly before and after this region greatly reduced the activity of HA22 immunotoxin (unpublished data). Indeed, modeling studies show that Thr100, His100a and Trp100b are highly exposed on the tip of heavy chain CDR3 loop (data not shown). Therefore, we decided to start by performing alanine scanning of all the VHCDR3 residues and then perform alanine scanning of VHCDR1 and VLCDR1 residues because these CDRs are often important in antigen binding. Several studies on the affinity maturation of antibodies suggest that the key to successful affinity maturation is to find and modify the residue(s) which affects binding significantly but are not essential, because direct alteration of an essential paratope usually results in the total loss of binding ability of the antibody.19–21 Because the definition of the CDR regions is slightly different between the Kabat and IMGT23 numbering schemes, we prepared alanine mutants of residues which either scheme defines as a VHCDR1, VHCDR3 or VLCDR1 residue (Fig. 1A and shown in purple).
Preparation of the immunotoxins.
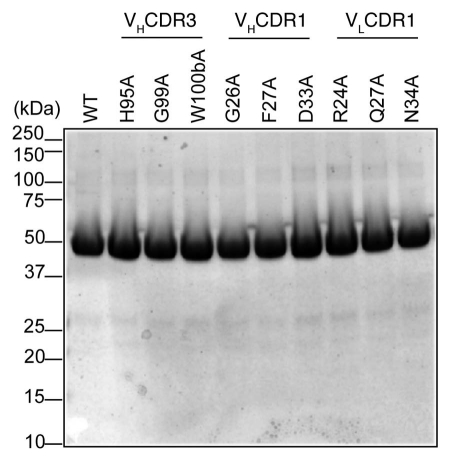
Wild type (WT) and alanine mutants of scdsFv-HA22-LR were expressed in E. coli BL21 (λDE3).15 The immunotoxins were refolded from solubilized inclusion bodies using a redox-shuffling buffer and were purified by ion-exchange chromatography on Q-Sepharose and Mono-Q columns followed by gel filtration chromatography on TSK (Toyo Soda Kogyo) column.15 Purified immunotoxins, migrated as a monomer on the TSK column, and had the expected size of 52 kDa when analyzed by SDS-PAGE (Fig. 2). The purity of each immunotoxin was over 90%.
Figure 2.
SDS-PAGE analysis of purified immunotoxins. Ten µg of purified immunotoxins were loaded per lane. Gel picture of 10 immunotoxins is shown as representative of the size and purity of all immunotoxins used in this study.
Alanine scanning of VHCDR1, VHCDR3 and VLCDR1 residues of scdsFv-HA22-LR.
Cytotoxic activities of the mutant immunotoxins were measured using WST-8 cell viability assays. The IC50 values were compared with that of WT scdsFv-HA22-LR to evaluate relative activities (Table 1). These relative activities correlated well with the KD values measured by Biacore (data not shown), although the variability was much smaller in cytotoxicity assays compared with Biacore measurements. Therefore, we used the relative cytotoxic activity values as an index to assess the contribution of each CDR residue toward antigen binding.
Table 1.
Specific cytotoxic activities of mutants in CDRs
| WST (Raji, IC50 ± SD (ng/ml)) | Relative activity* | ||
| VHCDR3 | |||
| WT | 0.56 ± 0.16 | 1.00 | |
| H95A | 1.07 ± 0.05 | 0.52 | |
| S96A | 0.61 ± 0.12 | 0.92 | |
| G97A | >1000 | <0.0005 | |
| Y98A | >1000 | <0.0005 | |
| G99A | >1000 | <0.0005 | |
| T100A | ND | ND | |
| H100aA | 0.84 ± 0.04 | 0.67 | |
| W100bA | 83.6 ± 7.6 | 0.0067 | |
| G100cA | ND | ND | |
| V100dA | 1.00 ± 0.26 | 0.56 | |
| L100eA | 0.53 ± 0.12 | 1.06 | |
| F100fA | 1.03 ± 0.12 | 0.54 | |
| A101A | 0.56 ± 0.16 | 1.00 | |
| Y102A | 0.68 ± 0.09 | 0.82 | |
| VHCDR1 | |||
| WT | 0.61 ± 0.12 | 1.00 | |
| G26A | 0.91 ± 0.05 | 0.67 | |
| F27A | 1.01 ± 0.19 | 0.60 | |
| A28A | 0.61 ± 0.12 | 1.00 | |
| F29A | 0.69 ± 0.11 | 0.88 | |
| S30A | ND | ND | |
| I31A | 0.67 ± 0.03 | 0.91 | |
| Y32A | ND | ND | |
| D33A | ND | ND | |
| M34A | 1.34 ± 0.07 | 0.46 | |
| S35A | 0.63 ± 0.06 | 0.97 | |
| VLCDR1 | |||
| WT | 0.61 ± 0.09 | 1.00 | |
| R24A | 0.55 ± 0.08 | 1.11 | |
| A25A | 0.61 ± 0.09 | 1.00 | |
| S26A | 0.87 ± 0.18 | 0.70 | |
| Q27A | 1.17 ± 0.11 | 0.52 | |
| D28A | 1.64 ± 0.05 | 0.37 | |
| I29A | 0.66 ± 0.04 | 0.92 | |
| S30A | 1.38 ± 0.08 | 0.44 | |
| N31A | 0.86 ± 0.23 | 0.71 | |
| Y32A | 0.99 ± 0.07 | 0.62 | |
| L33A | 0.66 ± 0.16 | 0.92 | |
| N34A | 0.11 ± 0.07 | 5.55 | |
| VLCDR1 Position 34 | |||
| WT | 0.59 ± 0.06 | 1.00 | |
| N34G | 0.26 ± 0.13 | 2.23 | |
| N34A | 0.12 ± 0.10 | 4.92 | |
| N34Q | 0.40 ± 0.07 | 1.48 | |
| N34E | 11.02 ± 0.18 | 0.05 | |
| N34Y | 6.63 ± 0.12 | 0.09 | |
| N34H | 5.48 ± 0.05 | 0.11 | |
| N34S | 0.63 ± 0.16 | 0.94 |
Relative activity is calculated by IC50 of WT/IC50 of mutant. ND, not done.
The relative activities of G97A, Y98A and G99A were exceptionally low (<0.0005), indicating that these residues make up the direct and functional paratope. W100bA showed a large reduction in relative activity (0.0067, Table 1), indicating that W100b contributes to binding but is not essential, and thus is an appropriate target for the modification in affinity. Since Trp100b has already been extensively examined in our previous study in which prototype BL22 Fv was affinity-maturated to HA22 Fv,10 this position was left intact in this study. In VHCDR1 and VLCDR1, most of the alanine mutants showed 0.4 ∼ 1.0 relative activities compared to WT (Table 1), indicating that the residues replaced by alanine do not contribute in a major way to binding to CD22. The exception is the N34A mutant of VLCDR1 (Fig. 3 and Table 1). N34A was ∼5-fold more active than WT on Raji cells (Table 1).
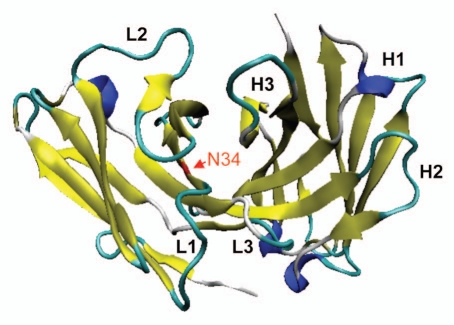
Figure 3.
Ribbon model of position VL34 of HA22-Fv. VL34 is buried and located at the interface of VL and VH.
Production and characterization of mutants of position 34 in VLCDR1.
As shown in Table 1, mutant N34A had about 5-fold increased activity relative to scdsFv-HA22-LR. The modeling of the Fv showed that VL34N of HA22-Fv is located at the VL/VH interface (Fig. 3). It is possible that the mutation in the VL/VH interface residue affects the affinity of the immunotoxin by influencing the interaction between the VL and the VH chain, thus altering the energetic stability of the VL/VH/antigen complex. Based on these information and speculation, we also mutated VL34N to Gly, Gln, Glu, Tyr, His and Ser, which are conserved at this position in mouse germ line antibody sequences, and tested activities of these immunotoxins (Table 1). All of these mutants were less active than WT, except N34G and N34Q. N34G and N34Q showed 2.2 and 1.5-fold activity than WT.
Affinity measurement on Daudi cells.
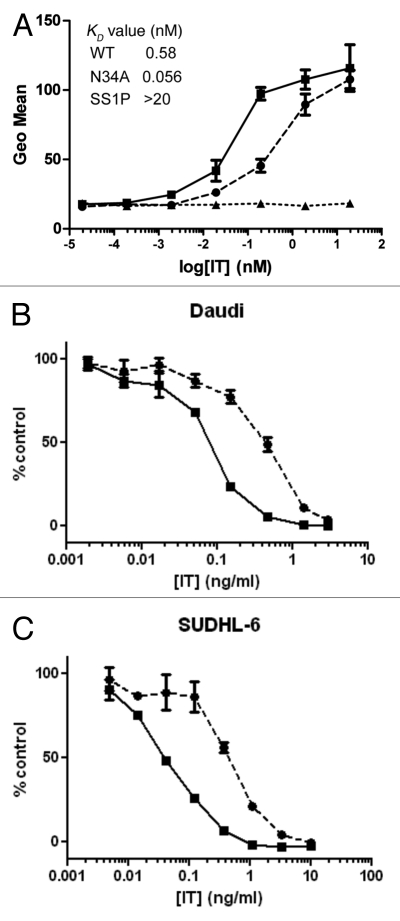
The relative affinities of the N34A mutant and WT scdsFv-HA22-LR were measured by fluorescence-activated cell sorting (FACS; Fig. 4A) because N34A mutant showed the most increased activity using Raji cells. As predicted from the increase in cytotoxic activity, N34A mutant showed ∼10 fold higher affinity to Daudi cells compared with WT. Estimated KD values of WT and N34A mutant are 0.58 nM and 0.056 nM, respectively. The KD value of the WT in this assay was consistent with the value calculated in the Biacore assay.10,13 We attempted to measure the affinity of the mutant by Biacore, but, because of the very slow off rate of the parent HA22 Fv, it was difficult to show a difference (unpublished data).
Figure 4.
Characterization of N34A mutant. (A) Affinities of WT scdsFv-HA22-LR (circles and dotted line) and its N34A mutant (squares and solid line) to CD22-positive Daudi cells. Affinities were measured by FACS. Briefly, pre-fixed Daudi cells were incubated with immunotoxins at 4°C overnight. Bound immunotoxins were detected with anti-LR-PE mouse polyclonal antibodies and PE-labeled goat anti-mouse IgG. Anti-mesothelin immunotoxin SS1P6 (triangles and dotted line) was used as a negative control. Mean fluorescence intensities are shown. Each assay was performed in triplicate. Data are expressed as the mean ± SD. (B) Specific cytotoxic activities of WT scdsFv-HA22-LR (circles and dotted line) and its N34A mutant (squares and solid line) on CD22-positive cells. The cytotoxicity was measured by WST-8 in triplicate at least nine times. Typical cytotoxic curves are shown. Data are expressed as the mean ± SD. We analyzed a total of 8 cell lines, and their IC50 concentrations are shown in Table 2.
Cytotoxic activities of N34A mutant on CD22-positive cell lines.
The activity of the N34A mutant was investigated on additional CD22-positive B-cell lymphoma and leukemia cell lines (Fig. 4B and Table 2). In Burkitt lymphoma cell lines, the N34A mutant was ∼4- to 5-fold more active than WT [IC50(WT) vs. IC50(N34A) (ng/ml): 0.59 ± 0.063 vs. 0.12 ± 0.10 (Raji), 0.36 ± 0.065 vs. 0.10 ± 0.042 (CA46) and 0.54 ± 0.26 vs. 0.11 ± 0.041 (Daudi)]. In ALL cell lines, the N34A mutant was ∼2- to 3-fold more active than WT [IC50(WT) vs. IC50(N34A) (ng/ml): 1.61 ± 0.25 vs. 0.56 ± 0.17 (Nalm-19) and 2.37 ± 0.62 vs. 1.32 ± 0.41 (HAL-1)]. The N34A mutant immunotoxin showed ∼3- and ∼10-fold increased cytotoxicity in ABC-DLBCL cell line SUDHL-5 (IC50(WT) vs. IC50(N34A) (ng/ml): 1.45 ± 0.15 vs. 0.43 ± 0.037) and GCB-DLBCL cell line SUDHL-6 (IC50(WT) vs. IC50(N34A) (ng/ml): 0.47 ± 0.090 vs. 0.048 ± 0.018), respectively. Thus, the N34A immunotoxin showed increased cytotoxicity compared with WT in all of the cell lines tested. The mutant immunotoxin was not cytotoxic to the CD22-negative cell line L55, which demonstrated that the cytotoxic effect is selective to antigen-positive cells.
Table 2.
Specific cytotoxic activities of WT and N34A mutant of scdsFvHA22LR on various cell lines
| IC50 ± SD (ng/ml) | |||
| Cells | Cell types** | WT | N34A |
| Raji* | Burkitt Lymphoma | 0.59 ± 0.063 | 0.12 ± 0.10 |
| CA46* | Burkitt Lymphoma | 0.36 ± 0.065 | 0.10 ± 0.042 |
| Daudi* | Burkitt Lymphoma | 0.54 ± 0.26 | 0.11 ± 0.041 |
| Nalm-19* | ALL | 1.61 ± 0.25 | 0.56 ± 0.17 |
| HAL-1* | ALL | 2.37 ± 0.62 | 1.32 ± 0.41 |
| SUDHL-5* | ABC DLBCL | 1.45 ± 0.15 | 0.43 ± 0.037 |
| SUDHL-6* | GCB DLBCL | 0.47 ± 0.090 | 0.048 ± 0.018 |
| L55 | NSCLC | >1000 | >1000 |
Significant difference (p < 0.05, in a non-parametric, 2-tailed t test) between the IC50 values of WT and N34A.
ALL, acute lymphoblastic leukemia; ABC, activated B-cell like; DLBCL, diffuse large B-cell lymphoma; GCB, germinal center B-cell like; NSCLC, non-small cell lung carcinoma.
Discussion
In the current study, we made a new immunotoxin in which the light and heavy chains are connected by both a peptide linker and a disulfide bond and used this protein as a starting point to map the location of important paratopes, and to improve the activity and the affinity of the immunotoxin.16 We found we could improve affinity and cytotoxic activity by mutating the VH/VL interface residue in VLCDR1. The mutant immunotoxin N34A showed increased cytotoxicity to CD22-positive cell lines. The mutant immunotoxin N34A had up to a 10-fold improvement in activity toward CD22-positive cells. Optimization of HA22-LR for large-scale production of clinical-grade protein by converting it to scdsFv-form should greatly facilitate the development of this molecule. Moreover, the increase in activity could be very useful in the treatment of malignancies that express CD22. The increased cytotoxic activity of the mutant immunotoxin should produce a clinical benefit with a lower dose and in turn lead to a decrease in nonspecific toxicities in patients. For these reasons, the N34A mutant of scdsFv-HA22-LR immunotoxin merits further preclinical development.
The approach used to increase the affinity of HA22 Fv was to identify functionally important residues for binding by alanine scanning of VHCDR1, 3 and VLCDR1. Other strategies that are used to increase the affinity of antibodies, such as codon-based mutagenesis,24 CDR walking,25,26 error-prone replication27 and synthetic CDR construction,28 require the construction of large libraries of phage or yeast because all CDRs residues are mutagenized. Such libraries are difficult to make and to handle.29 Also, panning on purified protein can isolate Fvs that bind poorly to the cell bound form of the target protein.30
Our approach was (1) to examine relative cytotoxicities of alanine mutants of immunotoxin and to find the candidate position to focus on and (2) to examine the residues conserved among the mouse germlines at the position identified. This approach is free from the problems of preparing and panning phage libraries. Also, measuring the cytotoxicity of each alanine mutant is more useful than first measuring affinity of purified CD22 protein whose comformation could be different than the conformation of the antigen on target cells.
Based on the alanine scanning of VHCDR3 residues, the relative activity of G97A, Y98A, and G99A were exceptionally low indicating that these residues make up the direct and functional paratope. In our previous phage-based study, these residues were always restored to starting residues after panning randomized phage libraries on CD22Fc protein (reviewed in refs. 10 and 13; unpublished data). This indicates that these residues are indispensable for binding and other amino acids cannot be allowed at these positions. W100bA showed a large reduction in relative activity (Table 1), indicating that W100b contributes to binding but is not essential, and thus is an appropriate target for the modification in affinity. Indeed, W100b was the key residue involved in increasing the affinity of prototype immunotoxin BL22 to HA22.10 The confirmation of previous results by our alanine scanning analysis of VHCDR3 residues indicates that this approach should be effective in finding other target residue to improve the affinity of scdsFv-HA22-LR.
Position VL34 is a part of VLCDR1 (L24–L34) as defined by Kabat et al. but not as defined by IMGT23 and Chothia and co-workers31,32 (Fig. 1). In the study of Chothia et al.31 VL34 is defined as a VL/VH interface residue belonging to the common structural core. The modeling of the Fv showed that VL34N of HA22-Fv is located at the VL/VH interface (Fig. 3). This modeling also predicted the possibility that VL34N could form a hydrogen bond with VH100dV, which is located close to the putative antigen binding site (data not shown). Alanine cannot form this hydrogen bond. It is possible that the elimination of this hydrogen bond by the N34A mutation affects the properties of the antigen binding pocket. Indeed, the elimination of the hydrogen bond by N34G also increased the activity of scdsFv-HA22-LR (Table 1).
Several studies have suggested that the interaction between the VH and VL chain of an antibody affects its binding affinity to antigen. Ueda et al. developed the “Open-Sandwich Elisa” method to quantify small molecules.33,34 This assay exploits the mechanism in which the packing of VH/VL is strengthened upon antigen binding. Hugo et al. focused on position L34 of antibodies from the standpoint of predictive engineering of antibodies.35 It is located at the VH/VL interface and therefore central to the paratope, but with only limited contacts with the antigen. This suggests a possible general, but not essential, contribution to paratope geometry, and consequently to binding affinity, making L34 a general target for predictive engineering of antibodies. Indeed, Hugo et al. have succeeded in the affinity maturation of two different antibodies by mutating the L34 residues of the molecules.
The relative activity of N34A mutant compared with WT varied greatly among the cell lines tested. This is probably attributable to different numbers of CD22 receptor sites on the cells but could also be caused by different variants of CD22 that are present on different cell types. Two different isoforms of CD22 have been described: CD22β is a full-length molecule with seven extracellular domains and CD22α, which lacks extracellular domains 3 and 4 because of alternative splicing.36,37 The epitope mapping study of HA22 Fv suggested that the N-terminal region of domain 3 and the C-terminal region of domain 4 of CD22 are parts of the conformational epitope.38 Because domains 3 and 4 are missing in CD22α we expect that the binding of the HA22 Fv will be weak. Differences in the rate of internalization and intracellular trafficking could also have caused the observed differences in activity.
In summary, we generated a scdsFv version of HA22-LR that may be more suitable for large scale production of clinical-grade protein, and showed it has comparable activity to conventional dsFv-HA22-LR. We also demonstrated that a rational approach, which identifies functional paratopes by non-library based alanine scanning can be a general method of affinity maturation of antibodies. We improved the activity and the affinity of an anti-CD22 immunotoxin HA22-LR by mutating the VH/VL interface residue in VLCDR1. The mutant immunotoxin N34A showed increased cytotoxicity to CD22-positive cell lines. These results indicate that the N34A mutant immunotoxin merits further preclinical development.
Materials and Methods
Construction and purification of immunotoxins.
scdsFv were PCR-amplified using mutagenesis primers and primers that introduced NdeI and HindIII restrictions sites. The products of the reaction were purified, digested with NdeI and HindIII and cloned into a T7 expression vector in which the scdsFv was fused to a truncated version of PE (LR-PE). The expression and purification of immunotoxins was performed as described previously in reference 15.
Cell lines.
CD22-positive human Burkitt lymphoma cell lines (CA46, Daudi and Raji) were obtained from ATCC (catalog #CCL-184, CCL-213 and CCL-86). Nalm-19 and HAL-1 ALL cell line was obtained from Dr. Alan Wayne (National Cancer Institute [NCI], Bethesda, MD). SUDHL-5 (ABC-DLBCL) and SUDHL-6 (GCB-DLBCL) were obtained from Dr. Louis M. Staudt (NCI, Bethesda, MD). All cell lines were grown at 37°C with 5% CO2 in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 1 mM of sodium pyruvate, 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen, catalog #15140163).
Cytotoxic activities.
Survival of cells treated with immunotoxins was measured by WST-8 assay using the Cell Counting Kit-8 (Dojindo Molecular Technologies, catalog #341-07761) essentially as described in reference 39 and 40. Briefly, 104 cells/well were incubated with various concentrations of immunotoxin in a 96-well plate for 72 h, after which 25 µL of the CCK-8 reagent was added to the wells. Plates were incubated until the wells with the maximum absorbance at 450 nm reached over values of approximately 1 optical density. Cycloheximide (10 µg/mL final concentration) was used as a control for 100% cell death. Values were normalized between the cycloheximide and phosphate buffered saline (PBS) controls and concentration of immunotoxin at which there was 50% cell death (IC50) was obtained.
Affinity measurement with FACS.
Daudi cells (1 × 105) pre-fixed with 2% PFA were incubated with various concentrations of immunotoxins in blocking buffer containing 5% bovine serum albumin plus 0.1% NaN3 in PBS in a 96-well plate at 4°C overnight. Cells were washed two times with PBS and resuspended in 100 µl blocking buffer containing 1 µl of mouse sera prepared against HA22-LR. The mixture was incubated at 4°C for 30 min and then washed twice with PBS. Cells were resuspended in 100 µl blocking buffer containing 1 µl of PE-labeled goat anti-mouse IgG (Jackson Immuno Research, catalog #115-295-003), and cells were incubated for 30 min at 4°C. Cells were washed two times, and analysis was performed in a Guava Easy cyto PLUS (Millipore, catalog #0500-1980). Affinity measurements were performed with flow cytometry following the protocols described previously in references 41 and 42, except that in this study Daudi cells and monomeric immunotoxins instead of the whole antibody or scFv molecules were used. Equilibrium constants and Scatchard plots were determined by using the Marquardt-Levenberg algorithm for nonlinear regression with the GraphPad Prism (version 2.00; GraphPad Software, San Diego, CA) program.
Statistics.
Mann-Whitney nonparametric method was used: p < 0.05 was considered statistically significant.
Acknowledgments
We thank Dr. Louis M. Staudt for providing SUDHL-5 and SUDHL-6 cells, and Dr. Alan Wayne for HAL-1 and Nalm-19 cells. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and in part by JSPS Research Fellowship to S.K. for Japanese Biomedical and Behavioral Research, Japan Society for the Promotion of Science.
Abbreviations
- ALL
acute lymphoblastic leukemia
- CDR
complementary determining region
- CLL
chronic lymphocytic leukemia
- HA22
moxetumomab pasudotox
- OD
optical density
- PBS
phosphate buffered saline
- PE
Pseudomonas exotoxin A
- RIT
recombinant immunotoxin
- scdsFv
single chain and disulfide bond-linked fragment variable
- TSK
toyo soda kogyo
- VH
variable domain of heavy chain
- VL
variable domain of light chain
- WT
wild type
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Reichert JM, Rosensweig CJ, Faden LB, Dewitz MC. Monoclonal antibody successes in the clinic. Nat Biotechnol. 2005;9:1073–1078. doi: 10.1038/nbt0905-1073. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Q, Chen G, Liu X, Qian Q. Monoclonal antibodies as therapeutic agents in oncology and antibody gene therapy. Cell Res. 2007;17:89–99. doi: 10.1038/sj.cr.7310143. [DOI] [PubMed] [Google Scholar]
- 3.Pastan I, Hassan R, Fitzgerald DJ, Kreitman RJ. Immunotoxin therapy of cancer. Nat Rev Cancer. 2006;6:559–565. doi: 10.1038/nrc1891. [DOI] [PubMed] [Google Scholar]
- 4.Pastan I, Hassan R, FitzGerald DJ, Kreitman RJ. Immunotoxin treatment of cancer. Annu Rev Med. 2007;58:221–237. doi: 10.1146/annurev.med.58.070605.115320. [DOI] [PubMed] [Google Scholar]
- 5.Kreitman RJ, Squires DR, Stetler-Stevenson M, Noel P, FitzGerald DJP, Wilson WH, et al. Phase I trial of recombinant immunotoxin RFB4(dsFv)-PE38 (BL22) in patients with B-cell malignancies. J Clin Oncol. 2005;23:6719–6729. doi: 10.1200/JCO.2005.11.437. [DOI] [PubMed] [Google Scholar]
- 6.Hassan R, Bullock S, Premkumar A, Kreitman RJ, Kindler H, Willingham MC, et al. Phase I study of SS1P, a recombinant anti-mesothelin immunotoxin given as a bolus I.V. infusion to patients with mesothelin-expressing mesothelioma, ovarian, and pancreatic cancers. Clin Cancer Res. 2007;13:5144–5149. doi: 10.1158/1078-0432.CCR-07-0869. [DOI] [PubMed] [Google Scholar]
- 7.Kreitman RJ, Wilson WH, White JD, Stetler-Stevenson M, Jaffe ES, Giardina S, et al. Phase I trial of recombinant immunotoxin anti-Tac(Fv)-PE38 (LMB-2) in patients with hematologic malignancies. J Clin Oncol. 2000;18:1622–1636. doi: 10.1200/JCO.2000.18.8.1622. [DOI] [PubMed] [Google Scholar]
- 8.Kreitman RJ, Margulies I, Stetler-Stevenson M, Wang QC, FitzGerald DJ, Pastan I. Cytotoxic activity of disulfide-stabilized recombinant immunotoxin RFB4(dsFv)-PE38 (BL22) toward fresh malignant cells from patients with B-cell leukemias. Clin Cancer Res. 2000;4:1476–1487. [PubMed] [Google Scholar]
- 9.Kreitman RJ, Fitzgerald DJ, Pastan I. Approach to the patient after relapse of hairy cell leukemia. Leuk Lymphoma. 2009;50:32–37. doi: 10.3109/10428190903142216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salvatore G, Beers R, Margulies I, Kreitman RJ, Pastan I. Improved cytotoxic activity toward cell lines and fresh leukemia cells of a mutant anti-CD22 immunotoxin obtained by antibody phage display. Clin Cancer Res. 2002;8:995–1002. [PubMed] [Google Scholar]
- 11.Lechleider R, Pastan I. Advances in the development of anti-CD22 immunotoxins containing Pseudomonas exotoxin for treatment of hematologic malignancies. J Cancer Sci Ther. 2011;3:050–052. [Google Scholar]
- 12.Weldon JE, Xiang L, Chertov O, Margulies I, Kreitman RJ, FitzGerald DJ, et al. A protease-resistant immunotoxin against CD22 with greatly increased activity against CLL and diminished animal toxicity. Blood. 2009;113:3792–3800. doi: 10.1182/blood-2008-08-173195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho M, Kreitman RJ, Onda M, Pastan I. In vitro antibody evolution targeting germline hot spots to increase activity of an anti-CD22 immunotoxin. J Biol Chen. 2005;280:607–617. doi: 10.1074/jbc.M409783200. [DOI] [PubMed] [Google Scholar]
- 14.Bostrom J, Yu SF, Kan D, Appleton BA, Lee CV, Billeci K, et al. Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site. Science. 2009;323:1610–1614. doi: 10.1126/science.1165480. [DOI] [PubMed] [Google Scholar]
- 15.Pastan I, Beers R, Bera TK. Recombinant immunotoxins in the treatment of cancer. Methods Mol Biol. 2004;248:503–518. doi: 10.1385/1-59259-666-5:503. [DOI] [PubMed] [Google Scholar]
- 16.Rajagopal V, Pastan I, Kreitman RJ. A form of anti-Tac(Fv) which is both single-chain and disulfide stabilized: comparison with its single-chain and disulfide-stabilized homologs. Protein Eng. 1997;12:1453–1459. doi: 10.1093/protein/10.12.1453. [DOI] [PubMed] [Google Scholar]
- 17.Reidl LS, Friedman DF, Goldman J, Hardy RR, Jefferies LC, Silberstein LE. Structural basis of a conserved idiotope expressed by an autoreactive human B cell lymphoma. Evidence that a VH. CDR3 mutation alters idiotypy and specificity. J Immunol. 1991;147:3623–3631. [PubMed] [Google Scholar]
- 18.Kimura H, Cook R, Meek K, Umeda M, Ball E, Capra JD, et al. Sequences of the VH and VL regions of murine monoclonal antibodies against 3-fucosyllactosamine. J Immunol. 1988;140:1212–1217. [PubMed] [Google Scholar]
- 19.James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 20.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 21.Kelley RF, O'Connell MP. Thermodynamic analysis of an antibody functional epitope. Biochemistry. 1993;32:6828–6835. doi: 10.1021/bi00078a005. [DOI] [PubMed] [Google Scholar]
- 22.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of Proteins of Immunological Interest. 5th Ed. Bethesda MD: National Institutes of Health Publication 91-3242; 1991. [Google Scholar]
- 23.Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2001;29:207–209. doi: 10.1093/nar/29.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yelton DE, Rosok MJ, Cruz G, Cosand WL, Bajorath J, Hellström KE, et al. Affinity maturation of the BR96 anti-carcinoma antibody by codon-based mutagenesis. J Immunol. 1995;155:1994–2004. [PubMed] [Google Scholar]
- 25.Barbas CF, Burton DR. Selection and evolution of high affinity human anti-viral antibodies. Trends Biotechnol. 1996;14:230–234. doi: 10.1016/0167-7799(96)10029-9. [DOI] [PubMed] [Google Scholar]
- 26.Yang WP, Green K, Pinzsweeney S, Briones AT, Burton DR, Barbas DF. CDR walking mutagenesis for the affinity maturation of a potent human anti-HIV-1 antibody into the picomolar range. J Mol Biol. 1995;254:392–403. doi: 10.1006/jmbi.1995.0626. [DOI] [PubMed] [Google Scholar]
- 27.Low NM, Holliger P, Winter F. Mimicking somatic hypermutation: affinity maturation of antibodies displayed on bacteriophage using a bacterial mutator strain. J Mol Biol. 1996;260:359–368. doi: 10.1006/jmbi.1996.0406. [DOI] [PubMed] [Google Scholar]
- 28.Dekruif J, Boel E, Logtenberg T. Selection and application of human single-chain Fv antibody fragments from a semisynthetic phage antibody display library with designed CDR3 regions. J Mol Biol. 1995;248:97–105. doi: 10.1006/jmbi.1995.0204. [DOI] [PubMed] [Google Scholar]
- 29.Chowdhury PS, Pastan I. Improving antibody affinity by mimicking somatic hypermutation in vitro. Nat Biotechnol. 1999;17:568–572. doi: 10.1038/9872. [DOI] [PubMed] [Google Scholar]
- 30.Schier R, Bye J, Apell G, McCall A, Adams GP, Malmqvist M, et al. Isolation of high-affinity monomeric human anti-c-erbB-2 single chain Fv using affinity-driven selection. J Mol Biol. 1996;255:28–43. doi: 10.1006/jmbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 31.Chothia C, Lesk AM, Tramontano A, Levitt M, Smith-Gill SJ, Air G, et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 32.Al-Lazikani B, Lesk AM, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J Mol Biol. 1997;273:927–948. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- 33.Ueda H, Tsumoto K, Kubota K, Suzuki E, Nagamune T, Nishimura H, et al. Open sandwich ELISA: a novel immunoassay based on the interchain interaction of antibody variable region. Nat Biotechnol. 1996;14:1714–1718. doi: 10.1038/nbt1296-1714. [DOI] [PubMed] [Google Scholar]
- 34.Sasajima Y, Iwasaki R, Tsumoto K, Kumagai I, Ihara M, Ueda H. Expression of antibody variable region-human alkaline phosphatase fusion proteins in mammalian cells. J Immunol Methods. 2010;361:57–63. doi: 10.1016/j.jim.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 35.Hugo N, Weidenhaupt M, Beukes M, Xu B, Janson JC, Vernet T, et al. VL position 34 is a key determinant for the engineering of stable antibodies with fast dissociation rates. Protein Eng. 2003;16:381–386. doi: 10.1093/protein/gzg042. [DOI] [PubMed] [Google Scholar]
- 36.Tedder TF, Tuscano J, Sato S, Kehrl JH. CD22, a B lymphocyte-specific adhesion molecule that regulates antigen receptor signaling. Annu Rev Immunol. 1997;5:481–504. doi: 10.1146/annurev.immunol.15.1.481. [DOI] [PubMed] [Google Scholar]
- 37.Mintz U, Sachs L. Changes in the surface membrane of lymphocytes from patients with chronic lymphocytic leukemia and Hodgkin's disease. Int J Cancer. 1975;15:253–259. doi: 10.1002/ijc.2910150211. [DOI] [PubMed] [Google Scholar]
- 38.Bannister D, Popovic B, Sridharan S, Giannotta F, Filée P, Yilmaz N, et al. Epitope mapping and key amino acid identification of anti-CD22 immunotoxin CAT-8015 using hybrid β-lactamase display. Protein Eng Des Sel. 2011;24:351–360. doi: 10.1093/protein/gzq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du X, Nagata S, Ise T, Stetler-Stevenson M, Pastan I. FCRL1 on chronic lymphocytic leukemia, hairy cell leukemia and B-cell non-Hodgkin lymphoma as a target of immunotoxins. Blood. 2008;111:338–343. doi: 10.1182/blood-2007-07-102350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berridge FV, Tan AS, Herst PM. Tetrazolium dyes as tools in cell biology: New insights into their cellular reduction. Biotechnol Annu Rev. 2005;11:127–152. doi: 10.1016/S1387-2656(05)11004-7. [DOI] [PubMed] [Google Scholar]
- 41.Benedict CA, MacKrell AJ, Anderson WF. Determination of the binding affinity of an anti-CD34 single-chain antibody using a novel, flow cytometry based assay. J Immunol Methods. 1997;2:223–231. doi: 10.1016/s0022-1759(96)00227-x. [DOI] [PubMed] [Google Scholar]
- 42.Krauss J, Arndt MA, Martin AC, Liu H, Rybak SM. Specificity grafting of human antibody frameworks selected from a phage display library: generation of a highly stable humanized anti-CD22 single-chain Fv fragment. Protein Eng. 2003;16:753–759. doi: 10.1093/protein/gzg096. [DOI] [PubMed] [Google Scholar]